Abstract
Transmission ratio distortion (TRD), the preferential inheritance of the t haplotype from t/+ males, is caused by the cooperative effect of four t-complex distorters (Tcd1–4) and the single t-complex responder (Tcr) on sperm motility. Here we show that Fgd2, encoding a Rho guanine nucleotide exchange factor, maps to the Tcd2 region. The t allele of Fgd2 is overexpressed in testis compared with wild type. A loss-of-function allele of Fgd2 generated by gene targeting reduces the transmission ratio of the t haplotype th49, directly demonstrating the role of Fgd2 as Distorter. Fgd2 identifies a second Rho G protein signaling pathway promoting TRD.
Keywords: Mouse, transmission ratio distortion, testis, sperm motility, Rho, G proteins
The t haplotype is a variant form of the proximal third of mouse chromosome 17, which was isolated from wild mouse populations (Schimenti 2000; Lyon 2003). Heterozygous t/+ males preferentially (up to 99%) transmit the t chromosome to their offspring. Meiotic recombination between t-haplotype and wild-type chromosomes is strongly suppressed due to four large inversions, ensuring cotransmission of all factors involved in transmission ratio distortion (TRD) (Fig. 1A; Lyon 2003). However, rare recombinants have been isolated and utilized to identify and map four Distorter loci (t-complex distorters 1–4, Tcd1–4) and the single Responder (t-complex responder, Tcr) to chromosomal subregions of several megabases (Lyon 1984; Silver and Remis 1987). Genetic data demonstrated that Tcd loci act additively in enhancing the transmission rate of Tcr. It was suggested that Distorter genes have a harmful effect on sperm function, brought about by an action on the Responder, and that the wild-type allele of the Responder is more sensitive than the t form (Lyon 1986). Motility tests showed that part of the sperm derived from t/+ males displays altered beat frequency and impaired forward movement, while sperm from males carrying two t haplotypes becomes immotile quickly (Katz et al. 1979; Olds-Clarke and Johnson 1993). These and other data have led to the conclusion that Tcds compromise the motility of all sperm, while Tcr specifically rescues sperm carrying this gene, giving t sperm an advantage in reaching the oocytes.
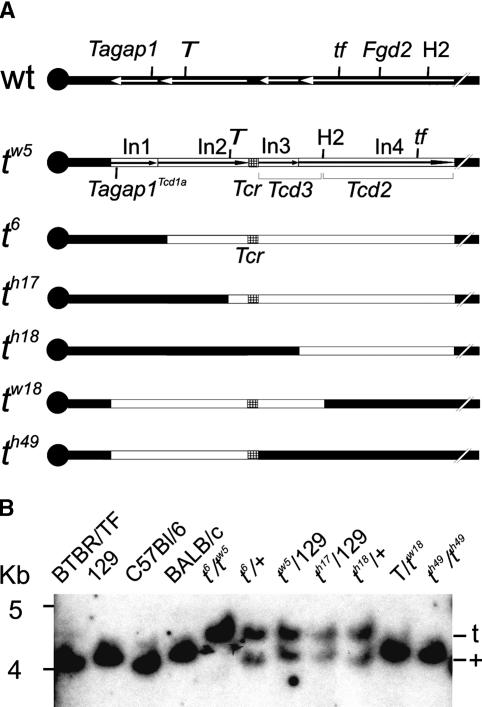
Figure 1.
Fgd2 maps to the Tcd2 region. (A) Structure of a complete (tw5) and various partial (tx) t haplotypes used for mapping Fgd2. The Tcd2 region is defined as the segment of t chromatin that is present in th18 and exchanged for wild-type chromatin in tw18. The centromere is shown as a filled circle at the left, wild-type chromatin is symbolized by filled bars, t chromatin is indicated by open bars, and inversions (In1–In4) are indicated by arrows. Markers and t haplotypes have been described (Lyon 1984); only three Distorters are indicated. (wt) Wild type. (B) Southern blot analysis of genomic DNA digested with PstI, using a full-length Fgd2 cDNA clone as probe, reveals a t-specific band of 4.5 kb that occurs in t haplotypes carrying Tcd2—but not in tw18 or th49—thus mapping Fgd2 to the Tcd2 region; only polymorphic fragments are shown.
