Abstract
GSTs (glutathione transferases) are multifunctional widespread enzymes. Currently there are 13 identified classes within this family. Previously most structural characterization has been reported for mammalian Alpha, Mu and Pi class GSTs. In the present study we characterize two enzymes from the insect-specific Delta class, adGSTD3-3 and adGSTD4-4. These two proteins are alternatively spliced products from the same gene and have very similar tertiary structures. Several major contributions to the dimer interface area can be separated into three regions: conserved electrostatic interactions in region 1, hydrophobic interactions in region 2 and an ionic network in region 3. The four amino acid side chains studied in region 1 interact with each other as a planar rectangle. These interactions are highly conserved among the GST classes, Delta, Sigma and Theta. The hydrophobic residues in region 2 are not only subunit interface residues but also active site residues. Overall these three regions provide important contributions to stabilization and folding of the protein. In addition, decreases in yield as well as catalytic activity changes, suggest that the mutations in these regions can disrupt the active site conformation which decreases binding affinity, alters kinetic constants and alters substrate specificity. Several of these residues have only a slight effect on the initial folding of each subunit but have more influence on the dimerization process as well as impacting upon appropriate active site conformation. The results also suggest that even splicing products from the same gene may have specific features in the subunit interface area that would preclude heterodimerization.
Keywords: Anopheles dirus, glutathione transferase (GST), hydrophobic interaction, subunit interface
Abbreviations: ANS, 8-anilino-1-naphthalene sulfonate; CDNB, 1-chloro-2,4-dinitrobenzene; DCNB, 1,2-dichloro-4-nitrobenzene; EA, ethacrynic acid; GST, glutathione transferase; PNBC, p-nitrobenzyl chloride; PNPB, p-nitrophenethyl bromide
INTRODUCTION
GSTs (glutathione transferases; EC 2.5.1.18) are a supergene family of multifunctional enzymes which are widely distributed in nature and found in most aerobic eukaryotes and prokaryotes. The dimeric cytosolic GSTs catalyse reactions with a broad range of substrates and play an essential role in detoxification of endogenous and xenobiotic compounds [1,2]. The dimerization of the GSTs not only contributes to stabilization of the subunit tertiary structure, but also allows for the construction of a fully functional active site [3]. Although tertiary structures of all classes of GSTs are similar, dimerization is highly specific and is thought to occur only between subunits within the same class [4,5].
The structural features at the dimer interface of the GSTs suggest at least two major subunit interaction areas [6]. The first area is the predominantly hydrophobic interaction at the outer ends of the interface called a hydrophobic ‘lock-and-key’ (also referred to as a ‘ball-and-socket’) motif which is formed by the insertion of an aromatic residue from domain I of one subunit into a ‘lock’ of five residues of domain II in the other subunit [6–9]. The second major subunit interaction area is the symmetry axis interactions at the 2-fold axis of the protein which show highly conserved electrostatic interactions at one edge of the subunit interface and a variety of interactions along the interface depending on the GST class. The Anopheles dirus mosquito is an important malaria vector in South East Asia. From an A. dirus genomic library, a 7.5 kb fragment containing the adgst1AS1 gene (A. dirus alternatively spliced GST gene) was identified [10]. This gene contains six exons that encode four Delta class GSTs, adGSTD1-1, 2-2, 3-3 and 4-4, which possess 61–77% amino acid identity compared among themselves. Previously these proteins had been named adGST1-1, 1-2, 1-3 and 1-4, according to insect GST nomenclature in use (that is, insect class 1-protein 1, 2, 3 and 4 respectively). However, to be in alignment with a proposed universal GST nomenclature, the proteins were renamed adGSTD1-1, adGSTD2-2, adGSTD3-3 and adGSTD4-4 respectively [11,12]. The subunit number remains the same, since subunits were enumerated as they were initially discovered, ‘D’ refers to GST Delta class and ‘4-4’ refers to the homodimeric isoenzyme. These four GSTs share an untranslated exon 1 and a translated exon 2 coding for 45 amino acids at the N-terminus but vary between four different exon 3 sequences (exons 3A–3D). The arrangement of each exon is similar to the aggst1α gene from A. gambiae, the major malaria vector in Africa, with approx. 79% nucleotide identity for the two genes [13]. In A. dirus, although four splice products are encoded from the same gene, the enzymes possess distinct enzyme kinetic properties for substrates, inhibitors, including insecticides, as well as physical properties such as stability [14,15]. Although two splice products, adgstD3-3 (PDB accession number: 1JLV) and adgstD4-4 (PDB accession number: 1JLW), have very similar tertiary structures when aligned, the amino acid identity is only 68% [16].
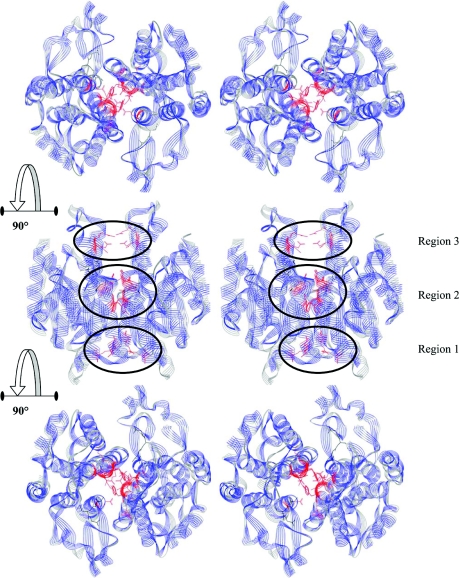
When compared with human Alpha, Mu and Pi classes, the dimer interfaces of both Delta class isoenzymes are more extensively hydrophilic. Although lacking the previously identified hydrophobic ‘lock-and-key’ motif (a conserved motif in human Alpha, Mu and Pi classes), the Delta class does possess a ‘Clasp’ motif with a similar function [17]. In addition to this motif, there are nine amino acids making major contributions to the interactions within this interface area, which can be separated into three regions: conserved electrostatic interactions in region 1, hydrophobic interactions in region 2 and an ionic network in region 3 (Figure 1).
Figure 1. The three regions of the interface characterized in the present study.
The two GST proteins are carbon backbone aligned with the adGSTD3-3 secondary structure wire ribbon shown in blue and the adGSTD4-4 secondary structure ribbon shown in grey. The GSH in the two active sites is shown as a black stick; the residues engineered in the present study are shown in red. The top panel is a stereo view looking at the 2-fold axis from the side opposite to the active sites. The middle panel shows the proteins rotated 90°, to show the position of the studied interface residues down the length of α-helices 3 and 4. The bottom panel shows the proteins rotated a further 90° and looking down upon the 2-fold axis on to the active sites. The Figure was created using Accelrys DS ViewerPro 5.0.
