Abstract
HDACs (histone deacetylases) are considered to be among the most important enzymes that regulate gene expression in eukaryotic cells acting through deacetylation of ϵ-acetyl-lysine residues within the N-terminal tail of core histones. In addition, both eukaryotic HDACs as well as their bacterial counterparts were reported to also act on non-histone targets. However, we are still far from a comprehensive understanding of the biological activities of this ancient class of enzymes. In the present paper, we studied in more detail the esterase activity of HDACs, focussing on the HDAH (histone deacetylase-like amidohydrolase) from Bordetella/Alcaligenes strain FB188. This enzyme was classified as a class 2 HDAC based on sequence comparison as well as functional data. Using chromogenic and fluorogenic ester substrates we show that HDACs such as FB188 HDAH indeed have esterase activity that is comparable with those of known esterases. Similar results were obtained for human HDAC1, 3 and 8. Standard HDAC inhibitors were able to block both activities with similar IC50 values. Interestingly, HDAC inhibitors such as suberoylanilide hydroxamic acid (SAHA) also showed inhibitory activity against porcine liver esterase and Pseudomonas fluorescens lipase. The esterase and the amidohydrolase activity of FB188 HDAH both appear to have the same substrate specificity concerning the acyl moiety. Interestingly, a Y312F mutation in the active site of HDAH obstructed amidohydrolase activity but significantly improved esterase activity, indicating subtle differences in the mechanism of both catalytic activities. Our results suggest that, in principle, HDACs may have other biological roles besides acting as protein deacetylases. Furthermore, data on HDAC inhibitors affecting known esterases indicate that these molecules, which are currently among the most promising drug candidates in cancer therapy, may have a broader target profile requiring further exploration.
Keywords: amidohydrolase, Bordetella/Alcaligenes, esterase, fluorogenic assay, histone deacetylase, lipase
Abbreviations: AMC, 7-amino-4-methylcoumarin; CypX, cyclopentyle-propionyle hydroxamic acid; HAT, histone acetyltransferase; HDAC, histone deacetylase; HDAH, histone deacetylase-like amidohydrolase (from strain FB188); HDLP, histone deacetylase-like protein; HMC, 7-hydroxy-4-methylcoumarin; MCA, 4-methylcoumarin-7-amide; NMM, N-methylmorpholine; SAHA, suberoylanilide hydroxamic acid
INTRODUCTION
HDACs (histone deacetylases) and HATs (histone acetyltransferases) are key enzymes in the regulation of gene expression in eukaryotic cells. HATs and HDACs catalyse the addition and the removal of acetyl moieties from the ϵ-amino groups of lysine residues near the N-termini of histones respectively. In general, increased levels of histone acetylation are associated with increased transcriptional activity, whereas decreased levels of acetylation are associated with repression of gene expression [1–3]. The activity of HATs and, in particular, of HDACs, both affect angiogenesis, cell-cycle arrest, apoptosis, terminal differentiation of different cell types and the pathogenesis of malignant disease [4]. Not surprisingly, a number of HDAC inhibitors are promising antitumour agents. Several drug candidates are currently in phase I–III clinical trials [5–8] with SAHA (Zolinza) being recently approved for the treatment of cutaneous T-cell lymphona (CTCL).
Four classes of deacetylase enzymes have been categorized on the basis of sequence homology and functional properties [9]: RPD3 (reduced potassium dependency 3)-like class 1 HDACs, HDA1 (histone deacetylase 1)-like class 2 HDACs and class 4 enzymes (HDAC11) are structurally related zinc-dependent enzymes, whereas class 3 enzymes comprise the structurally unrelated NAD-dependent sirtuins [10].
For a long time research on HDACs had focussed mainly on their role in transcriptional regulation. More recently, however, data became available indicating that HDACs may also act on non-histone proteins and small molecules. Examples include HDAC6, which has tubulin deacetylase activity [11–13] and bacterial HDAHs (histone deacetylase-like amidohydrolases) [14,15].
