Abstract
The Mad3/BubR1, Mad2, Bub1, and Bub3 proteins are gatekeepers for the transition from metaphase to anaphase. Mad3 from Saccharomyces cerevisiae has homology to Bub1 but lacks a corresponding C-terminal kinase domain. Mad3 forms a stable heterodimer with Bub3. Negative-stain electron microscopy shows that Mad3 is an extended molecule (≈200 Å long), whereas Bub3 is globular. The Gle2-binding-sequence (GLEBS) motifs found in Mad3 and Bub1 are necessary and sufficient for interaction with Bub3. The calorimetrically determined dissociation constants for GLEBS-motif peptides and Bub3 are ≈5 μM. Crystal structures of these peptides with Bub3 show that the interactions for Mad3 and Bub1 are similar and mutually exclusive. In both structures, the GLEBS peptide snakes along the top surface of the β-propeller, forming an extensive interface. Mutations in either protein that disrupt the interface cause checkpoint deficiency and chromosome instability. We propose that the structure imposed on the GLEBS segment by its association with Bub3 enables recruitment to unattached kinetochores.
Keywords: β-propeller, crystal structures, Rae1/Nup98, GLEBS motif
The spindle checkpoint controls the transition from metaphase to anaphase by monitoring attachment of kinetochores to spindle microtubules and tension between sister chromatids (reviewed in ref. 1). The proteins that monitor and transmit the delay signal in response to attachment defects include Mad1, Mad2, Mad3 (BubR1 in higher eukaryotes), the kinase Bub1, and the adaptor protein Bub3. Entry into anaphase requires a cascade of events. Ubiquitination and degradation of Pds1 (securin), an inhibitor of the protease Esp1 (separase), allow Esp1 to sever the cohesins that hold sister chromatids together. The spindle checkpoint prevents ubiquitination of Pds1/securin by the anaphase-promoting complex (APC) until bipolar attachment and tension have been achieved at all chromosome pairs. Both checkpoint proteins Mad2 and BubR1/Mad3, the effectors of this checkpoint, inhibit APCCdc20-mediated ubiquitination of Pds1/securin in vitro, presumably through direct interaction with the APC-associated specificity factor Cdc20 (2, 3). Regulation of Mad2 involves an unusual conformational switch (4) that enables it to sequester Cdc20 by binding a conserved sequence N-terminal to the WD40 domain (5). The mechanisms for BubR1/Mad3-mediated inhibition are not yet understood.
Mad3, which is homologous to the N-terminal, nonkinase domain of the checkpoint kinase Bub1, is believed to have arisen during an ancient whole-genome duplication event that occurred in fungi (6). Mad3 presumably originated from a duplicated copy of Bub1 and subsequently evolved a new or related function that did not require kinase activity. Mad3 and Bub1 both form constitutive complexes with Bub3 throughout the cell cycle, suggesting that this interaction is important for the function of both proteins (7, 8). Primitive fungi that did not undergo whole-genome duplication have a Bub1 gene but lack a corresponding Mad3 gene. In higher eukaryotes, BubR1 likewise arose during a separate whole-genome duplication event but retained its kinase domain. Thus, Mad3 and BubR1 have evolved a related function along distinct evolutionary branches. One key difference between Mad3 and BubR1 is the retention of the kinase domain in higher eukaryotes. This kinase domain is activated by the microtubule-associated protein CENP-E, which has no homolog in lower eukaryotes (9).
Mad3 and the corresponding domain in Bub1 have high helical content and may contain tetracotripeptide repeats (10, 11). Deletion mapping and pull-down experiments have shown that the Mad3–Bub3 and Bub1–Bub3 interactions are probably restricted to a conserved Gle2-binding sequence (GLEBS) motif (Fig. 1A) (12, 13). The GLEBS motif was first characterized in the nuclear pore complex protein Nup98 and found to be sufficient for binding the mRNA export factor Gle2 (also known as Rae1) (14). This Gle2–Nup98 complex specifically inhibits APCCdh1-mediated ubiquitination of securin (15). Gle2/Rae1 and Bub3 are both WD40 proteins with significant homology to each other, and we therefore anticipate that the interactions between Nup98-Gle2 will resemble those between Mad3–Bub3 (8).
Fig. 1.