The conserved electrostatic interactions in region 1 of the subunit interface are formed by two amino acid residues from one subunit (a glutamate residue in α-helix 3 and an arginine residue in α-helix 4) interacting with the same two amino acids from the other subunit. These interactions are of interest because the four amino acid side chains interact with each other as a planar rectangle with distances of 3.83–4.26 Å (1 Å=0.1 nm). These interactions are highly conserved among the GST classes, Delta, Sigma and Theta, however at present there are no reports of these interactions for any of the three GST classes. In adGSTD4-4, each arginine residue not only interacts with both of the negatively charged Glu75 residues in both subunits, but also forms cation–π interactions with the aromatic ring of Tyr89 and an ionic interaction with the carboxy group of Pro90. In adGSTD3-3, the equivalent residues to Glu75 and Arg96 of adGSTD4-4 are Glu74 and Arg90. These residues also form the same planar rectangle arrangement within adGSTD3-3 and are stabilized in a similar manner by interactions with Tyr83 and Pro84 (Tyr89 and Pro90 in adGSTD4-4). Therefore as the motif appears to be highly conserved in both proteins it was only studied in adGSTD4-4. In addition, these interactions appear to be highly conserved among the insect GST classes. Therefore the aim of the present study was to determine whether the inter- and intra-subunit electrostatic interactions of these residues are important contributions that help to maintain tertiary and quaternary structures.
Region 2 at the subunit interface shows the most variation in amino acid residues at equivalent positions between the two isoenzymes, Tyr98, Met101 and Gly102 of adGSTD3-3 and Phe104, Val107 and Ala108 of adGSTD4-4 respectively. These residues are of interest because they are not only subunit interface residues but also active site residues with several of them involved in both active sites of the dimeric enzyme. Therefore the amino acids at the equivalent positions of the two isoenzymes were studied by switching the amino acids of the two proteins; Y98F, M101V, G102A and Y98F/M101V/G102A of adGSTD3-3 and F104Y, V107M, A108G and F104Y/V107M/A108G of adGSTD4-4.
The last region is the hydrophilic area in region 3 of the subunit interface, Asp110 of adGSTD3-3 and Glu116 of adGSTD4-4. Not only are these subunit interface residues, but these two equivalent positions also are involved in the active site as part of the H-site (hydrophobic substrate-binding site). For adGSTD4-4, Glu116 forms hydrogen bonds with Arg134 in both inter- and intra-subunit interactions; however, these interactions do not appear in adGSTD3-3. For adGSTD3-3, there are hydrogen bonds only in the same subunit between Asp110 and the highly conserved residue Glu106. To study the influence of the ionic network in region 3 of the subunit interface and whether it impacts upon the catalytic activity and stability of the enzymes, mutations at the equivalent positions of these two isoenzymes were generated, that is, D110A of adGSTD3-3 and E116A of adGSTD4-4.
MATERIALS AND METHODS
Site-directed mutagenesis and protein purification
The adGSTD3-3 and adGSTD4-4 plasmid DNA templates were prepared from previous constructs [14]. The construction of the mutants was based on the Stratagene QuikChange® site-directed mutagenesis kit using pfu DNA polymerase. The expression constructs of recombinant plasmid that were obtained were sequenced at least twice and transformed into Escherichia coli BL21DE3plysS. The soluble recombinant GSTs were purified by GSTrap affinity chromatography (Amersham Phamacia) as previously described [14]. After purification, proteins were homogeneous as judged by SDS/PAGE, and the protein concentration was determined using the Bradford protein reagent with bovine serum albumin as a standard [18].
Catalytic activity and kinetic studies
Steady-state kinetics were studied at various concentrations of CDNB (1-chloro-2,4-dinitrobenzene) and GSH in 0.1 M phosphate buffer (pH 6.5). The reaction was monitored at 340 nm, ϵ=9600 M−1·cm−1. Apparent kinetic parameters, kcat, Km and kcat/Km were determined by fitting the collected data to a Michaelis–Menten equation by non-linear regression analysis using GraphPad Prism version 4 (GraphPad software; www.graphpad.com) [19,20]. Specific activities of the enzymes were determined with five different substrates; CDNB, DCNB (1,2-dichloro-4-nitrobenzene), EA (ethacrynic acid), PNPB (p-nitrophenethyl bromide) and PNBC (p-nitrobenzyl chloride) as previously described [21]. Specific activities reported are the means±S.D. from at least three independent experiments. One-way ANOVA with Dunnett's post test was performed using GraphPad InStat version 3.06 for Windows (GraphPad software; www.graphpad.com).
Structural studies
Enzymes at 0.1 mg/ml final concentration were incubated at 45 °C. Inactivation time courses were determined by withdrawing suitable aliquots at different time points for an assay of remaining activity to calculate the half-life of the enzyme [22]. Data are the means±S.D. from at least three independent experiments.
Spectroscopic properties of wild-type and mutant proteins were measured for evidence of conformational changes. Intrinsic fluorescence emission spectra were measured at an excitation wavelength of 295 nm and λmax and fluorescence intensity of emission spectra were analysed at a protein concentration of 0.2 mg/ml [23].
A refolding experiment was performed with enzymes first being denatured in 4 M guanidinium chloride in renaturation buffer [0.2 M phosphate, 1 mM EDTA and 5 mM dithiothreitol (pH 7.0)] at room temperature (25 °C) for 1 h and then rapidly diluted (defining time zero) 1:40 into renaturation buffer. Therefore the final guanidinium chloride concentration was 0.1 M during refolding. Recovered activity was monitored as a function of time by withdrawal of appropriate aliquots of renaturation mixture and immediately assaying for activity. Refolding rate constants were determined by non-linear regression analysis using a single exponential equation [23].
The ANS (8-anilino-1-naphthalenesulfonate) binding assay was monitored using a PerkinElmer Luminescence spectrometer LS50B. ANS [200 μM in 0.1 M sodium phosphate buffer (pH 6.5)] was added to a final concentration of 2 μM enzyme in a 400 μl reaction mixture [24]. The spectrum of ANS in phosphate buffer (pH 6.5) was subtracted from the spectrum of protein-binding ANS. A total of three scans each for blank and sample were recorded and averaged for each enzyme. Reported spectra are the means from at least three independent experiments. Enzyme activity measurements in the presence of ANS were assessed immediately after adding ANS by using the standard reaction assay.
RESULTS AND DISCUSSION
The investigations of GST dimerization performed in mammalian GSTs indicated that the subunit interactions are a significant source of stabilization not only for the association of subunits but also for tertiary structures of the individual subunits [9,25,26]. In the present study, most mutations gave similar purification yields as the wild-type with the exception of the E75A and R96A mutants of adGSTD4-4, which had yields approx. 2 and 0.4% of the wild-type respectively. As the proteins were expressed in similar amounts, it appears that the alteration of these residues disrupts the intra-subunit interaction between helix 3 and helix 4 which impacts upon the active site architecture thereby affecting binding to the affinity matrix.