Recently, the crystal structures of two class 1 enzymes, HDLP (histone deacetylase-like protein) from Aquifex aeolicus [16] and HDAC8 [17,18], as well as that of one class 2 enzyme, FB188 HDAH [19], have been solved. Based particularly on enzyme–inhibitor co-complex structures (see for example Figures 3A and 3B), a mechanism has been proposed which includes features of those from metallo and serine proteases [16]. By this mechanism (Figures 3C–3E), the active site zinc ion would bind to the carbonyl oxygen of the acetyl moiety, polarizing the carbonyl group and thereby increase the electrophilicity of the carbon. The zinc ion also binds to the oxygen of a water molecule such that the nucleophilicity of the water oxygen is increased. Analogous to the mechanism of serine proteases, the nucleophilicity of the water molecule is further increased by the negative charge of a buried Asp–His charge-transfer relay system, to which the water molecule is hydrogen bonded. The nucleophilic attack of the water molecule on the carbonyl carbon would lead to a tetrahedral oxyanion transition state which would be stabilized by the aforementioned zinc–oxygen contacts and by a potential hydrogen bond to the hydroxyl group of a tyrosine residue (Tyr312 in HDAH). Finally, the acetate would be released and the ϵ-nitrogen of the lysine residue would accept a proton from a second Asp–His charge-transfer relay system not present in FB188 HDAH or any other class 2 enzymes [20]. Confirmation of the important role played by the aforementioned active site amino acid residues came from mutagenesis studies [16,19,21,22]. The proposed mechanism has been challenged in part by: (i) ab initio calculation studies [23], (ii) experiments with transition state analogue inhibitors designed to mimic the proposed oxyanion intermediate [10] and, (iii) experiments with substrates containing different acyl leaving groups [20]. Despite the huge body of data generated throughout many years of HDAC research, relatively little is known about natural substrates of different HDACs. Indeed, studies so far have focussed exclusively on amides as possible substrates. In the present study, we demonstrate that HDAC enzymes such as HDAH, HDAC1, HDAC3 and HDAC8 also have a very pronounced esterase activity that can be inhibited by known HDAC inhibitors. On the other hand, HDAC inhibitors are also active against known esterases. Specificity towards acyl moieties is similar for both amidohydrolase and esterase activities. However, mutation of the active site tyrosine residue (Tyr312 in HDAH) into a phenylalanine residue impairs only amidohydrolase activity but actually improves esterase activity. Taken together, our experimental results improve our understanding of the catalytic mechanism of HDACs. Furthermore, we provide the first evidence suggesting that at least certain members of the HDAC family may assume the biological role of an esterase.
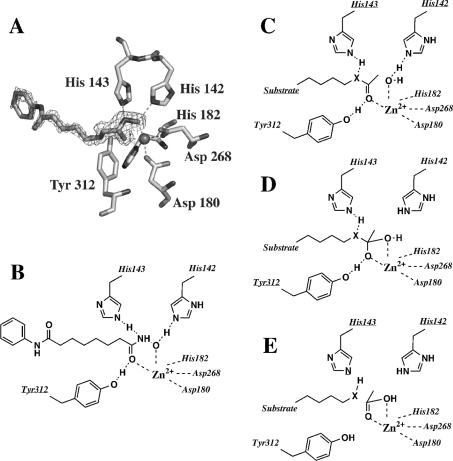
Figure 3. Structure of the active site of FB188 HDAH and proposed catalytic mechanism.
(A) Crystallographic structure of the inhibitor SAHA bound to the active site. (B) Schematic representation of the interactions between SAHA and the active site residues of FB188 HDAH. (C–E) Proposed mechanism for the deacetylation of amides (X=NH) and esters (X=O). HDAH residues are labelled.
EXPERIMENTAL
Synthesis of fluorogenic substrates
MCA (4-methylcoumarin-7-amide) and Boc-L-Lys(ϵ-acetyl)-MCA were purchased from Bachem. 4-Nitrophenyl acetate and all other reagents for organic synthesis were obtained from Sigma. The ϵ-propionyl-derivative of Boc-L-Lys(ϵ-acyl)-MCA was synthesized as described in [20]. The acetyl and propionyl esters of HMC (7-hydroxy-4-methylcoumarin) were synthesized using standard protocols. Briefly, 0.5 mmol of HMC was suspended in 2 ml of dioxane. After the addition of 90 μl of NMM (N-methylmorpholine) and 180 μl of acetyl chloride (or propionyl chloride) the suspension was incubated for 10 min at room temperature. After the addition of 2 ml of acetonitrile and 180 μl of NMM the solution was shaken overnight at room temperature. The solvent was removed under reduced pressure and the residue was redissolved in ethyl acetate. After extraction with 3×20 ml of water the solvent was removed under reduced pressure. The product was analysed by reversed phase HPLC/MS using a Waters Alliance/MicromassZQ system and a 250 mm×10 mm-C18 column (Jupiter, Phenomenex). Purities were >90% with no educt present. LC-MS (ESI): 4-methylcoumarin-7-acetate calculated for C12H10O4 (M+H): 219.2. Found: 219.2. 4-methylcoumarin-7-propionate calculated for C13H12O4 (M+H): 233.2. Found: 233.2.
Assays
HDAC1 and HDAC3/N-COR2 were obtained from BPS Bioscience. HDAC8 was expressed in Escherichia coli and purified as described in [20]. Porcine liver esterase (EC 3.1.1.1.) was from Sigma and Pseudomonas fluorescens lipase was purchased from Fluka. Mutant HDAH genes were generated by the QuikChange® method using the manufacturer's protocol (Stratagene). FB188 HDAH and its mutants were prepared as described in [14]. For fluorometric assays, all pipetting and fluorescence detection steps were performed with the help of a robotic workstation (CyBi™-Screen-Machine, CyBio AG) including a Polarstar fluorescence reader (BMG). Reactions were conducted in black 96-well microplates. For assays with chromogenic substrates, a UV-1601 spectrophotometer (Shimadzu) was used.