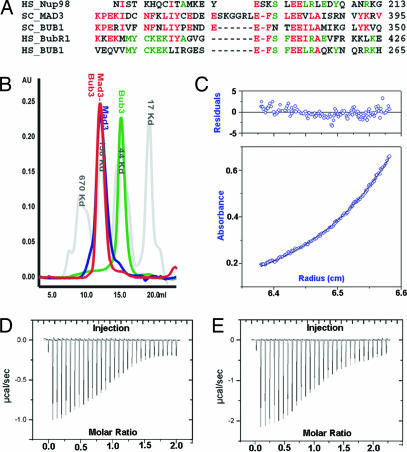
Interaction of Bub3 with Mad3 and Bub1. (A) Sequence alignments of GLEBS motifs. The yeast sequences shown correspond to the peptides used in crystallization. HS, human, SC, baker's yeast. (B) Analytical gel filtration. Molecular weight standards are in gray. Mad3 and the complex Mad3–Bub3 have hydrodynamic properties characteristic of elongated molecules. (C) Sedimentation equilibrium analytical ultracentrifugation of Mad3–Bub3 complex. A representative fit in which Mad3–Bub3 was modeled as a heterodimer. (D) ITC for Mad3 GLEBS peptide binding Bub3. The heat of complex formation for each injection is shown. (E) Likewise, a peptide corresponding to the Bub1 GLEBS motif titrated into Bub3.
The proteins Mad2, Mad3, and Bub3 form an inhibitory complex with Cdc20 called the mitotic checkpoint complex (2, 3, 7, 16, 17). The mitotic checkpoint complex constituents are present throughout the cell cycle but inhibit phosphorylated APC/C in mitotic cells more strongly than unphosphorylated APC/C in interphase cells (17). It is not known whether Bub1 substitutes for Mad3 in more primitive yeast. Despite the sequence homology, Bub1 does not appear to inhibit APCCdc20 by stoichiometric binding like Mad3 (2) but rather inhibits by phosphorylating Cdc20 in vivo (7, 18). The interactions between Bub1 and Bub3 and between Mad3 and Bub3 are probably important for localization to unattached kinetochores (12, 19, 20).
We have characterized by x-ray crystallography the interactions between Mad3 and Bub3 and between Bub1 and Bub3. Our crystal structures show that Mad3 and Bub1 bind the top face of the Bub3 β-propeller and that their interaction is mutually exclusive. The GLEBS motif forms a helix–loop–helix on the top surface of the β-propeller, anchored by conserved sequence-specific contacts.
Results
Bub3 Forms a 1:1 Complex with Mad3.
Yeast two-hybrid, coimmunoprecipitation, and deletion-mapping experiments have shown that Bub3 interacts with Mad3 (7, 12). We confirmed that Mad3 associates tightly with untagged Bub3 and that the purified complex elutes as a narrow peak on size-exclusion chromatography. The elution volume is consistent with either an extended molecule with a large Stokes radius or a heterooligomer (Fig. 1B). Sedimentation equilibrium analytical ultracentrifugation confirms that Mad3 and Bub3 form a 1:1 complex with a measured molecular mass of 99 ± 2 Kd, in excellent agreement with the calculated mass of 98.8 Kd (Fig. 1C).
We characterized a proteolytically stable region of Mad3 (46–273) lacking the GLEBS motif and found that this domain did not interact with Bub3. In contrast, the His6-tagged GLEBS motif from Mad3 was sufficient to pull down untagged Bub3. Moreover, the peptide remained stoichiometrically bound to Bub3 after size-exclusion chromatography. We measured the strength of this interaction with isothermal titration calorimetry (ITC) by using purified Bub3 and chemically synthesized peptides corresponding to the Mad3 GLEBS motif. Likewise, we compared this interaction to an analogous peptide from the Bub1 GLEBS motif. The measured Kd values were 2.7 μM and 3.6 μM, respectively (Fig. 1 D and E). We regard these Kd values as upper limits for interaction with the full-length protein, and we anticipate that the values for the full-length protein could be lower.
Mad3 Is an Extended Molecule with Predicted Tetratricopeptide Repeat Motifs.