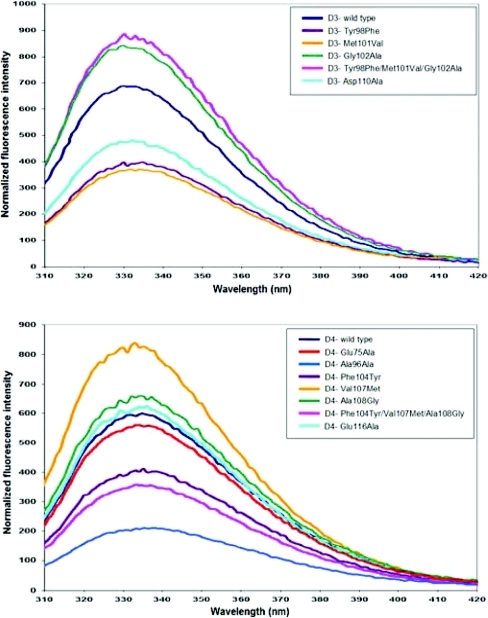
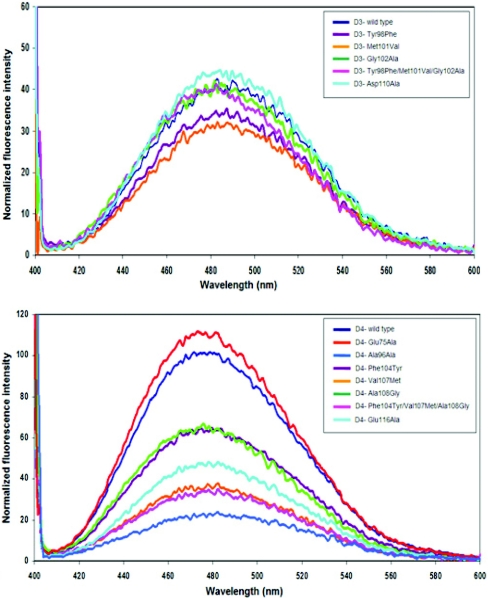
Both adGSTD3-3 and adGSTD4-4 have two tryptophan residues in each subunit that are located in β-sheet 4 in domain I and α-helix 7 in domain II. The tryptophan residue located in β-sheet 4 in domain I (Trp63) is in close proximity to the active site, with an involvement in sequestering the substrate glutathione, as well as being in the subunit interface. This makes it a sensitive fluorescence probe to monitor conformational changes at/near the active site. The normalized fluorescence spectra of adGSTD3-3 and adGSTD4-4 wild-type compared with the mutants were obtained to study the effect of mutations on the enzyme tertiary structure (Figure 2). The results showed that although every mutant had the same λmax as the wild-type, several mutants presented differences in the normalized intensities of fluorescence, implying that the mutations caused significant conformational changes in the environment of the tryptophan residues located near the subunit interface and the mutation site. The mutants also had different intensities from their wild-type especially R96A, with the intensity of fluorescence decreased by approx. 66%. This finding suggests that there are significant conformational changes in the tryptophan residue neighbourhood that distinguish the final structure of the mutant from that of the wild-type. This decrease demonstrates that movements have occurred in the tryptophan residues or in the surrounding fluorescence quenching groups.
Figure 2. Normalized intrinsic tryptophan residue fluorescence spectra of the adGSTD3-3 and adGSTD4-4 (wild-type) and the recombinant mutant GSTs.
The same colour represents the equivalent position. The data are means of three independent experiments.
The half-life of the enzymes corresponds to the time of incubation when there is 50% remaining activity (Table 1). The conserved cation–π interaction residue, Arg96, and the conserved electrostatic interaction residue, Glu75, have important roles in stabilizing enzyme structure as shown by a much decreased stability for R96A and E75A of approx. 15- and 7.4-fold respectively. For region 2, several mutants possessed similar stability as the wild-type enzymes. However, there were three mutants which had very different thermal stabilities. Two of the mutations were at an equivalent position, adGSTD3-3 Gly102 and adGSTD4-4 Ala108. Replacement of a glycine residue by an alanine residue in adGSTD3-3 at this position caused decreases of about 8-fold in half-life, whereas a glycine residue substitution of adGSTD4-4 showed an increase in half-life of about 4.7-fold. The other mutant that showed stability changes was the V104M protein which decreased the half-life by approx. 3-fold. For region 3, the half-life of D110A of adGSTD3-3 was similar to the wild-type. However, the equivalent adGSTD4-4 mutation E116A disrupted the charge–charge network and showed a 64% decreased stability for the enzyme. The results showed that the positively charged residue at the edge of the subunit interface of adGSTD4-4 participates in stabilizing the enzyme structure while the equivalent residue of adGSTD3-3 appeared to have only a minor contribution.
Table 1. Half-life, refolding rate constants, activity recovered (%) and the effect of ANS on CDNB specific activity of the adGSTD3-3 and adGSTD4-4 (wild-type) and the recombinant engineered GSTs.
The data are means±S.D. for at least three independent experiments. One-way ANOVA with Dunnett's post test was performed to show statistical significance with *P<0.05 and †P<0.01. D3 and D4 indicate adGSTD3-3 and adGSTD4-4 respectively. Inside parentheses the numbers indicate the subunit interface region: 1 is region 1, 2 is region 2 and 3 is region 3; and the same lowercase letter indicates an equivalent residue position for the two GST isoenzymes. nd, not detectable.