Fluorogenic assays
Amidohydrolase assays were performed similarly to the standard fluorogenic HDAC assay [24]. Briefly, 10 μl of diluted enzyme solution (corresponding to 50 ng HDAH), 50 μl of substrate solution in HD buffer {15 mM Tris/HCl (pH 8.0), 250 μM EDTA, 250 mM NaCl and 0.1% (w/v) PEG [poly(ethylene glycol)] 8000} including 50 mM potassium phosphate and 80 μl of trypsin solution (1 mg/ml in HD buffer) were mixed and incubated at 30 °C. The release of AMC (7-amino-4-methylcoumarin) was monitored by measuring the fluorescence at 460 nm (λex=390 nm). Fluorescence intensity was calibrated using free AMC. For HDAC1 and HDAC3, the two-step variant of the assay was used. Briefly, 10 μl of diluted enzyme solution and 50 μl of substrate solution in HD buffer were mixed and incubated at 30 °C. After 45 min incubation, 80 μl of trypsin solution was added. After a further incubation for 15 min at 30 °C, the released AMC was monitored by measuring the fluorescence at 460 nm. Km and Vmax values as well as IC50 concentrations were determined as previously described [25]. Substrate concentrations between 0.5 and 50 μM were used for standard Km and Vmax determinations. For IC50 determinations a substrate concentration of 2 μM was used.
Fluorescence-based esterase assays were performed by mixing 10 μl of enzyme solution, 180 μl of HD buffer (pH 8.0) and 10 μl of substrate solution (from a 40 μM stock of either substrate C or D, Figure 1) on ice. After switching the reaction temperature to 20 °C the release of HMC was monitored by measuring the fluorescence at 460 nm. The fluorescence intensity was calibrated using free HMC. Note that the esterase substrates (substrate C and D, Figure 1) showed substantial hydrolysis at higher temperatures.
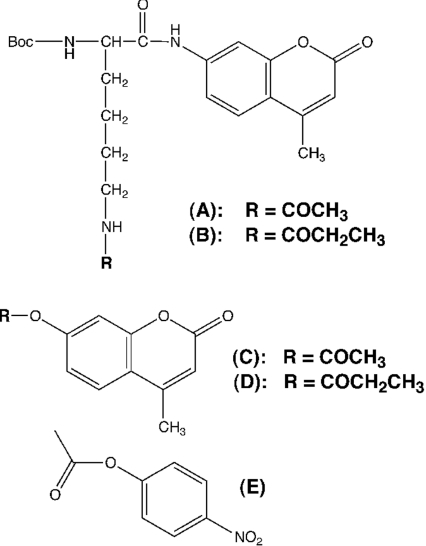
Figure 1. Small molecule substrates of FB188 HDAH.
(A) Boc-L-Lys(ϵ-acetyl)-MCA, (B) Boc-L-Lys(ϵ-propionyl)-MCA, (C) 4-methylcoumarin-7-acetate, (D) 4-methylcoumarin-7-propionate, and (E) 4-nitrophenyl-acetate.
Chromogenic esterase assay
In the standard chromogenic assay, 10 μl of enzyme was mixed on ice with 980 μl of HD buffer and 10 μl of substrate solution. After switching the reaction temperature to 25 °C the release of 4-nitrophenol was monitored by measuring the absorbance at 405 nm. Absorbance was calibrated using free 4-nitrophenol (ϵ405=18500 M−1·cm−1).
RESULTS
The present study was initiated to examine a possible esterase activity of HDACs. Esters and amides are rather similar in structure and indeed a number of enzymes have been reported to exhibit dual functionalities accepting either substrate classes [26–29]. The work presented herein concentrates particularly on the HDAH from Bordetella Alcaligenes strain FB188 [14] as an HDAC homologue which can be easily produced in large quantities. FB188 HDAH exhibits 30% identity over 291 residues with the first domain of human HDAC6 (HDAC6a), 35% identity with the second domain (HDAC6b), and both enzymes share a number of functional similarities. They are zinc-dependent amidohydrolases that are able to efficiently deacetylate chicken histones, and they show the same substrate specificity towards small peptidic substrates [20,30]. In addition, FB188 HDAH is also inhibited by common HDAC inhibitors such as SAHA (suberoylanilide hydroxamic acid). Furthermore, the crystal structure of FB188 HDAH reveals the canonical fold of HDACs [19].
HDAH is an efficient esterase
Previously, we have shown that FB188 HDAH is an amidohydrolase that catalyses the release of acetate from ϵ-acetylated lysine residues in the context of various substrates including fluorogenic peptides ([14,20,25,31] and D. Riester, C. Hildmann, S. Grünewald, T. Beckers and A. Schwienhorst, unpublished work) as well as eukaryotic histones [14]. In addition, we demonstrated that the enzyme also converted non-peptidic amides such as cis-(+/−)-N-[4-(hydroxymethyl)cyclopent-2-enyl] acetamide [14]. For Boc-L-Lys(ϵ-acyl)-MCA substrates (Figures 1A and 1B) the acetyl derivative was the superior substrate with Km and Vmax values of 14.4±3.0 μM and 136±22 pmol/s/mg respectively [20]. Boc-L-Lys(ϵ-propionyl)-MCA, however, did not show a significant conversion [20]. To test the possibility that FB188 HDAH could also act as an esterase we studied esterase activity in comparison to that of porcine liver esterase and P. fluorescens lipase using 4-methylcoumarin-7-acetate as a substrate (Figure 1C; Table 1). Cleavage of this substrate resulted in the release of fluorescent HMC, which was monitored by measuring the fluorescence at λem=460 nm and λex=390 nm. HDAH indeed proved to exhibit specific esterase activity that was comparable with or even superior to those of porcine liver esterase and P. fluorescens lipase (Table 1), which were used as controls. For 4-methylcoumarin-7-acetate Km and Vmax values of 0.90±0.13 μM and 14100±2100 pmol/s/mg were measured respectively. For porcine liver esterase the corresponding values were 2.84±0.84 μM and 2150±630 pmol/s/mg. For P. fluorescens lipase we obtained Km and Vmax values of 1.66±0.30 μM and 183±41 pmol/s/mg respectively.