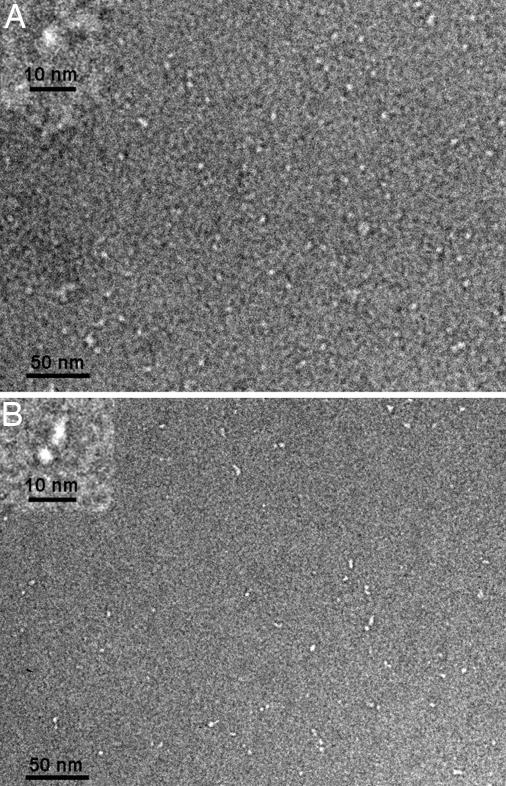
We visualized Bub3 and Mad3 by negative-stain electron microscopy. Our images show that Bub3 is a small globular ball with a diameter of ≈50 Å, as expected from the crystal structure (Fig. 2A). In contrast, Mad3 appears to be extended, ≈150–250 Å long (Fig. 2B). An extended structure would be consistent with the observed gel filtration profile and its known helical composition (10), including predicted tetratricopeptide repeats (11), which are multiple helix–turn–helix motifs packed together into an extended spiral of antiparallel α-helices (21).
Fig. 2.
Negative-stain electron microscopy. Shown are wide fields with higher magnification insets. (A) Bub3 appears as isolated punctate objects. (B) Mad3 resembles elongated beads on a string.
The GLEBS Motif Interacts with the Top Face of Bub3.
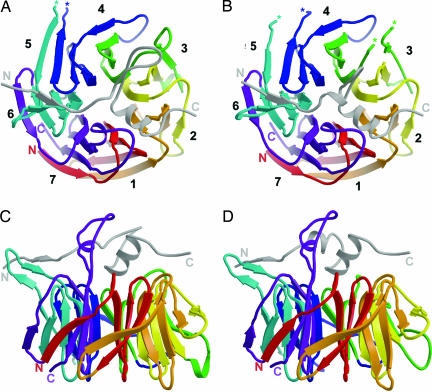
We have shown previously that apo-Bub3 is a canonical seven-bladed β-propeller (8). Each blade is a four-stranded β-sheet arranged in pseudo7-fold rotational symmetry about a central axis (Fig. 3A and B). The top surface of the propeller contains the loops that join strands βB and βC within each blade and strands βD and βA of consecutive blades. The width of Bub3 from the top face to the bottom face is ≈27 Å; the diameter is ≈42 Å. From analysis of sequence conservation, we predicted that binding partners of Bub3 would associate along the top surface of the propeller (8). Our peptide-bound structures validate that prediction and allow us to describe the interactions. Analysis of the interface further allows us to interpret the phenotypes of Bub3, Mad3, and Bub1 mutants.
Fig. 3.
Overview of Bub3 bound to GLEBS motif. (A) Top view of Bub3 bound to the GLEBS motif from Mad3. The GLEBS peptide is colored in gray and lies along the top face of the propeller. N and C termini are labeled, and breaks in main-chain density are denoted with asterisks. (B) Top view of Bub3 bound to GLEBS motif from Bub1. Overall, the structures are quite similar, except that the Bub1 GLEBS motif has a shorter loop between helices α1 and α2. (C and D) Side views of Bub3 bound to Mad3 (C) and Bub1 (D) GLEBS motifs. The three-stranded β-sheet that includes the DA loop between blades 5 and 6 of Bub3 projects leftward in these views.