| Enzymes | Half-life (min) | Refolding rate constant (min−1) | Activity recovered (%) | Inhibition by ANS (%) |
|---|---|---|---|---|
| D3-wild-type | 2.71±0.35 | 0.87±0.09 | 56.9±0.23 | 24.6±1.41 |
| D3-Y98F (2/a) | 3.12±0.33 | 0.58±0.05† | 66.1±2.83* | 13.1±0.54† |
| D3-M101V (2/b) | 2.77±0.43 | 0.42±0.05† | 40.5±1.93† | 11.9±0.80† |
| D3-G102A (2/c) | 0.34±0.03† | 0.38±0.07† | 94.4±5.77† | 13.7±2.66† |
| D3-Y98F/M101V/G102A (2/d) | 1.88±0.01* | 0.31±0.03† | 85.8±4.39† | 18.4±0.65† |
| D3-D110A (3/e) | 2.21±0.24 | 0.58±0.11† | 80.5±3.48† | 23.4±1.83 |
| D4-wild-type | 14.0±1.70 | 0.59±0.03 | 19.7±0.70 | 18.4±1.20 |
| D4-G75A(1) | 1.89±0.13† | nd | 0.00 | 22.5±0.82 |
| D4-R96A(1) | 0.91±0.07† | nd | 0.00 | 18.8±3.89 |
| D4-F104Y (2/a) | 17.1±1.76* | 0.28±0.02† | 23.5±0.51 | 31.9±1.96† |
| D4-V107M (2/b) | 4.76±0.15† | nd | 0.00 | 25.4±1.41† |
| D4-A108G (2/c) | 65.4±1.45† | 0.32±0.04† | 69.6±7.55† | 32.8±1.03† |
| D4-F104Y/V107M/A108G (2/d) | 16.7±1.23 | 0.60±0.10 | 25.0±0.18 | 21.1±1.83 |
| D4-E116A (3/e) | 5.16±0.93† | 0.68±0.11 | 9.93±0.68* | 17.3±2.05 |
Three mutations in adGSTD4-4, E75A, R96A and V107M, did not recover activity after being unfolded by 4 M guanidinium chloride (Table 1). This implies that these residues play a critical role in the folding process of the enzymes. For the remaining mutants, the refolding rate constants were similar. The activity recovered illustrates the ability of the enzymes to recover their appropriate active site conformation for catalytic activity. After unfolding, the enzymes were able to recover activity ranging from 19% to 94%. To study the influence of mutations on protein folding and to assess whether the changes affected tertiary folding of each subunit or dimerization of the two subunits, intrinsic tryptophan residue fluorescence spectroscopy was performed. The fluorescence spectra of native, unfolded and refolded enzymes were monitored to compare the tertiary structure of the proteins in each state (Figures 3 and 4). The λmax values of the native (335 nm) and the unfolded form (355 nm) of the protein were observed for every enzyme. The data showed that the enzymes were refolded in a similar manner as the native form, as shown by similar λmax values, including the E75A, R96A and V107M mutants, although these enzymes could not recover catalytic activity. As shown by the similar patterns of the adGSTD3-3 mutant spectra, the mutations did not affect the tertiary folding of each subunit but influenced the dimerization process, which is necessary to achieve an appropriate active site conformation. Therefore both catalytic and structural data suggest that the single mutation of Gly102 and the triple mutation had significant effects on the dimerization of the enzymes by increasing the activity recovered from 56.9% for wild-type to approx. 94.4% and 85.8% for the two proteins respectively (Table 1). In both catalytic and structural experiments, the refolding data showed that loss of the conserved electrostatic interactions, E75A and R96A, and the hydrophobic residue, V107M, had a dramatic effect on the dimerization process, which is critical to formation of the complete active site pocket. The results for the Arg96 position indicated that not only does this residue have a critical role in the dimerization process, but it is also important for the protein folding pathway of each monomeric subunit.
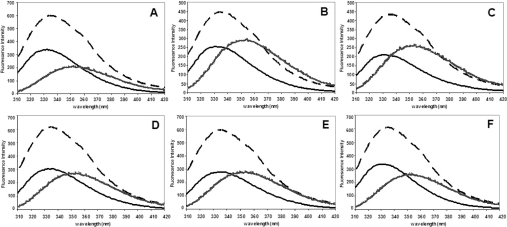
Figure 3. Normalized intrinsic tryptophan residue fluorescence spectra compared with the native, refolded and unfolded forms of the adGSTD3-3 (wild-type) and the recombinant mutant GSTs.
(A) Wild-type, (B) Y98F, (C) M101V, (D) G102A, (E) Y98F/M101V/G102A and (F) D110A. The data are means for at least three independent experiments. Solid line, native form; dashed line, refolded form; and hashed line, unfolded form of the enzyme.
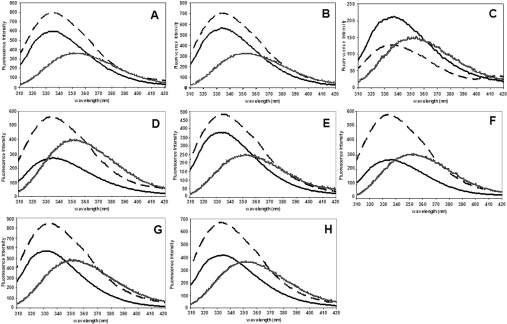
Figure 4. Normalized intrinsic tryptophan residue fluorescence spectra compared with the native, refolded and unfolded forms of the adGSTD4-4 (wild-type) and the recombinant mutant GSTs.
(A) Wild-type, (B) G75A, (C) R96A, (D) F104Y, (E) V107M, (F) A108G, (G) F104Y/V107M/A108G and (H) E116A. The data are means for at least three independent experiments. Solid line, native form; dashed line, refolded form; and hashed line, unfolded form of the enzyme.
The anionic dye ANS has been shown to bind in the solvent-exposed cleft in the subunit interface of class Alpha and Pi enzymes [27–29]. Therefore ANS was utilized as a probe to monitor the appearance/disappearance of hydrophobic patches or surfaces on the proteins that were undergoing structural changes. When ANS was bound to the proteins the fluorescence was enhanced, accompanied by a blue shift in its emission maximum from 514 nm (free ANS in buffer) to 498 nm for adGSTD3-3 and 482 nm for wild-type adGSTD4-4, indicating that the polarity of the binding site had become more hydrophobic, with decreasing polarity the greater the blue shift.
When compared with the wild-type proteins, there was no change in the emission maximum wavelength of ANS bound to all mutants (Figure 5). For the ANS fluorescence intensity, which reflects the degree of solvent quenching of ANS bound to the protein, the adGSTD4-4 mutants showed variations in the amount of ANS bound, except E75A, which showed a similar intensity to the wild-type. However, there was no relative intensity change for the adGSTD3-3 mutants. The results showed that the residue substitutions of adGSTD4-4 with the adGSTD3-3 amino acids had a dramatic effect at the subunit interface, much more than for adGSTD3-3.
Figure 5. ANS binding spectra of the adGSTD3-3 and adGSTD4-4 (wild-type) and the recombinant mutant GSTs.
The same colour represents the equivalent position. The spectra were measured immediately after addition of ANS. As ANS fluorescence is quenched by water, alteration of the fluorescence intensity of protein-bound ANS is highly dependent upon its accessibility to water. The data are means for at least three independent experiments.
To study the impact of ANS on the enzyme conformation, the enzyme activity in the presence and absence of ANS was measured using the standard reaction assay. The results showed that for the mutants, ANS molecules can bind and alter the active site architecture in a manner similar to the wild-type (Table 1). It implied that differences in ANS spectra intensities in Figure 5 occurred only from an alteration of a hydrophobic patch at the subunit-binding site.
