Abstract
Background: DNA extraction from paraffin wax embedded tissue requires special protocols, and most described methods report an amplification success rate of 60–80%.
Aims: To propose a simple and inexpensive protocol consisting of xylene/ethanol dewaxing, followed by a kit based extraction.
Method: Xylene/ethanol dewaxing was followed by a long rehydration step and a kit based DNA extraction step.
Results: This method produced a 100% amplification success rate for fragments of 121 to 227 bp for tamponated formalin fixed paraffin wax embedded tissue.
Conclusion: This cost effective and non-laborious protocol can successfully extract DNA from tamponated formalin fixed paraffin wax embedded tissue and should facilitate the molecular analysis of a large number of archival specimens in retrospective studies.
Keywords: DNA extraction, paraffin wax embedded tissue, polymerase chain reaction
Tissues removed from the human body for diagnostic or treatment purposes can be fixed, paraffin wax embedded, and stored for years. These specimens are an endless source of material for research. There are several protocols described for the extraction of DNA from fresh tissue, blood, and cells in culture, but extraction from paraffin wax embedded tissue requires special protocols.1–7
“Tissues removed from the human body for diagnostic or treatment purposes can be fixed, paraffin wax embedded, and stored for years”
Obtaining good quality polymerase chain reaction (PCR) products from DNA extracted from paraffin wax embedded tissue is a difficult task because, in general, this material is scarce, degraded, and often contains remnants of substances that inhibit the amplification reaction, such as formalin,2 or inhibit proteinase K used in the extraction procedure, such as xylene.
Here, we sought an alternative procedure that is simpler and cheaper than the usually described methods, and that produces a high rate of amplification and high quality amplification products from small amounts of tamponated paraffin wax embedded tissue.
MATERIALS AND METHODS
Material
DNA was extracted from a total of 79 paraffin wax embedded tissues derived from the pathology service of Hospital de Clínicas de Porto Alegre, Brazil, and from three private laboratories. All samples were surgical specimens of rectal carcinoma. The 42 samples from Hospital de Clínicas de Porto Alegre and the 15 from one of the private laboratories were fixed in 4% tamponated formalin for 24 hours and then submitted to 95% ethanol and xylene washes before being embedded in paraffin wax. The archival material had been stored for a mean of 11.83 years (range, 8–16). The 22 samples from the other two laboratories were fixed in non-tamponated formalin.
Procedure
One 20 μm slice of paraffin wax embedded tissue was collected and manually microdissected with sterile scalpels under an inverted optical microscope to separate normal from tumorous tissue.
The sections were dewaxed with two xylene washes (30 minutes at 55°C each), two 100% ethanol washes, two 70% ethanol washes, and two distilled water washes (five minutes at 55°C each step).
Samples were then rehydrated with 1× Tris/EDTA for five minutes at 55°C and Tris (1M, pH 7.5) overnight at 55°C.
After rehydration, samples were digested with proteinase K (20 mg/ml) at 55°C for one to three hours.
DNA was extracted with the Ultraclean bloodspin kit (MoBio Laboratories Inc, Carlsbad, California, USA) and resuspended in Tris/EDTA (200 μl).
The amplification of fragments of 121 to 227 bp (microsatellites BAT-25, BAT-26, D2S123, and D17S250) was carried out using PCR containing 5 μl of DNA (mean of 84 ng/μl), 50mM KCl, 20mM Tris/HCl (pH 8.4), 400μM of each deoxyribonucleotideotriphosphate, 20 pmol of each primer4 and, after testing different concentrations, 5mM MgCl2 and 2.5 U of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, California, USA) in a final volume of 25 μl. PCR was performed using a DNA thermal cycler (MiniCyclerTM; MJ Research PTC150; Bio-Rad, Hercules, California, USA) with 35 amplification cycles at annealing temperatures of 58°C and 60°C (D2S123). PCR products were analysed by electrophoresis on 2% agarose gels.