The Mad3 and Bub1 GLEBS peptides form similar interactions with Bub3, and binding of one partner therefore excludes the other. Both peptides snake along the top face beginning at blade 6, intersect the central axis, and continue outward over blade 2 (Fig. 3). The orientation of the peptide is approximately transverse to the strands that make up each blade. The peptide has clear secondary structure beginning with a short strand segment over blade 6, a helix–coil–helix motif, and a final coil over blade 2 (Fig. 3). In the Bub1 peptide, the helix–coil–helix motif is very well defined, but in the Mad3 peptide the intervening coil sequence has a six-residue loop insertion that is not as well ordered, and we could model it in only one of the four molecules in the asymmetric unit.
In the apo-Bub3 structure, the DA loop between blades 5 and 6 is extended with no secondary structure elements. In the complexes, the peptides induce a conformational change in this DA loop that creates a well ordered, three-stranded β-sheet with two strands from Bub3 and one strand from the peptide (Fig. 3 C and D). The length and composition of this DA loop are not well conserved across species (8), but it is possible that the three-stranded β-sheet structure is conserved, because the observed interactions are through the backbone. This sheet could serve as a platform for interactions with additional protein binding partners, either at the kinetochore or within the mitotic checkpoint complex.
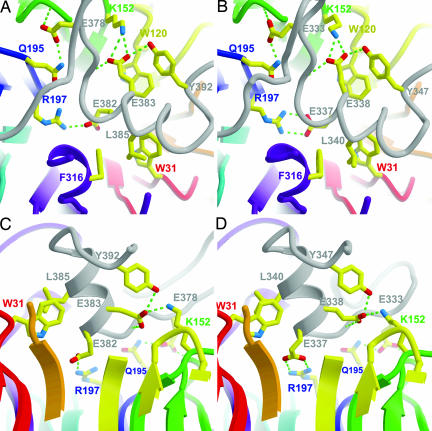
The more conserved interactions occur in the helix–coil–helix motif close to the axis of Bub3. A conserved Glu-333Bub1/Glu-378Mad3 before the second helix α2 forms H-bonds with the backbone amide Asn-169Bub3 and side chain of Gln-195Bub3, creating a H-bonding bridge that links the BC loop of blade 4 with the DA loop of blade 5 (Fig. 4). The Glu-333Bub1Lys mutation causes a chromosome instability and checkpoint deficient phenotype in vivo as severe as bub1Δ (22, 23). This phenotype may be reversed by overexpression of Bub3 (22, 23). From our model, we expect that this mutation would weaken the Bub1–Bub3 interaction. The mutant Lys-333 could adopt a less favorable, solvent-exposed rotamer and still allow association with Bub3. Thus, overexpression could drive the equilibrium toward association.
Fig. 4.
Conserved interactions at the interface of Bub3 and the GLEBS motifs. H-bonds are illustrated by broken green lines. (A and B) Top views of Bub3 bound to Mad3 (A) and Bub1 (B) GLEBS motifs. The helices were not rendered in this view because they obscured the underlying residues. (C and D) Side views of Bub3 bound to Mad3 (C) and Bub1 (D) GLEBS motifs. The slab has been positioned to reveal the interactions near the pore of Bub3. For clarity, Trp-120 has not been rendered in this view.
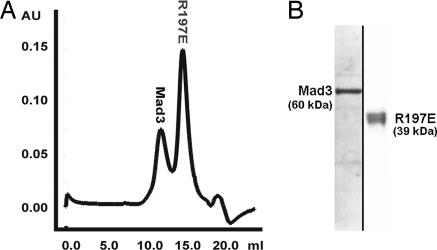
Another set of important interactions occurs where the N-terminal end of the GLEBs α2 helix dips into the central pore of Bub3 and inserts a pair of conserved Glu residues like a pair of tweezers (Fig. 4). The first Glu 382Mad3/337Bub1 is completely buried and forms a salt bridge with Arg-197Bub3 from blade 5. The second Glu 383Mad3/338Bub1 forms a salt bridge with Lys-152Bub3 from blade 4. The electrostatic interactions described are totally conserved in aligned sequences of homologs from other species. We found that mutation of either Glu-382Mad3Lys or Arg-197Bub3Glu disrupts any detectable interaction between Mad3 and Bub3 in vitro (Fig. 5). The in vivo consequence of the Glu-382Mad3Lys mutation is a benomyl-sensitive phenotype as severe as the mad3Δ strain (7).
Fig. 5.