In general, the engineered enzyme reactions followed Michaelis–Menten kinetics as did the wild-type except for E75A, F104Y and E116A of adGSTD4-4 which showed positive co-operativity upon substrate binding. Positive co-operativity is shown by a sigmoidal velocity curve which reflects the substrate binding in the first active site, that then facilitates a second substrate molecule binding in the second active site by increasing the binding affinity of the vacant binding site [30]. For CDNB, two of the enzymes had significantly different Hill coefficients (shown by one-way ANOVA with Dunnett's post test) compared with wild-type enzyme [Hill coefficients for wild-type 1.08±0.08 versus E75A 1.79±0.03 (P<0.01) and E116A 1.40±0.20 (P<0.05)]. For GSH only F104Y had a significantly different Hill coefficient (shown by one-way ANOVA with Dunnett's post test) compared with wild-type enzyme [Hill coefficients for wild-type 1.06±0.14 versus F104Y 2.01±0.09 (P<0.01)]. These data show that two of the mutant enzymes, E75A and F104Y, had very strong positive co-operativity, with the Hill coefficients approaching the number of substrate-binding sites. In addition, comparison of the kinetic constants of the mutants with the wild-type values showed that the residue changes affected additional enzymatic properties (Table 2). These effects on the active site were also reflected in changes in substrate specificity with changes in the equivalent residue position showing different effects on the two isoforms (Table 3). For example, D110A activity was significantly decreased with CDNB and DCNB in adGSTD3-3 but E116A activity was significantly increased with CDNB and showed no significant activity with DCNB in adGSTD4-4. For another position, M101V in adGSTD3-3 equivalent to V107M in adGSTD4-4, both enzymes showed significant decreases in activity with CDNB. However, the adGSTD4-4 enzyme showed significant increases in activity with DCNB and EA, whereas the adGSTD3-3 enzyme showed a significant decrease with DCNB and no significant change with EA.
Table 2. Yield of purification and kinetic parameters of the adGSTD3-3 and adGSTD4-4 (wild-type) and the recombinant engineered GSTs.
The data are means±S.D. for at least three independent experiments. One-way ANOVA with Dunnett's post test was performed to show statistical significance with *P< 0.05 and †P<0.01. D3 and D4 indicate adGSTD3-3 and adGSTD4-4 respectively. Inside parentheses the numbers indicate the subunit interface region: 1 is region 1, 2 is region 2 and 3 is region 3; and the same lowercase letter indicates an equivalent residue position for the two GST isoenzymes.
| Enzymes | CDNB kcat (s−1) | Km (mM) | kcat/Km (mM−1•s−1) | GSH Km (mM) | kcat/Km (mM−1•s−1) | Purification yield (%) |
|---|---|---|---|---|---|---|
| D3-wild-type | 39.2 | 0.15±0.01 | 258 | 0.29±0.04 | 114 | 53.7 |
| D3-Y98F (2/a) | 24.9† | 0.12±0.01 | 213 | 0.47±0.05 | 53.1 | 49.2 |
| D3-M101V (2/b) | 26.0† | 0.72±0.06† | 36.3 | 1.04±0.06† | 25.0 | 60.0 |
| D3-G102A (2/c) | 23.5† | 0.37±0.02† | 63.3 | 2.94±0.38† | 7.99 | 47.6 |
| D3-Y98F/M101V/G102A (2/d) | 40.7 | 0.28±0.03† | 147 | 1.12±0.07† | 36.7 | 58.3 |
| D3-D110A (3/e) | 51.7† | 0.49±0.07† | 106 | 0.54±0.04 | 95.8 | 34.3 |
| D4-wild-type | 22.5 | 0.63±0.09 | 35.7 | 0.67±0.05 | 33.6 | 61.3 |
| D4-G75A (1) | 23.3 | 8.26±2.63† | 2.82 | 0.54±0.06 | 43 | 5.5 |
| D4-R96A (1) | 16.2† | 0.80±0.06 | 22.0 | 1.08±0.02 | 15.0 | 1.4 |
| D4-F104Y (2/a) | 15.5† | 0.52±0.03 | 29.8 | 24.01±2.94† | 0.65 | 42.2 |
| D4-V107M (2/b) | 13.7† | 1.6±0.08 | 8.56 | 0.76±0.11 | 18.0 | 60.0 |
| D4-A108G (2/c) | 26.8 | 0.87±0.07 | 26.8 | 0.73±0.12 | 36.7 | 59.2 |
| D4-F104Y/V107M/A108G (2/d) | 32.5† | 0.72±0.04 | 45.2 | 0.25±0.02 | 130 | 59.2 |
| D4- E116A (3/e) | 46.1† | 1.55±0.01 | 29.7 | 1.24±0.06 | 37.1 | 37.1 |
Table 3. Specific activities of the adGSTD3-3 and adGSTD4-4 (wild-type) and the recombinant engineered GSTs.
The data are means±S.D. for at least three independent experiments. One-way ANOVA with Dunnett's post test was performed to show statistical significance with *P< 0.05 and †P<0.01. The substrate concentrations used were 1 mM CDNB for adGSTD3 and 3 mM CDNB for adGSTD4, 1 mM DCNB, 0.1 mM PNBC, 0.1 mM PNPB and 0.2 mM EA. D3 and D4 indicate adGSTD3-3 and adGSTD4-4 respectively. Inside parentheses the numbers indicate the subunit interface region: 1 is region 1, 2 is region 2 and 3 is region 3; and the same lowercase letter indicates an equivalent residue position for the two GST isoenzymes. nd is not detectable.
| Specific Activity (μmol/min/mg) | |||||
|---|---|---|---|---|---|
| Enzymes | CDNB | DCNB | EA | PNPB | PNBC |
| D3-wild-type | 85.3±3.23 | 0.25±0.01 | 0.10±0.05 | nd | 0.13±0.01 |
| D3-Y98F (2/a) | 46.9±6.38† | 0.23±0.03 | 0.03±0.01* | nd | 0.05±0.00† |
| D3-M101V (2/b) | 41.1±0.81† | 0.05±0.01† | 0.08±0.02 | nd | 0.09±0.00† |
| D3-G102A (2/c) | 46.8±0.25† | 0.08±0.01† | nd | nd | 0.05±0.00† |
| D3-Y98F/M101V/G102A (2/d) | 84.8±5.47 | 0.21±0.01* | 0.15±0.01 | nd | 0.08±0.00† |
| D3-D110A (3/e) | 71.6±2.69† | 0.16±0.01† | 0.01±0.01† | 0.03±0.01 | 0.07±0.01† |
| D4-wild-type | 48.0±1.98 | 0.03±0.00 | 0.27±0.00 | 0.06±0.01 | 0.03±0.01 |
| D4-E75A (1) | 36.6±5.03† | 0.06±0.01 | 0.12±0.01 | 0.02±0.01 | 0.06±0.00 |
| D4-R96A (1) | 26.0±0.13† | 0.27±0.04† | 0.18±0.01 | 0.23±0.02 | nd |
| D4-F104Y (2/a) | 28.5±2.24† | 0.06±0.01 | 0.14±0.01 | 0.05±0.01 | 0.11±0.05 |
| D4-V107M (2/b) | 22.0±2.29† | 0.08±0.004* | 2.06±0.29† | 0.31±0.08 | nd |
| D4-A108G (2/c) | 55.3±2.07 | 0.06±0.02 | 0.32±0.01 | nd | 0.08±0.02 |
| D4-F104Y/V107M/A108G (2/d) | 53.8±1.43 | 0.05±0.01 | 0.02±0.00* | nd | 0.04±0.01 |
| D4- E116A (3/e) | 56.3±6.83* | 0.06±0.01 | 0.01±0.01* | 0.04±0.01 | 0.04±0.01 |
The insect GST class Delta has a conserved planar rectangular electrostatic motif formed by four amino acid residues from different helices of both subunits (Glu75 in α-helix 3 and Arg96 in α-helix 4) (Figure 6). These interactions are highly conserved among GST classes Delta, Sigma and Theta. This is the first report of these interactions for any of these three classes. To study the function of this motif two mutants were generated, E75A and R96A, which break the conserved electrostatic interactions and ionic network at the dimer interface of adGSTD4-4. The results showed that the motif provides an important contribution to the stabilization and folding of the protein. The mutants were expressed in both inclusion and soluble forms, with the R96A mutant mostly being expressed as an inclusion body. This evidence and the results from the refolding assay (Table 1; Figures 3 and 4) indicated that the folding process was altered by the mutations. In addition, decreases in yield as well as catalytic activity changes (Table 2), suggest that the mutations disrupt the active site conformation which decreases binding affinity, alters kinetic constants and substrate specificities.