RESULTS AND DISCUSSION
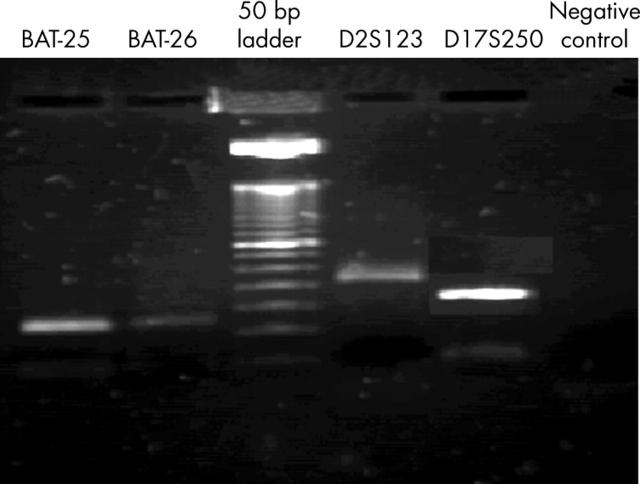
Figure 1 depicts an agarose gel. The amplification products were suitable for microsatellite instability determination by single stranded conformational polymorphism (data not shown).
Figure 1.
Agarose gel (2%) stained with ethidium bromide, showing the polymerase chain reaction products of the four markers analysed.
“This simple, cost effective, and non-laborious procedure should facilitate the molecular analysis of a large number of archival specimens in retrospective studies”
We had a 100% amplification success rate in the range of 121 to 227 bp (table 1) for the samples fixed in tamponated formalin, in contrast to the 60–88% amplification success rate described using other methods. However, paraffin wax embedded tissues fixed in non-tamponated formalin did not amplify, and showed only a smear, which probably corresponds to degraded DNA. A previous study comparing several different methods reported better results with the use of a Qiagen extraction kit (60% amplification success rate).3 Another study reported best results with a simple boiling method (88%), followed by a phenolchloroform extraction method (85%) and a Qiagen extraction kit (53%).2 Other studies report a 70–88% amplification success rate with dewaxing only,5 or with dewaxing and protease digestion,6,7 with the sections or lysates used directly for PCR. The procedure described here allows the efficient amplification of DNA extracted from archival paraffin wax embedded tissues fixed in tamponated formalin. The main difference in our protocol is the long rehydration step, which probably contributed to the better extraction performance.
Table 1.
Polymerase chain reaction (PCR) amplification of different sized DNA fragments after extraction from tamponated, formalin fixed, paraffin wax embedded tissue
| Microsatellite marker | PCR product (bp) | Total number of samples amplified | PCR amplification success (1 round PCR) |
| BAT 25 | 123 | 114 | 100% |
| BAT 26 | 121 | 84 | 100% |
| D2S123 | 227 | 114 | 100% |
| D17S250 | 162 | 84 | 100% |
This simple, cost effective, and non-laborious procedure should facilitate the molecular analysis of a large number of archival specimens in retrospective studies.
Abbreviations
PCR, polymerase chain reaction
REFERENCES
- 1.Wright DK, Manos MM. Sample preparation from paraffin-embedded tissues. In: Innis MA, Gelfand DH, Sninsky JJ, et al, eds. PCR protocols: a guide to methods and applications. London: Academic Press, 1990:153–8.
- 2.Cao W, Hashibe M, Rao J - Y, et al. Comparison of methods for DNA extraction from paraffin-embedded tissues and buccal cells. Cancer Detect Prev 2003;27:397–404. [DOI] [PubMed] [Google Scholar]
- 3.Coombs NJ, Gough AC, Primrose JN. Optimization of DNA and RNA extraction from archival formalin-fixed tissue. Nucleic Acids Res 1999;27:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dietmaier W, Hofstädter F. Detection of microsatellite instability by real time PCR and hybridization probe melting point analysis. Lab Invest 2001;81:1453–6. [DOI] [PubMed] [Google Scholar]
- 5.Torlakovic E, Cherwitz DL, Jessurun J, et al. B-cell gene rearrangement in benign and malignant lymphoid proliferations or mucosa-associated lymphoid tissue and lymph nodes. Hum Pathol 1997;28:166–73. [DOI] [PubMed] [Google Scholar]
- 6.Ritter JH, Wick MR, Adesokan PN, et al. Assessment of clonality in cutaneous lymphoid infiltrates by polymerase chain reaction analysis of immunoglobulin heavy chain gene rearrangement. Am J Clin Pathol 1997;108:60–8. [PubMed] [Google Scholar]
- 7.Inghirami G, Szabolcs MJ, Yee HT, et al. Detection of immunoglobulin gene rearrangement of B cell non-Hodgkin’s lymphomas and leukemias in fresh, unfixed and formalin-fixed, paraffin-embedded tissue by polymerase chain reaction. Lab Invest 1993;68:746–57. [PubMed] [Google Scholar]