Mad3 and R197E Bub3 do not associate. (A) Mad3 was mixed with excess Bub3 mutant and analyzed by gel filtration. (B) Coomassie-stained gel of peak fractions from A showing that the Bub3 mutant does not coelute with Mad3.
The GLEBS α2 helix is buttressed on one side by two conserved Trp residues: Trp-31Bub3 from blade 1 and Trp-120Bub3 from blade 3 (Fig. 4). These Trp residues provide a hydrophobic platform on the top face of Bub3 that helps orient GLEBS helix α2. The double mutation Trp31Gly and Trp120Gly leads to a benomyl-sensitive phenotype similar to that seen in bub3Δ cells (16).
Structural Homology with Gle2 (Rae1).
Bub3 and Rae1 have 34% sequence identity and 52% similarity in humans (8). Although Bub3 appears to be specific for the GLEBS sequences of Bub1 and Mad3, murine Rae1 (mRae1) has been reported to bind the GLEBS motifs of mNup98 as well as mBub1 and mBubR1 (13). Moreover, mRae1 complements the haploinsufficiency of Bub3 in mice (24). The complex between Rae1 and Nup98 inhibits the APCCdh1-mediated ubiquitination of Pds1/securin (15). It is possible that the mechanism of inhibition is similar to the Mad3–Bub3-mediated inhibition of APCCdc20. Comparison of key conserved contact residues in the Bub3–Mad3 interface shows nearly identical predicted contacts between Rae1 and Nup98. The residues contributing to the conserved interaction between Glu-333Bub1/Glu-378Mad3 and Gln-195Bub3 correspond to Glu-195hNup98 and Gln-214hRae1. The central salt bridges also are conserved. Specifically, the Glu-382Mad3/337Bub1 and Arg-197Bub3 pair corresponds to Glu-200hNup98 and Arg-216hRae1, and the Glu-383Mad3/338Bub1 and Lys-152Bub3 pair corresponds to Glu-201hRae1 and Arg-172hRae1. In addition, the hydrophobic Trp platform on the top face also is present in Rae1, where Trp-31Bub3 and Trp-120Bub3 correspond to Trp-62hRae1 and Trp-149hRae1, respectively. Thus, the high sequence conservation and preservation of key contacts is consistent with the cross-interaction between Rae1 and checkpoint proteins Bub1 and BubR1.
Discussion
Bub3-Mad3/BubR1 and Bub3–Bub1 are present as complexes throughout the cell cycle. Our data show that Bub3 and Mad3 associate stably with a stoichiometry of 1:1. The conserved GLEBS motif is necessary and sufficient for complex formation with Bub3. This conclusion is based on the following observations. First, the GLEBS motif alone is sufficient to pull down Bub3 in a 1:1 complex in vitro. Second, a proteolytically stable region of Mad3 lacking the GLEBS motif fails to interact with Bub3. Third, a single point mutation in the GLEBS motif or on the top face of Bub3 is sufficient to abolish association between the two proteins. Thus, a restricted segment of Mad3 mediates its interaction with Bub3, suggesting that Bub3 is not a direct switch for closing or opening Mad3 but rather a template for presenting the GLEBS region to another surface.
When the GLEBS motif interacts with Bub3, it undergoes a transition from an unfolded conformation to a well defined and ordered structure. Indeed, the relatively short polypeptide segments that are often the targets of “protein interaction domains” generally assume specific conformations only upon binding. For example, the Cdc4-Sic1 and WDR5-histone tail structures both reveal a short peptide fixed by its interactions across the central pore on the top face of a WD40 propeller (25, 26). The GLEBS motif is unusual, however, in its length and in the complexity of the structure imposed on it. The Mad3 and Bub1 peptides traverse the entire top face of Bub3, beginning with the short strand that forms a three-stranded β-sheet with the DA loop between blades 5 and 6 of Bub3 and continuing with the helix–coil–helix structure that covers the central pore. The more conserved interactions are in the latter region, where two Glu side chains from the GLEBS motifs form invariant salt bridges with Bub3.