A molecular dissection of the TRD phenomenon became possible through cloning of the Responder gene Smok1Tcr (Herrmann et al. 1999), which encodes a dominant-negative form of the protein kinase Smok1. The biochemical function of Tcr suggested that Tcd genes encode signaling molecules acting upstream of Smok1. In accordance with this model, we demonstrated that Tcd1a encodes the GTPase-activating protein Tagap1, a control factor of Rho G proteins (Bauer et al. 2005). This result prompted us to search the distal t-haplotype region for Distorter candidates, using the following criteria: The gene must (1) be located in the genomic interval comprising Tcd2, (2) encode a protein involved in signaling, (3) be expressed in testis, and (4) show alterations in the t haplotype as compared with the wild-type allele.
Results and Discussion
We identified several genes encoding signaling molecules in the distal t-haplotype region. One of them, Fgd2, fulfilled all criteria for a Distorter candidate. Fgd2 encodes a guanine nucleotide exchange factor (GEF) for Rho small G proteins (Pasteris and Gorski 1999; Rossman et al. 2005). GEFs promote the active state of small G proteins by catalyzing the exchange of GDP for GTP (Schmidt and Hall 2002). According to its map position in the ENSEMBL genome database, Fgd2 should be located within the fourth inversion of the t complex, which contains Tcd2. We confirmed that Fgd2 is located in the Tcd2 region by mapping it to partial t haplotypes, previously used to define the location of Tcd2 (Fig. 1A,B; Lyon 1984).
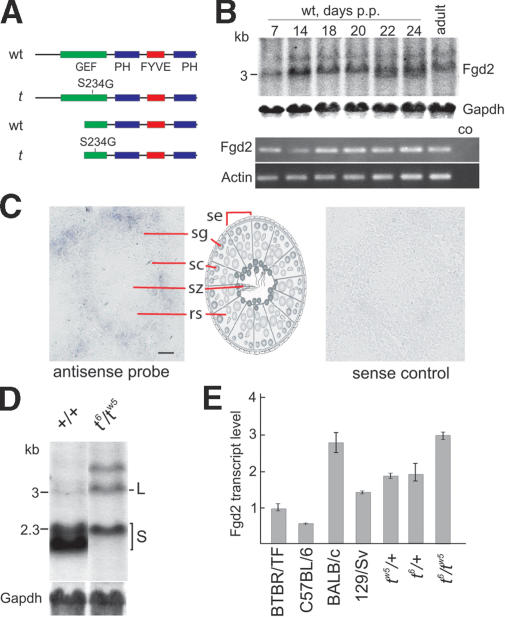
Fgd2 contains an N-terminal GEF domain followed by a Pleckstrin homology (PH) domain, a FYVE domain, and an additional C-terminal PH domain (Fig. 2A). The t form of the deduced Fgd2 protein differs from the wild-type sequence in a single amino acid residue. Ser 234 in the GEF domain was replaced in the t form by a glycine residue (S234G). Fgd2 is expressed in testis (Fig. 2B–E) and in a number of tissues and organs (Pasteris and Gorski 1999). Northern and RT–PCR analyses showed that, in testis, Fgd2 is already transcribed at 7 d post-partum, which corresponds to early meiotic stages of the first cycle of spermatogenesis (Fig. 2B). In situ hybridization analysis of testis sections confirmed this result and furthermore showed that Fgd2 transcription is down-regulated during spermiogenesis, the haploid phase of spermatogenesis (Fig. 2C). Expression in the diploid phase of spermatogenesis may facilitate the distribution of gene products from both alleles to all haploid sperm cells, a prerequisite for a Distorter (Willison and Ashworth 1987). Early expression during spermatogenesis was previously also shown for the Distorter Tagap1Tcd1a (Bauer et al. 2005).
Figure 2.
Fgd2 expression from the t-haplotype allele is strongly enhanced as compared with wild-type alleles. (A) Domain structure of Fgd2 proteins encoded by the long and short transcript variants derived from this gene. (GEF) Catalytical domain of guanosine nucleotide exchange factors; (PH) Pleckstrin homology domain; (FYVE) FYVE domain. (B) Temporal expression profile of the long Fgd2 transcript in postnatal testes representing the first cycle of spermatogenesis. (Top panels) Northern blot. (Bottom panels) RT–PCR analysis. (p.p.) Post-partum, after birth. (C) In situ hybridization of Fgd2 antisense (left) or sense (right) control transcripts to testis cryosections showing expression of Fgd2 in early stages of spermatogenesis. (Middle) Schematic view of a transversal section through a seminiferous tubule. (se) Sertoli cell; (sg) spermatogonia; (sc) spermatocytes; (rs) round spermatid; (sz) spermatozoa. Bar, 20 μm. (D) Northern blot analysis of Fgd2 expression in testis derived from wild-type (+/+) and t6/tw5 males demonstrates strongly enhanced expression of the long (L)—and simultaneous strongly reduced expression of the short (S)—Fgd2 transcript in t haplotypes. (Gapdh) Gapdh loading control. (E) Quantitative RT–PCR analysis of the long Fgd2 testis transcript in various strains and t haplotypes, demonstrating up to sixfold higher expression of Fgd2 in t haplotypes compared with wild-type strains, which show considerable differences. (+) Wild-type strain BTBR/TF.