Table 1. Kinetic parameters for chromogenic and fluorogenic substrates.
NA, no activity (background); nd, not determined.
| HDAH | Y312F HDAH | P. fluorescens lipase | Porcine liver esterase | |||||
|---|---|---|---|---|---|---|---|---|
| Substrate | Km (μM) | Vmax (pmol/s/mg) | Km (μM) | Vmax (pmol/s/mg) | Km (μM) | Vmax (pmol/s/mg) | Km (μM) | Vmax (pmol/s/mg) |
| A Boc-L-Lys(ϵ-acetyl)-MCA* | 14.4±3.0 | 136±22 | NA | NA | NA | NA | NA | NA |
| B Boc-L-Lys(ϵ-propionyl)-MCA | NA | NA | nd | nd | nd | nd | nd | nd |
| C 4-methyl-coumarin-7-acetate | 0.90±0.13 | 14100±2100 | 1.68±0.32 | 513000±97700 | 1.66±0.30 | 183±41 | 2.84±0.84 | 2150±630 |
| D 4-methyl-coumarin-7-propionate | 31.2±3.7 | 2220±260 | 0.22±0.02 | 52400±4700 | 2.54±0.23 | 313±28 | nd | nd |
| E 4-nitrophenyl-acetate | 11.8±1.8 | 33000±4900 | nd | nd | 47.0±0.8 | 4850±1220 | nd | nd |
*named as in Figure 1.
Esterase activity is dependent on the acyl moiety
In the aforementioned experiments with amide substrates, FB188 HDAH exhibited a clear preference for acetamides over the corresponding propionyl derivatives. We therefore also tested 4-methylcoumarin-7-propionate as a substrate (Table 1). For the latter substrate Km and Vmax values of 31.2±3.7 μM and 2220±260 pmol/s/mg were measured respectively. Thus the enzyme again showed a preference for the acetyl derivative, reflected by both a 34-fold increase in Km and a 6-fold decrease in Vmax for 4-methylcoumarin-7-propionate.
Esterase activity is modulated by the nature of the alcoholic moiety
To show whether the alcohol moiety of the ester has a significant influence on the kinetic parameters of the ester substrates we compared the conversions of the aforementioned 4-methylcoumarin-7-acetate and 4-nitrophenyl acetate (Figure 1E; Table 1). For the latter, we measured Km and Vmax values of 11.8±1.8 μM and 33000±4900 pmol/s/mg respectively, indicating a 13-fold increase in Km and a 2-fold increase in Vmax.
Esterase activity can be blocked by HDAC inhibitors
To confirm that the esterase activity resides in the same active site of FB188 HDAH that is used to catalyse amidohydrolase reactions we tested esterase activity in the presence of known HDAC inhibitors (Table 2). For SAHA and CypX (cyclopentyle-propionyle hydroxamic acid), IC50 values of 0.43 μM and 0.59 μM were obtained using 4-methylcoumarin-7-acetate as a substrate. Comparing these IC50 values with that obtained in the fluorogenic HDAC assay ([24]; Table 2) it appeared that the inhibition of amidohydrolase and esterase activities were comparable for both hydroxamate inhibitors.
Table 2. IC50 values of HDAC inhibitors monitored by effects on amidohydrolase and esterase activity.
NA, no activity (background); nd, not determined.
| IC50 (μM) | ||||
|---|---|---|---|---|
| HDAC assay* | Fluorogenic esterase assay† | |||
| Inhibitor | HDAH | HDAH | P. fluorescens lipase | Porcine liver esterase |
| SAHA | 0.95 | 0.43 | 3.82 | NA |
| CypX | 0.29 | 0.59 | nd | NA |
*[24].
†Based on 4-methylcoumarin-7-acetate as a substrate.
Interestingly, HDAC inhibitors also showed a significant inhibition of control esterases. For P. fluorescens lipase, SAHA showed an IC50 value of 3.82. For porcine liver esterase no inhibitory activity was observed for the hydroxamic acids (SAHA, CypX). In this case, however, MS275 proved to be a rather good inhibitor (results not shown). Furthermore, for a trifluoromethylketone derivative which we recently had identified in a screening program as a submicromolar inhibitor of HDAH and eukaryotic HDACs, we could also detect submicromolar inhibition of porcine liver esterase (results not shown).