Yeast cells with various mutations at the GLEBS/Bub3 interface exhibit benomyl sensitivity and chromosome missegregation phenotypes. That is, destabilizing the GLEBS/Bub3 contact compromises the spindle checkpoint. Moreover, as shown here, disrupting one of the conserved salt bridges completely abolishes any detectable interaction between Bub3 and Mad3 in vitro, corresponding to the benomyl sensitivity phenotype in vivo. Overexpression of the GLEBS motif in HeLa cells disrupts the checkpoint, but overexpression of the same motif with a single point mutation does not (27). That is, extra copies of the GLEBS motif can saturate Bub3, rendering it unavailable to endogenous Bub1 and BubR1, whereas the mutated GLEBS motif cannot.
Acquired or inherited mutation in genes of the spindle checkpoint have been linked to several cancers (30–33). For example, insertions and point mutations in both the kinetochore localization domains and the kinase domains of Bub1 and BubR1 have been found in leukemias and lymphomas (34–36). The results described here show how specific mutations at the Bub3–Bub1 or Bub3–Mad3/BubR1 interface can destabilize the complexes, defeating the spindle checkpoint and leading to chromosome missegregation phenotypes.
How do the Bub3 complexes regulate the spindle checkpoint? Bub3 is necessary for association of Bub1 and BubR1/Mad3 with unattached kinetochores (12, 27, 28). Bub3 appears not to be necessary, however, for inhibiting ubiquitination of Pds1/securin in vitro by APCCdc20 (2, 3). Rather, segments of Mad3/BubR1 other than the GLEBS motif bind Cdc20 to prevent the latter from activating APC. Thus, a critical property of the Bub3/GLEBS complex is probably specific association with some kinetochore component, an “upstream” event rather than an “effector” step. We can imagine two mechanisms. In one mechanism, Bub3 binds an unattached kinetochore and, in turn, recruits Mad3/BubR1. In the other mechanism, it is the structure imposed on the GLEBS segment by its association with Bub3 that enables the complex to recognize a kinetochore site. The relatively elaborate presentation of the GLEBS segment across the top surface of the Bub3 β-propeller leads us to favor the latter picture.
Materials and Methods
Expression and Purification of Saccharomyces cerevisiae Checkpoint Proteins.
Bub3 was expressed as described in ref. 8. Bub3 and the GLEBs motif of Mad3 (residues 354–401) were subcloned by using NdeI/XhoI and NcoI/EcoRI sites, respectively, in the pET-duet(+) vector (Novagen, Madison, WI). Bub3 and full-length Mad3 were subcloned similarly with a primer-encoded N-terminal His-tag on Mad3. The Bub1 GLEBs motif (residues 315–356) was chemically synthesized and loaded onto purified Bub3. Mutants were constructed by PCR overlap extension. All checkpoint proteins (except Mad3 alone) were otherwise expressed and purified as described (8). All proteins could be frozen in 20% sucrose and stored indefinitely at −80°C.
Expression and purification of Mad3 or Mad3E382K alone failed. Thus, untagged Bub3 and His-tagged Mad3 were coexpressed and incubated on Ni-NTA. The bound complex was then washed extensively with 2 M KI, a mild chaotrope found to dissociate the complex. Bub3 was thereby separated from Mad3. Mad3 was then eluted from the resin in 250 mM imidazole, 250 mM NaCl, and 50 mM Bis-Tris-propane, pH 6.5. Later purification steps were carried out at this mildly acid pH, 1 unit below the calculated pI of the protein, to circumvent aggregation. Mad3E382K does not associate with Bub3 and, therefore, could not be purified with this method.
Limited Proteolysis of Mad3–Bub3.
Optimized chymotryptic digests were performed on ice with substrate-to-protease ratios of 100:1. The reactions were terminated at various time intervals with 100 μM PMSF. Stable fragments were characterized by N-terminal sequencing and MALDI MS.
ITC.
Bub3 was purified on a S200 sizing column with PBS (pH 7.4) as the running buffer. The Mad3 and Bub1 GLEBs motif peptides were chemically synthesized and dissolved in 100% DMSO and diluted 1:10 (vol:vol) into PBS. Bub3 also was diluted to give the same final concentration of DMSO. The final concentrations for Bub3, Mad3 peptide, and Bub1 peptide were 9, 170, and 240 μM, respectively. ITC experiments were conducted with a MCS ITC (Microcal, Northampton, MA) at 23°C. Approximately 2 ml of the Bub3 solution was loaded into the sample cell, and peptide was loaded into the injection syringe. Subsequently, 25 injections of 10 μl were added to the sample cell to give a final injectant-to-sample-cell molar ratio of ≈2:1. Sufficient time was allowed between injections for the heat generated to reequilibrate. Titration data were fit to a single binding site model (29) by using Origin ITC software (version 5.0, Microcal Software).