Fgd2 also expresses a shorter transcript, which is of variable size (2.0–2.3 kb) in wild-type strains, from a promoter located within the gene (Fig. 2D). The shorter transcript encodes a truncated protein lacking the N-terminal half of its GEF domain (Fig. 2A). The GEF domain of this protein most likely is not functional, since a splicing variant of the related human protein FGD1 lacking 36 amino acid residues from the N-terminal part of its GEF domain (FGD1Δ) has been shown to lack GEF activity (Olson et al. 1996). However, the truncated Fgd2 variant may form nonproductive interactions with its target Rho protein and function as dominant inhibitory protein, as has been suggested for FGD1Δ (Whitehead et al. 1998).
Northern blot analysis of testis RNA derived from wild-type and t6/tw5 compound heterozygous mice showed that t-haplotype mice express significantly higher levels of the long Fgd2 mRNA than various wild-type strains. This observation was confirmed by quantitative RT–PCR (Fig. 2D,E). Tagap1 also shows various levels of RNA expression in different wild-type strains (Bauer et al. 2005). These findings are in line with the observation of different penetrance of the TRD phenotype on various wild-type backgrounds (Gummere et al. 1986).
We found that, in testis, t6/tw5 males express threefold higher levels of Fgd2 mRNA than BTBR/TF, the strain we used for testing Distorter candidates for TRD. A similar result was obtained in the analysis of the Distorter Tagap1Tcd1a, which was shown to represent a hypermorph. The high levels of Tagap1Tcd1a transcripts were shown to be caused at least in part by amplification of the Tagap1 locus (Bauer et al. 2005). We excluded this mechanism for Fgd2 (data not shown). Instead, it seems that in t6/tw5 testis the level of the large Fgd2 transcript is highly increased at the expense of the smaller transcript, which is strongly reduced compared with wild type. If the truncated Fgd2 form acted as dominant negative, as discussed above, and was coexpressed with the full-length protein, coupled up-regulation of the functional GEF and down-regulation of the dominant inhibitory form in t/+ spermatocytes would even further increase Fgd2 GEF activity in sperm derived from t/+ males as compared with +/+ sperm. The mechanism of possibly coupled up- and down-regulation of the two transcripts remains elusive.
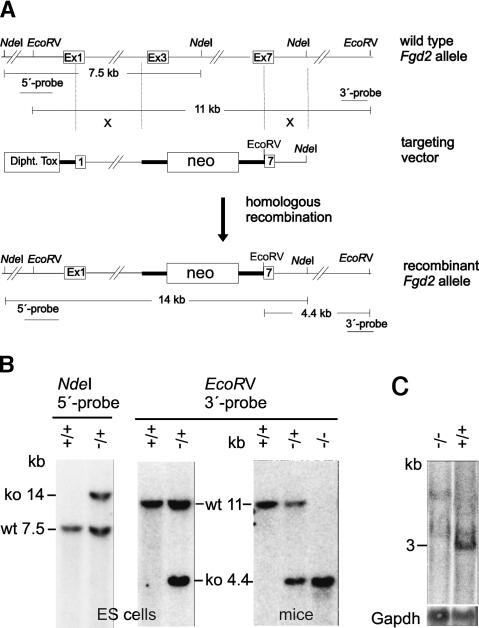
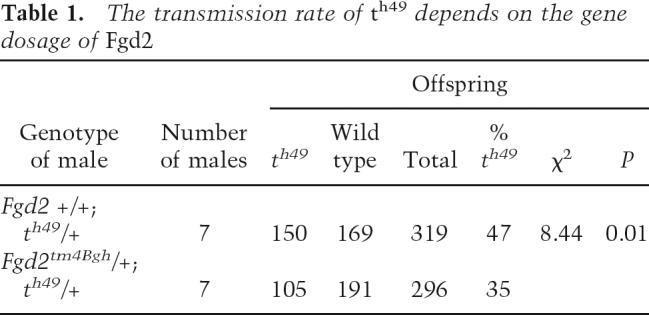