Y312F mutation impairs amidohydrolase but not esterase activity
To further confirm that both amidohydrolase and esterase activities reside in the same active site and to study possible similarities in the catalytic mechanism between FB188 HDAH acting as an amidohydrolase or esterase we analysed the effect of mutations on enzyme activity (Table 1). Whereas the mutation H142N (inner charge-transfer relay system) proved to be inactive for both enzyme activities, Y312F only decreased amidohydrolase activity. Surprisingly, esterase activity was even improved in the mutant as compared with the wild-type enzyme. Using 4-methylcoumarin-7-acetate as a substrate, Km and Vmax values of 1.68±0.32 μM and 513000±97700 pmol/s/mg respectively, were measured. This is less than a 2-fold increase in Km but a 36-fold increase in Vmax compared with the wild-type enzyme. Using 4-methylcoumarin-7-propionate as a substrate, Km and Vmax values of 0.22±0.02 μM and 52400±4700 pmol/s/mg were measured respectively. This is a 4-fold decrease in Km and a 23-fold increase in Vmax as compared with the wild-type enzyme and the same substrate.
Human HDAC1, HDAC3 and HDAC8 also exhibit esterase activity
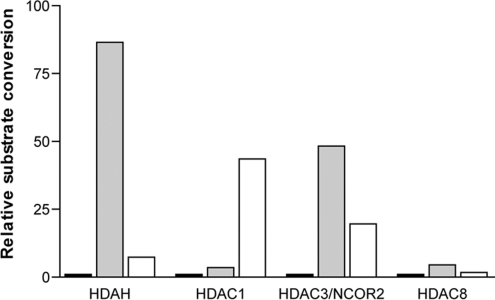
Next, we were interested to know whether human HDACs are also capable, in principle, of executing esterase activity. Human HDAC1, 3 and 8 substrate conversions (in pmol/min) were compared using the same enzyme and substrate concentrations but different substrates. In Figure 2 the relative substrate conversions are depicted by arbitrarily setting the conversion of the HDAC substrate [Boc-L-Lys(ϵ-acetyl)-MCA] to 1. Using both 4-methylcoumarin-7-acetate and 4-methylcoumarin-7-propionate as substrates, we clearly demonstrated that HDAC1, HDAC3/N-COR2 and HDAC8 also have esterase activities. However, different specificities concerning the acyl moiety were observed. HDAC1 appeared to have a preference for the propionate derivative of the ester. As compared with Boc-L-Lys(ϵ-acetyl)-MCA, the ester substrates were more efficiently converted by a factor of 3 and 43 for the acetate and the propionate derivative respectively. For HDAC3/N-COR2 and HDAC8, similar to FB188 HDAH, a preference for the acetate derivative of the ester was observed. Here, as compared with the conversion of Boc-L-Lys(ϵ-acetyl)-MCA, the ester substrates were more efficiently converted by a factor of 48 and 20 for the acetate and the propionate derivatives respectively.
Figure 2. Histogram of amidohydrolase and esterase activities of different HDACs.
Fluorogenic amide Boc-L-Lys(ϵ-acetyl)-MCA (black bars) or esters 4-methylcoumarin-7-acetate (grey bars) and 4-methylcoumarin-7-propionate (white bars) were used as substrates respectively. Human HDAC1, 3 and 8 substrate conversions in pmol/min were compared using the same enzyme and substrate concentrations but different substrates. Relative substrate conversions are depicted by arbitrarily setting the conversion of the HDAC substrate Boc-L-Lys(ϵ-acetyl)-MCA to 1 for each enzyme.
DISCUSSION
Esterase activity of FB188 HDAH
The major biological function of HDACs is believed to be the deacetylation of ϵ-acetylated lysines in histones and other proteins [33]. For their bacterial counterparts the biological role is much less clear. Experimental data available so far supports the idea that these enzymes may also function as deacetylases of proteins with ϵ-acetylated lysine residues [14] as well as polyamines [15]. In the present paper we present experimental evidence that HDAC-like enzymes are also very catalytically efficient esterases. Using chromogenic and fluorogenic substrates we demonstrated that FB188 HDAH indeed has esterase activity comparable with or superior to that of well-known lipases and esterases. In terms of specific substrate conversion, the catalytic properties of HDAC-like enzymes conform to the general reactivity of amides and esters: in contrast with esters, amides are known to be resonance-stabilized, as reflected by a more than 103 times slower reaction in OH−-catalysed reactions [34–36]. In the present study we observed that the bacterial FB188 HDAH hydrolysed specific ester substrates with deacylation rates that exceeded those of amides by up to two orders of magnitude. Other researchers have reported on similar results obtained with certain serine proteases which also show amidohydrolase and esterase activity [34–38].
In both ester and amide substrates, FB188 HDAH has a significant preference for acetyl over propionyl moieties, presumably due to space limitations in the active-site cleft [25]. In line with this assumption, the larger 4-methylcoumarin-7-propionate is only bound well in the larger active site of the Y312F mutant, as reflected by a 142-fold decrease in the Km value. On the other hand, comparing substrates based on 4-nitrophenol or HMC it appears that enzyme activity is less dependent on the alcohol moiety. However, as shown for amide substrates [14,16], it cannot be excluded that there may be more dramatic effects with other alcohol moieties.