Sedimentation Equilibrium Analytical Ultracentrifugation.
Sedimentation equilibrium measurements of Mad3 and Mad3–Bub3 were made at 4°C in an analytical ultracentrifuge (Optima XLA; Beckman Coulter, Fullerton, CA) as described in ref. 30. Mad3 was prepared in 25 mM Bis-Tris-Pro, pH 6.5/250 mM NaCl/1 mM 2-mercaptoethanol, and Mad3–Bub3 was prepared in 10 mM Tris, pH 8.0/250 mM NaCl/1 mM 2-mercaptoethanol. The molecular weights were obtained from multiple fit alignment of data from three speeds and three protein concentrations using the XL-1 software (Beckman Coulter).
Negative-Stain Electron Microscopy.
Proteins were diluted to ≈2 μg·ml−1 and incubated on glow-discharged, continuous carbon support film on 400 copper mesh grids for 1 min. The grids were washed with buffer, stained with 0.5% uranyl acetate, and dried. Sample grids were imaged at ×52,000 magnification on a Techni-12 microscope (Phillips, Hillsboro, CO) operated at 120 kV with a low dose kit. Images were collected on an AQ4 CCD camera by using Digital Micrograph software (Gatan, Pleasanton, CA).
Crystallization, Structure Determination, and Refinement.
Optimized Bub3–Mad3 peptide complex crystals grew overnight from 17% PEG 4000/10% isopropanol/100 mM Hepes, pH 7.5. Bub3–Bub1 peptide crystals grew from 18% PEG 3000/200 mM NaCl/100 mM cacodylate pH 6.4. Crystals were frozen after immersion in a 20% (vol:vol) glycerol-substituted cryoprotectant. Data were collected to 2.8 Å and 1.9 Å, respectively, at the Advanced Light Source Beamline 8.2.2 (Table 1). The data were processed with HKL2000 (31). Bub3–Mad3 crystals belong to space group P21, with a = 51 Å, b = 173 Å, c = 90 Å, β = 95.3°, and four complexes per asymmetric unit. Bub3–Bub1 crystals also belong to space group P21 with a = 51 Å, b = 79 Å, c = 143 Å, β = 98.3°, and three complexes per asymmetric unit.
Table 1.
Data and refinement statistics
| Bub3–Mad3 | Bub3–Bub1 | |
|---|---|---|
| Wavelength | 1.18070 | 0.97780 |
| Resolution | 2.8 (2.85–2.80) | 1.9 (1.97–1.90) |
| Space group | P21 | P21 |
| Unique observations | 40,970 | 79,711 |
| Redundancy | 3.6 | 3.3 |
| Completeness | 95.2 (76.1) | 90.6 (68.9) |
| Rsym* | 9.4 (36.7) | 5.4 (36.1) |
| I/σ | 16.4 (1.9) | 23.1 (2.3) |
| Refined residues | 1,484 | 1,102 |
| Refined waters | NA | 646 |
| Rcryst† | 23.9 | 21.8 |
| Rfree‡ | 26.8 | 26.2 |
| Average B-values, Å2 | ||
| Bub3 | 57 | 39 |
| GLEBs peptide | 66 | 40 |
| Water | NA | 45 |
Numbers in parentheses refer to the highest-resolution shell. NA, not applicable.
*Rsym = [ΣhΣi∣Ii(h) − 〈I(h)〉∣/ΣhΣI Ii(h)] × 100, where 〈I(h)〉 is the average intensity of i symmetry related observations of reflections with Bragg index h.
†Rcryst = [Σhkl∣Fo − Fc∣/Σhkl ∣Fo∣] × 100, where Fo and Fc are the observed and calculated structure factors.
‡Rfree was calculated as for Rcryst, but on 5% of data excluded before refinement.