Figure 6. Conserved electrostatic interaction in region 1 of the subunit interface.
(A) The planar rectangle electrostatic interaction of Glu75 and Arg96 from both subunits. Distances between the charged atoms are shown in Å. Also shown are the conserved anion–cation–π interactions between Tyr89, Pro90 and Arg96. (B) Amino acid alignment of insect GST classes. Dirus is A. dirus and Gam is A. gambiae. The arrows point to the highly conserved tyrosine, proline and arginine residues. D is Delta, u is unclassified, E is Epsilon, T is Theta, S is Sigma, Z is Zeta and O is Omega GST class. GenBank® accession numbers: adGSTD1 (AF273041), adGSTD2 (AF273038), adGSTD3 (AF273039), adGSTD4 (AF273040), adGSTD5 (AF251478), adGSTD6 (AY014406), agGSTD1-3 (Protein ID AAC79992), agGSTD1-4 (Protein ID AAC79994), agGSTD1-5 (Protein ID AAC79993), agGSTD1-6 (Protein ID AAC79995), agGSTD2 (Z71480), agGSTD3 (AF513638), agGSTD4 (AF513635), agGSTD5 (AF513634), agGSTD6 (AF513636), agGSTD7 (AF071161), agGSTD8 (AF316637), agGSTD9 (AY255857), agGSTD10 (AF515527), agGSTD11 (AF513637), agGSTD12 (AF316638), agGSTu1 (AF515521), agGSTu2 (AF515523), agGSTu3 (AF515524), agGSTE1 (AF316635), agGSTE2 (AF316636), agGSTE3 (AY070234), agGSTE4 (AY070254), agGSTE5 (AY070255), agGSTE6 (AY070256), agGSTE7 (AF491816), agGSTE8 (AY070257), agGSTT1 (AF515526), agGSTT2 (AF515525), agGSTS1-1 (L07880), agGSTS1-2 (AF513639), agGSTZ1 (AF515522), agGSTO1 (AY255856). agGSTD6 and agGSTD9 were suggested to be pseudogenes [33]. The Figure in (A) was created using Accelrys DS ViewerPro 5.0.
Although both Glu75 and Arg96 are located in the same area, in the present study region 1 of the subunit interface, there is a conserved amino acid sequence around Arg96 which forms a pocket around the arginine residue. Arg96 is stabilized by several highly conserved residues in a cation–π interaction with Tyr89 and Pro90, whereas Glu75 does not have amino acids surrounding it, except Arg96, as it is located within a hole in the subunit interface edge (Figure 6). Therefore substitution of an alanine residue for Arg96 appears to have more impact on the tertiary and quaternary structure of the protein compared with Glu75, as shown by decreases in the fluorescence intensities of both intrinsic tryptophan spectroscopy (Figure 2) and the ANS binding assay (Figure 5). This also suggests that there are significant conformational changes in amino acid side chains near the subunit interface and the mutation site. However, disappearance of the salt bridges with the loss of either Glu75 or Arg96 affected both initial protein folding, as shown in the refolding experiment, and the protein stability as shown by decreased half-life. Although Arg96 is not located in the active site pocket, structural changes that occurred due to this mutation also affected the active site conformation, probably through packing effects (Tables 2 and 3).
Region 2 of the subunit interface, which shows the most variation in amino acid residues at the equivalent positions between adGSTD3-3 and adGSTD4-4, is of interest because the residues are not only in the interface but also in the active site, with several residues involved in both active sites of the dimer. The effects of the different hydrophobic amino acids at the equivalent positions of the two isoenzymes were studied by switching the equivalent amino acids with the amino acid from the other protein; that is, Y98F, M101V, G102A and Y98F/M101V/G102A for adGSTD3-3 and F104Y, V107M, A108G and F104Y/V107M/A108G for adGSTD4-4. The refolding experiments demonstrated that every enzyme could be refolded, although the activity recovered varied (Table 1). This indicates that the mutations have only a slight effect on the initial folding of each subunit but have more influence on the dimerization process through subunit interface conformation, as well as other structural aspects which impact upon appropriate active site conformation. When comparing the two isoforms adGSTD3-3 and adGSTD4-4, in terms of changes in catalytic activity, the mutations affected the proteins in different ways for the equivalent residues, as shown in Table 2. The crystal structures show that all selected equivalent positions are located in the active site pocket suggesting that the whole electrostatic field in the active site pocket was disturbed by the mutations, which thereby altered catalytic parameters of the enzymes. Moreover, these residues are also located at the interface of the two active site areas which provide different substrate-binding sites for GSH and the hydrophobic substrate. Therefore these positions would allow the residues to influence binding of both substrates as shown by the Km values.
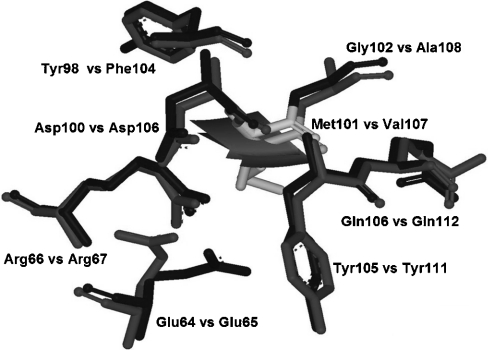
The first sphere milieu of the equivalent positions, Met101 of adGSTD3-3 and Val107 of adGSTD4-4, consists of seven amino acids of which only two residues are different between the two isoforms, adGSTD3-3 Tyr98 compared with adGSTD4-4 Phe104 and adGSTD3-3 Gly102 compared with adGSTD4-4 Ala108 (Figure 7). However, the triple mutations which changed all three amino acids at these equivalent positions to the amino acid of the other isoform, showed that they had only a slight effect on the half-life. Steric interactions and van der Waals forces are important interactions for the stability of proteins [31,32]. The side chain size of the three amino acids at these equivalent positions had a major impact on subunit interface packing, with differences between the two splice forms, especially Met101 and Gly102 of adGSTD3-3 and Val107 and Ala108 of adGSTD4-4. Although the amino acids possess similar properties, the packing effects were great enough to alter the protein and enzyme properties.