Implications for the catalytic mechanism
Since standard HDAC/HDAH inhibitors such as SAHA and CypX inhibited not only amidohydrolase but also esterase activity, we tend to believe that both catalytic activities reside in the same active-site cleft. Although differences between IC50 values measured in different assays in general can be fairly large [39], for SAHA and CypX amidohydrolase and esterase assays the IC50 data differed only by a factor of 2. In addition, both the amidohydrolase and the esterase activities show the same preference concerning acyl moieties, thus arguing against the possibility of two independent and rather different mechanisms. The view that both catalytic activities reside in the same active site gained further support from the fact that a H142N mutation in the ‘inner charge-transfer relay system’ was detrimental to both amidohydrolase activity and esterase function.
Consistent with the aforementioned facts we propose a mechanism for ester cleavage (Figures 3C–3E) that very much resembles the proposed mechanism for amide cleavage. In both cases a water molecule as the attacking nucleophile is activated by interaction with an Asp–His142 charge-transfer relay system and a zinc ion (Figure 3C). To increase the electrophilicity of the carbonyl carbon, the carbonyl bond of the ester or amide is polarized by interactions of the carbonyl oxygen with the zinc ion and with the hydroxyl group of Tyr312 (Figure 3C). Given the higher reactivity of esters compared with that of amides it seems plausible to assume that part of this carbonyl polarization is dispensable in the case of ester cleavage. This would explain the fact that a Y312F mutation did impair the energetically more demanding amidohydrolase activity but still allowed the enzyme to work as an esterase. Furthermore, in the originally proposed mechanism it was stressed that tyrosine would stabilize the tetrahedral oxyanion transition-state by hydrogen bonding to the carbonyl oxygen (Figure 3D). At least in the case of ester cleavage this function of tyrosine is obviously also non-essential. Surprisingly, however, a Y312F mutation did stimulate esterase activity, particularly Vmax. As mentioned earlier, FB188 HDAH shows space limitations in the active-site cleft. These structural constraints may force the substrate and side chains of the enzyme into unfavourable conformations. In line with this assumption, the absence of a hydroxyl group in mutant Y312F would provide the active site with additional space, i.e. conformational freedom that could stimulate catalytic activity at least in the case of ester cleavage. Future crystallographic studies will have to specify the structural differences between wild-type HDAH and the Y312F mutant that presumably causes the observed differences in catalytic activity.
The fact that amidohydrolase activity is more sensitive to small structural perturbations in the active site as compared with esterase activity has also been observed in other systems. It is for example a well-known fact that amidohydrolase activity of serine proteases can be easily damaged by site-directed mutagenesis [40] or chemical modifications [41–45]. In contrast, esterase activity was much less affected and even increased in some cases, although never to the extent reported in the present paper for the Y312F mutant of FB188 HDAH. Thus it appears that hydrolases, in general, require a more finely tuned and stronger catalytic activity to hydrolyse energetically more demanding amides as compared with ester cleavage.
Implications for possible biological roles of HDAC-like enzymes and HDACs
In line with previous findings indicating that FB188 HDAH structurally and functionally resembles eukaryotic HDACs, we observed that not only FB188 HDAH but particularly also recombinant human HDAC1 and HDAC3 exhibited pronounced esterase activity that far exceeded their amidohydrolase activity in terms of molar substrate conversion. In regard to specificity, however, we noticed some differences. Whereas HDAC3/N-COR2 and HDAC8 resembled FB188 HDAH in favouring the acetyl derivative of the ester substrate, HDAC1 showed a clear preference for the propionyl derivative. It can be speculated that esterase activity may have preceded amidohydrolase activity, which later in evolution may have been acquired through mutation of an active-site residue (Tyr312 in HDAH). The mere existence of a pronounced esterase activity neither proves nor disproves that members of the HDAC family at present have biological roles as esterases. The discovery of esterase activity, however, could be taken as an opportunity to reconsider our current view of HDACs as specialized amidohydrolases and to motivate further research to identify biologically important esters as possible biological substrates of HDACs.
Implications for HDAC inhibitor development
It is a well-known fact that inhibitors of HDACs affect angiogenesis, cell cycle arrest, apoptosis, terminal differentiation of different cell types and the pathogenesis of malignant disease [4]. The development of HDAC inhibitors has become a very promising therapeutic approach in modern cancer research [8]. However, mass screening for HDAC inhibitors, particularly in miniaturized formats, is still impeded by the relatively modest activities of the recombinant enzymes available. Furthermore, the most common fluorogenic assay in primary screening [24] usually has to be performed as a two-step endpoint assay due to the susceptibility of most HDACs to trypsin [39]. In the light of our results it may be worthwhile to consider the esterase assay as a possible alternative [46]. In the esterase assay, substrate conversion rates are much more favourable for highly sensitive detection of small amounts of enzyme, particularly in miniaturized sample carriers. Furthermore, the esterase assay e.g. with substrates C or D (Figure 1), permits continuous monitoring of enzyme activity. At least for all hydroxamate and trifluoromethylketone inhibitors tested so far the results of both assays, for example the IC50 values, are comparable. However, classes of compound may exist for which the transferability of results is not guaranteed. Whatever assays are used for HDAC inhibitor screening though, it appears mandatory to include major esterases in the inhibitor profiling. Our results indicate that at least certain hydroxamates and trifluoromethylketone inhibitors of HDACs are also very efficient lipase or esterase inhibitors and therefore could cause unwanted side-effects in the treatment of patients.