The structures were determined by molecular replacement with the program MOLREP (32) by using native Bub3 [Protein Data Bank ID code 1U4C (8)] as the search model. Initial simulated annealing refinement using the CNS program suite (33) with noncrystallographic symmetry restraints followed by difference map calculation showed extra interpretable electron density for the GLEBS peptide snaking across the top surface of Bub3. A crude model was traced into this difference density in O (34) and used to generate a mask in CCP4 (35), which was used in turn to calculate a noncrystallographic symmetry averaged map in DM (36). Subsequently, the peptides were retraced, and additional refinement yielded final R = 24.1% and Rfree = 27.0% for the Bub3–Mad3 complex and R = 21.8% and Rfree = 26.2% for the Bub3–Bub1 complex.
Acknowledgments
We thank Scott Schuyler and Andrew Murray for many helpful discussions and Piotr Sliz for computational assistance with noncrystallographic symmetry averaging. N.A.L. was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research and a senior fellow award from the Leukemia and Lymphoma Society. J.A.-B. was supported by a postdoctoral fellowship from the American Cancer Association. S.C.H. is an Investigator in the Howard Hughes Medical Institute.
Abbreviations
- APC
anaphase-promoting complex
- GLEBS
Gle2-binding-sequence
- ITC
isothermal titration calorimetry.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2I3S and 2I3T for Bub3–Mad3 and Bub3–Bub1, respectively).
References
- 1.Pinsky BA, Biggins S. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Tang Z, Bharadwaj R, Li B, Yu H. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 3.Fang G. Mol Biol Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu H. J Cell Biol. 2006;173:153–157. doi: 10.1083/jcb.200601172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner J. Nat Struct Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- 6.Kellis M, Birren BW, Lander ES. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 7.Hardwick KG, Johnston RC, Smith DL, Murray AW. J Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsen NA, Harrison SC. J Mol Biol. 2004;344:885–892. doi: 10.1016/j.jmb.2004.09.094. [DOI] [PubMed] [Google Scholar]
- 9.Mao Y, Abrieu A, Cleveland DW. Cell. 2003;114:87–98. doi: 10.1016/s0092-8674(03)00475-6. [DOI] [PubMed] [Google Scholar]
- 10.Yoon J, Kang Y, Kim K, Park J, Kim Y. Protein Express Purif. 2005;44:1–9. doi: 10.1016/j.pep.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 11.Bharadwaj R, Yu H. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- 12.Taylor SS, Ha E, McKeon F. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Babu JR, Harden JM, Jablonski SA, Gazi MH, Lingle WL, de Groen PC, Yen TJ, van Deursen JM. J Biol Chem. 2001;276:26559–26567. doi: 10.1074/jbc.M101083200. [DOI] [PubMed] [Google Scholar]
- 14.Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E. EMBO J. 1998;17:1107–1119. doi: 10.1093/emboj/17.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeganathan KB, Malureanu L, van Deursen JM. Nature. 2005;438:1036–1039. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- 16.Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, Piatti S. EMBO J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudakin V, Chan GK, Yen TJ. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Z, Shu H, Oncel D, Chen S, Yu H. Mol Cell. 2004;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 19.Sharp-Baker H, Chen R-H. J Cell Biol. 2001;153:1239–1249. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerscher O, Crotti LB, Basrai MA. Mol Cell Biol. 2003;23:6406–6418. doi: 10.1128/MCB.23.18.6406-6418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Andrea LD, Regan L. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Hoyt MA, Totis L, Roberts BT. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 23.Warren CD, Brady DM, Johnston RC, Hanna JS, Hardwick KG, Spencer FA. Mol Biol Cell. 2002;13:3029–3041. doi: 10.1091/mbc.E02-04-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 26.Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, Min J. EMBO J. 2006;25:4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris L, Davenport J, Neale G, Goorha R. Exp Cell Res. 2005;308:85–100. doi: 10.1016/j.yexcr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 28.Gillett ES, Espelin CW, Sorger PK. J Cell Biol. 2004;164:1–12. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiseman T, Williston S, Brandts JF, Lin LN. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 30.Al-Bassam J, van Breugel M, Harrison SC, Hyman A. J Cell Biol. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Vagin A, Teplyakov A. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 33.Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang J.-S., Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 35.Collaborative Computation Project, Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 36.Cowtan KD, Main P. Acta Crystallogr D. 1996;52:43–48. doi: 10.1107/S090744499500761X. [DOI] [PubMed] [Google Scholar]