Figure 7. In region 2 the amino acid milieu of adGSTD3-Met101 and adGSTD4-Val107.
The dark grey represents adGSTD3-3 and medium grey is adGSTD4-4. The Figure was created using Accelrys DS ViewerPro 5.0.
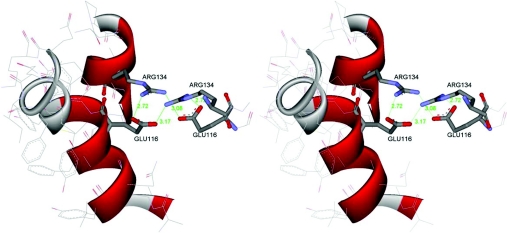
From the conserved electrostatic interactions in region 1 to the hydrophobic area in region 2 of the subunit interface, every residue appears to contribute to either maintaining structure or subunit binding. The last area to be examined was the hydrophilic area in region 3 of the subunit interface, Asp110 of adGSTD3-3 and Glu116 of adGSTD4-4. Both mutations, D110A of adGSTD3-3 and G116A of adGSTD4-4, affected catalysis as shown by changes in both specific activity and kinetic constants. The equivalent residue in the human Alpha class A1-1 is Glu104 whereas in the human Pi class GST it is a polar Ser105. In insects, the acidic residue appears to be conserved within the Delta class, however in the Epsilon class it is generally an aromatic amino acid and in the other insect classes, such as Sigma, Theta and Zeta, the residue is hydrophobic. This residue therefore appears to be conserved within a class and would contribute to class specific dimerization motifs. Additionally, in adGSTD4-4 the Glu116 interacts with Arg134 in a similar fashion to Glu75 and Arg96 in region 1 (Figure 8). That is, electrostatic interactions occur between the two residues within the same subunit as well as with the identical residues from the other subunit. So in adGSTD4-4 but not in adGSTD3-3 similar intra- and inter-subunit electrostatic interactions occur in both regions 1 and 3 of the subunit interface. In adGSTD3-3 the equivalent residue to Arg134 is Asn126 and so, because of the shorter side chains of the Asp110 and the Asn126, the distances preclude electrostatic interaction across the subunits.
Figure 8. The electrostatic interaction in region 3 of Glu116 and Arg134 in adGSTD4-4.
The two residues from each subunit are in proximity to interact electrostatically with each other as well as with the same residues from the other subunit. For the first subunit parts of α-helices 4 and 5, containing Glu116 and Arg134 respectively, are shown. For clarity only Glu116 and Arg134 from the second subunit are shown. Distances between the charged atoms are shown in Å. The Figure was created using Accelrys DS ViewerPro 5.0.
In conclusion, the conserved electrostatic interactions between the charged residues from α-helix 3 and α-helix 4 show important roles for protein folding, stabilization and dimerization of the alternatively spliced enzymes. However, the subunit interface region with the most variation in amino acid residues at equivalent positions between adGSTD3-3 and adGSTD4-4 showed that although the mutations did not alter the overall protein folding, the enzyme properties were changed, especially the catalytic activity, thermal stability and subunit interface. Even highly conservative amino acid replacements changed the protein properties. The results suggest that even splicing products from the same gene may have specific features in the subunit interface area that would preclude heterodimerization.
Acknowledgments
This work was funded by the TRF (Thailand Research Fund). J.P. held a DPST (Development and Promotion of Science and Technology Talent) scholarship. J.W. was supported by a Royal Golden Jubilee scholarship.
References
- 1.Ketterer B. A bird's eye view of the glutathione transferase field. Chem.-Biol. Interact. 2001;138:27–42. doi: 10.1016/s0009-2797(01)00277-0. [DOI] [PubMed] [Google Scholar]
- 2.Hayes J. D., Flanagan J. U., Jowsey I. R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 3.Luo J.-K., Hornby J. A. T., Wallace L. A., Chen J., Armstrong R. N., Dirr H. W. Impact of domain interchange on conformational stability and equilibrium folding of chimeric class μ glutathione transferases. Protein Sci. 2002;11:2208–2217. doi: 10.1110/ps.0208002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hornby J. A. T., Luo J.-K., Stevens J. M., Wallace L. A., Kaplan W., Armstrong R. N., Dirr H. W. Equilibrium folding of dimeric class μ glutathione transferases involves a stable monomeric intermediate. Biochemistry. 2000;39:12336–12344. doi: 10.1021/bi000176d. [DOI] [PubMed] [Google Scholar]
- 5.Luo J.-K., Hornby J. A. T., Armstrong R. N., Dirr H. W. Equilibrium unfolding and enzyme kinetics of chimeric Mu class glutathione transferases. Chem.-Biol. Interact. 2001;133:58–59. [Google Scholar]
- 6.Sinning I., Kleywegt G. J., Cowan S. W., Reinemer P., Dirr H. W., Huber R., Gilliland G. L., Armstrong R. N., Ji X., Board P. G., et al. Structure determination and refinement of human Alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes. J. Mol. Biol. 1993;232:192–212. doi: 10.1006/jmbi.1993.1376. [DOI] [PubMed] [Google Scholar]
- 7.Ji X., Zhang P., Armstrong R. N., Gilliland G. L. The three-dimensional structure of a glutathione S-transferase from the Mu gene class: structural analysis of the binary complex of isoenzyme 3-3 and glutathione at 2.2 Å resolution. Biochemistry. 1992;31:10169–10184. doi: 10.1021/bi00157a004. [DOI] [PubMed] [Google Scholar]
- 8.Reinemer P., Dirr H. W., Ladenstein R., Huber R., Lo Bello M., Federici G., Parker M. W. Three-dimensional structure of class π glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 Å resolution. J. Mol. Biol. 1992;227:214–226. doi: 10.1016/0022-2836(92)90692-d. [DOI] [PubMed] [Google Scholar]
- 9.Hornby J. A. T., Codreanu S. G., Armstrong R. N., Dirr H. W. Molecular recognition at the dimer interface of a class Mu glutathione transferase: role of a hydrophobic interaction motif in dimer stability and protein function. Biochemistry. 2002;41:14238–14247. doi: 10.1021/bi020548d. [DOI] [PubMed] [Google Scholar]