Acknowledgments
This research was in part supported by grant BioFuture 0311852 from the Bundesministerium für Forschung und Technologie, Germany. Technical support from Cybio and BMG is greatly appreciated. HDAC1, HDAC3/N-COR2 were a gift from H. Zhu (BPS Bioscience Inc, San Diego, U.S.A.). We would like to thank T.K. Nielsen for helpful discussions and help in Figure preparation. A.S. thanks G. Garrettson for critically reading the manuscript.
References
- 1.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 2.Peterson C. L. HDAC's at work: everyone doing their part. Mol. Cell. 2002;9:921–929. doi: 10.1016/s1097-2765(02)00534-8. [DOI] [PubMed] [Google Scholar]
- 3.Wade P. A., Pruss D., Wolffe A. P. Histone acetylation: chromatin in action. Trends Biochem. Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 4.Chung D. Histone modification: the ‘next wave’ in cancer therapeutics. Trends Mol. Med. 2002;8:S10–S11. doi: 10.1016/s1471-4914(02)02303-1. [DOI] [PubMed] [Google Scholar]
- 5.Johnstone R. W. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nature Rev. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 6.Rosato R. R., Grant S. Histone deacetylase inhibitors in cancer therapy. Cancer Biol. Ther. 2003;2:30–37. doi: 10.4161/cbt.190. [DOI] [PubMed] [Google Scholar]
- 7.Villar-Garea A., Esteller M. Histone deacetylase inhibitors: understanding a new wave of anticancer agents. Int. J. Cancer. 2004;112:171–178. doi: 10.1002/ijc.20372. [DOI] [PubMed] [Google Scholar]
- 8.Minucci S., Pelicci P. G. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nature Rev. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 9.Gregoretti I. V., Lee Y.-M., Goodson H. V. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Grozinger C. M., Schreiber S. L. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 11.Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.-F., Yao T.-P. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 12.Matsuyama A., Shimazu T., Sumida Y., Saito A., Yoshimatsu Y., Seigneurin-Berry D. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Li N., Caron C., Matthias G., Hess D., Khochbin S., Matthias P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003;22:1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildmann C., Ninkovic M., Dietrich R., Wegener D., Riester D., Zimmermann T., Birch O. M., Bernegger C., Loidl P., Schwienhorst A. A new amidohydrolase from Bordetella/Alcaligenes strain FB188 with similarities to histone deacetylases. J. Bacteriol. 2004;8:2328–2339. doi: 10.1128/JB.186.8.2328-2339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakurada K., Ohta T., Fujishiro K., Hasegawa M., Aisaka K. Acetylpolyamine amidohydrolase from Mycoplana ramosa: gene cloning and characterization of the metal-substituted enzyme. J. Bacteriol. 1996;178:5781–5786. doi: 10.1128/jb.178.19.5781-5786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finnin M. S., Donigian J. R., Cohen A., Richon V. M., Rifkind R. A., Marks P. A., Breslow R., Pavletich N. P. Structures of a histone deacetylase homologue bound to TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 17.Vannini A., Volpari C., Filocamo G., Casavola E. C., Brunetti M., Renzoni D., Chakravarty P., Paolini C., De Francesco R., Gallinari P., Steinkühler C. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Nat. Acad. Sci. U.S.A. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somoza J. R., Skene R. J., Katz B. A., Mol C., Ho J. D., Jennings A. J., Luong C., Arvai A., Buggy J. J., Chi E., et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen T. K., Hildmann C., Dickmanns A., Schwienhorst A., Ficner R. Crystal structure of a bacterial class 2 histone deacetylase homologue. J. Mol. Biol. 2005;354:107–120. doi: 10.1016/j.jmb.2005.09.065. [DOI] [PubMed] [Google Scholar]
- 20.Riester D., Wegener D., Hildmann C., Schwienhorst A. Members of the histone deacetylase superfamily differ in substrate specificity towards small synthetic substrates. Biochem. Biophys. Res. Commun. 2004;324:1116–1123. doi: 10.1016/j.bbrc.2004.09.155. [DOI] [PubMed] [Google Scholar]
- 21.Hassig C. A., Tong J. K., Fleischer T. C., Owa T., Grable P. G., Ayer D. E., Schreiber S. L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadosh D., Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanommeslaeghe K., VanAlsenoy C., DeProft F., Martins J. C., Tourwé D., Geerlings P. Ab initio study of the binding of trichostatin A (TSA) in the active site of histone deacetylase like protein (HDLP) Org. Biomol. Chem. 2003;1:2951–2957. doi: 10.1039/b304707e. [DOI] [PubMed] [Google Scholar]
- 24.Wegener D., Wirsching F., Riester D., Schwienhorst A. A fluorogenic histone deacetylase assay well suited for high-throughput activity screening. Chem. Biol. 2003;10:61–68. doi: 10.1016/s1074-5521(02)00305-8. [DOI] [PubMed] [Google Scholar]
- 25.Hildmann C., Wegener D., Riester D., Hempel R., Schober A., Merana J., Giurato L., Guccione S., Nielsen T. K., Ficner R., Schwienhorst A. Substrate and inhibitor specificity of class 1 and class 2 histone deacetylases. J. Biotechnol. 2006;124:258–270. doi: 10.1016/j.jbiotec.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 26.Thompson R. C., Blout E. R. Evidence for an extended active center in elastase. Proc. Natl. Acad. Sci. U.S.A. 1970;67:1734–1740. doi: 10.1073/pnas.67.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hui D. Y., Hayakawa K., Oizumi J. Lipoamidase activity in normal and mutagenized pancreatic cholesterol esterase (bile salt-stimulated lipase) Biochem. J. 1993;291:65–69. doi: 10.1042/bj2910065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alkema W. B. L., Dijkhuis A.-J., de Vries E., Janssen D. B. The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of pencillin acylase. Eur. J. Biochem. 2002;269:2093–2100. doi: 10.1046/j.1432-1033.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- 29.Komeda H., Asano Y. A DmpA-homologous protein from Pseudomonas sp. is a dipeptidase specific for -alanyl dipeptides. FEBS J. 2005;272:3075–3084. doi: 10.1111/j.1742-4658.2005.04721.x. [DOI] [PubMed] [Google Scholar]
- 30.Heltweg B., Dequiedt F., Marshall B. L., Brauch C., Yoshida M., Nishino N., Verdin E., Jung M. Subtype selective substrates for histone deacetylases. J. Med. Chem. 2004;47:5235–5243. doi: 10.1021/jm0497592. [DOI] [PubMed] [Google Scholar]
- 31.Wegener D., Hildmann C., Riester D., Schwienhorst A. Improved fluorogenic histone deacetylase assay for high-throughput-screening applications. Anal. Biochem. 2003;321:202–208. doi: 10.1016/s0003-2697(03)00426-3. [DOI] [PubMed] [Google Scholar]
- 32. Reference deleted.
- 33.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bender M. L., Schonbaum G. R., Zerner B. Spectrophotometric investigation of the mechanism of α-chymotrypsin catalysed hydrolases. J. Am. Chem. Soc. 1962;84:2540–2550. [Google Scholar]
- 35.Zerner B., Bond R. P. M., Bender M. L. Kinetic evidence for the formation of acyl-enzyme intermediates in the α-chymotrypsin-catalyzed hydrolyses of specific substrates. J. Am. Chem. Soc. 1964;113:2259–2263. [Google Scholar]
- 36.Bender M. L., Kezdy J. Mechanism of action of proteolytic enzymes. Ann. Rev. Biochem. 1965;34:49–76. doi: 10.1146/annurev.bi.34.070165.000405. [DOI] [PubMed] [Google Scholar]
- 37.Walsh C. San Francisco: Freeman; 1979. Enzymatic reaction mechanisms. [Google Scholar]
- 38.Bennet A. J., Brown R. S. Comprehensive Biological Catalysis: A Mechanistic Reference, vol. 1. In: Sinnott M., editor. San Diego: Academic Press; 1998. pp. 293–326. [Google Scholar]
- 39.Wegener D., Hildmann C., Schwienhorst A. Recent progress in the development of assays suited for histone deacetylase inhibitor screening. Mol. Gen. Metab. 2003;80:138–147. doi: 10.1016/j.ymgme.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 40.Abrahamsén L., Tom J., Burnier J., Butcher K. A., Kossiakoff A., Wells J. A. Engineering subtilisin and its substrates for efficient ligation of peptide bonds in aqueous solution. Biochemistry. 1991;30:4151–4159. doi: 10.1021/bi00231a007. [DOI] [PubMed] [Google Scholar]
- 41.Nakatsuka T., Sasaki T., Kaiser E. T. Peptide segment coupling catalyzed by the semisynthetic enzyme thiolsubtilisin. J. Am. Chem. Soc. 1987;109:3808–3810. [Google Scholar]
- 42.West J. B., Scholten J. D., Stolowich N. J., Hogg J. L., Scott A. I., Wong C.-H. Modification of protease to esterase for peptide synthesis: methyl chymotrypsin. J. Am. Chem. Soc. 1988;110:3709–3710. [Google Scholar]
- 43.Zhong Z., Bibbs J. A., Yuan W., Wong C. H. Active-site-directed modification of subtilisin. J. Am. Chem. Soc. 1991;113:2259–2263. [Google Scholar]
- 44.Plettner E., DeSantis G., Stabile M. R., Jones J. B. Modulation of esterase and amidase activity of subtilisin Bacillus lentus by chemical modification of cysteine mutants. J. Am. Chem. Soc. 1999;121:4977–4981. [Google Scholar]
- 45.Lloyd A. C., Davis B. G., Jones J. B. Site-selective glycosylation of subtilisin Bacillus lentus causes dramatic increases in esterase activity. Bioorg. Med. Chem. 2000;8:1537–1544. doi: 10.1016/s0968-0896(00)00084-5. [DOI] [PubMed] [Google Scholar]
- 46.Gronlund J. L., Yu M. K., Concepcion A. M., He S., Venkataramani R., Marmorstein R., McCafferty D. G. Peptides: The Wave of the Future. In: Lebl M., Houghten R. A., editors. Norwell: Kluwer Academic Publishers; 2002. pp. 541–542. [Google Scholar]