- 10.Pongjaroenkit S., Jirajaroenrat K., Boonchauy C., Chanama U., Leetachewa S., Prapanthadara L., Ketterman A. J. Genomic organization and putative promotors of highly conserved glutathione S-transferases originating by alternative splicing in Anopheles dirus. Insect Biochem. Mol. Biol. 2001;31:75–85. doi: 10.1016/s0965-1748(00)00107-7. [DOI] [PubMed] [Google Scholar]
- 11.Chelvanayagam G., Parker M. W., Board P. G. Fly fishing for GSTs: a unified nomenclature for mammalian and insect glutathione transferases. Chem.-Biol. Interact. 2001;133:256–260. [Google Scholar]
- 12.Wongsantichon J., Harnnoi T., Ketterman A. J. A sensitive core region in the structure of glutathione S-transferases. Biochem. J. 2003;373:759–765. doi: 10.1042/BJ20030394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranson H., Collins F., Hemingway J. The role of alternative mRNA splicing in generating heterogeneity within the Anopheles gambiae class I glutathione S-transferase family. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14284–14289. doi: 10.1073/pnas.95.24.14284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jirajaroenrat K., Pongjaroenkit S., Krittanai C., Prapanthadara L., Ketterman A. J. Heterologous expression and characterization of alternatively spliced glutathione S-transferases from a single Anopheles gene. Insect Biochem. Mol. Biol. 2001;31:867–875. doi: 10.1016/s0965-1748(01)00032-7. [DOI] [PubMed] [Google Scholar]
- 15.Ketterman A. J., Prommeenate P., Boonchauy C., Chanama U., Leetachewa S., Promtet N., Prapanthadara L. Single amino acid changes outside the active site significantly affect activity of glutathione S-transferases. Insect Biochem. Mol. Biol. 2001;31:65–74. doi: 10.1016/s0965-1748(00)00106-5. [DOI] [PubMed] [Google Scholar]
- 16.Oakley A. J., Harnnoi T., Udomsinprasert R., Jirajaroenrat K., Ketterman A. J., Wilce M. C. J. The crystal structures of glutathione S-transferases isozymes 1-3 and 1-4 from Anopheles dirus species B. Protein Sci. 2001;10:2176–2185. doi: 10.1110/ps.ps.21201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wongsantichon J., Ketterman A. J. An intersubunit lock-and-key ‘Clasp’ motif in the dimer interface of Delta class glutathione transferase. Biochem. J. 2006;394:135–144. doi: 10.1042/BJ20050915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 19.Prapanthadara L., Koottathep S., Promtet N., Hemingway J., Ketterman A. J. Purification and characterization of a major glutathione S-transferase from the mosquito Anopheles dirus (species B) Insect Biochem. Mol. Biol. 1996;26:277–285. doi: 10.1016/0965-1748(95)00090-9. [DOI] [PubMed] [Google Scholar]
- 20.Udomsinprasert R., Ketterman A. J. Expression and characterization of a novel class of glutathione S-transferase from Anopheles dirus. Insect Biochem. Mol. Biol. 2002;32:425–433. doi: 10.1016/s0965-1748(01)00119-9. [DOI] [PubMed] [Google Scholar]
- 21.Habig W. H., Pabst M. J., Jakoby W. B. Glutathione S-transferases: the first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- 22.Vararattanavech A., Ketterman A. Multiple roles of glutathione binding-site residues of glutathione S-transferase. Protein Pept. Lett. 2003;10:441–448. doi: 10.2174/0929866033478654. [DOI] [PubMed] [Google Scholar]
- 23.Stenberg G., Dragani B., Cocco R., Mannervik B., Aceto A. A conserved ‘hydrophobic staple motif’ plays a crucial role in the refolding of human glutathione transferase P1-1. J. Biol. Chem. 2000;275:10421–10428. doi: 10.1074/jbc.275.14.10421. [DOI] [PubMed] [Google Scholar]
- 24.Stevens J. M., Hornby J. A. T., Armstrong R. N., Dirr H. W. Class Sigma glutathione transferase unfolds via a dimeric and a monomeric intermediate: impact of subunit interface on conformational stability in the superfamily. Biochemistry. 1998;37:15534–15541. doi: 10.1021/bi981044b. [DOI] [PubMed] [Google Scholar]
- 25.Dirr H. Folding and assembly of glutathione transferases. Chem.-Biol. Interact. 2001;133:19–23. [Google Scholar]
- 26.Sayed Y., Wallace L. A., Dirr H. W. The hydrophobic lock-and-key intersubunit motif of glutathione transferase A1-1: implications for catalysis, ligandin function and stability. FEBS Lett. 2000;465:169–172. doi: 10.1016/s0014-5793(99)01747-0. [DOI] [PubMed] [Google Scholar]
- 27.Sluis-Cremer N., Naidoo N., Dirr H. Class-Pi glutathione S-transferase is unable to regain its native conformation after oxidative inactivation by hydrogen peroxide. Eur. J. Biochem. 1996;242:301–307. doi: 10.1111/j.1432-1033.1996.0301r.x. [DOI] [PubMed] [Google Scholar]
- 28.Sayed Y., Hornby J. A. T., Lopez M., Dirr H. Thermodynamics of the ligandin function of human class Alpha glutathione transferase A1-1: energetics of organic anion ligand binding. Biochem. J. 2002;363:341–346. doi: 10.1042/0264-6021:3630341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sluis-Cremer N., Naidoo N. N., Kaplan K. H., Manoharan T. H., Fahl W. E., Dirr H. W. Determination of a binding site for a nonsubstrate ligand in mammalian cytosolic glutathione S-transferases by means of fluorescence-resonance energy transfer. Eur. J. Biochem. 1996;241:484–488. doi: 10.1111/j.1432-1033.1996.00484.x. [DOI] [PubMed] [Google Scholar]
- 30.Segel I. H. New York: John Wiley & Sons; 1993. Enzyme kinetics, Behavior and Analysis of Rapid Equilibrium and Steady State Enzyme Systems. [Google Scholar]
- 31.Otzen D. E., Rheinnecker M., Fersht A. R. Structural factors contributing to the hydrophobic effect: the partly exposed hydrophobic minicore in chymotrypsin inhibitor 2. Biochemistry. 1995;34:13051–13058. doi: 10.1021/bi00040a016. [DOI] [PubMed] [Google Scholar]
- 32.Xu J., Baase W. A., Baldwin E., Matthews B. W. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 1998;7:158–177. doi: 10.1002/pro.5560070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding Y., Ortelli F., Rossiter L. C., Hemingway J., Ranson H. The Anopheles gambiae glutathione transferase supergene family: annotation, phylogeny and expression profiles. BMC Genomics. 2003;4:35–50. doi: 10.1186/1471-2164-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]