The enhanced expression of Fgd2 in t haplotypes suggested that the t allele of Fgd2, like Tagap1Tcd1a, represents a gain-of-function allele. Distorters act in a dosage-dependent manner (Lyon 1992; Bauer et al. 2005). If a gain of Fgd2 function promotes t-haplotype transmission, one would expect that a reduction in Fgd2 activity would decrease the transmission ratio of a t haplotype. Therefore, in order to test Fgd2 for Distorter activity directly we generated a loss-of-function allele by gene targeting of the wild-type locus (Fig. 3A). A selection cassette was inserted into the genomic Fgd2 locus, replacing exons 3–6 and part of exon 7 (which code for most of the GEF domain) and causing premature termination of Fgd2 transcription. The targeted allele, Fgd2tm4Bgh, was introduced into the germline and tested for transcriptional activity (Fig. 3B,C). Northern blot analysis showed that mice homozygous for the targeted allele lack the large Fgd2 transcript, suggesting that the mutant represents a null allele. To determine the effect of the mutant allele on TRD, females heterozygous for Fgd2tm4Bgh were crossed to males homozygous for the partial t haplotype th49 (see Fig. 1A). Littermates carrying th49 and either the wild-type Fgd2 allele or Fgd2tm4Bgh on the homologous chromosome were tested for the transmission rate of th49, using Southern blot analysis of embryos derived from matings with outbred females (see Materials and Methods). While the former (th49/+; +/+) transmitted th49 at the expected rate of 47% to their offspring, the latter (th49/+; +/Fgd2tm4Bgh) transmitted the t haplotype to only 35% of their offspring. Thus, the reduction of the Fgd2 gene dosage by one-half changed the transmission ratio of th49 to wild-type offspring from almost equal (1:1.1) to a 1.8-fold excess of wild-type offspring (1:1.8; p < 0.01; Table 1). These data directly demonstrate that Fgd2 acts as Distorter and strongly suggest that the t allele of Fgd2 represents Tcd2. Therefore, this locus is designated Fgd2Tcd2.
Figure 3.
Gene targeting of Fgd2 by homologous recombination. (A) Scheme of gene targeting. A neo selection cassette was introduced into the Fgd2 locus replacing exons 3–6 and part of exon 7. Probe locations and predicted diagnostic restriction fragments are indicated. (B) Southern blot analysis of DNA derived from targeted and control embryonic stem (ES) cells (left) and of mice (right) carrying the mutant allele (−), with the 5′ and 3′ probes. (C) Northern blot analysis of testis RNA derived from wild-type and mutant animals. The long Fgd2 transcript is not detected in homozygous mutant animals. (Gapdh) Gapdh loading control; (ko) band derived from targeted allele; (wt) wild-type fragment; (+) wild type; (−) mutant.
Table 1.
The transmission rate of th49 depends on the gene dosage of Fgd2
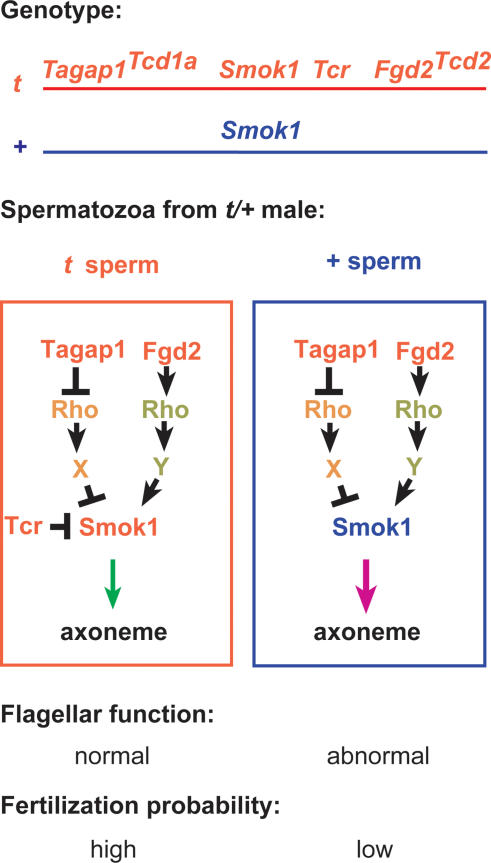
The identification of Fgd2 as Distorter allows us to refine our model of the molecular basis of TRD (Fig. 4; Bauer et al. 2005). Previously we have shown that overexpression of Tagap1, an inhibitor of Rho small G proteins, increases the transmission rate of the t haplotype, while a loss-of-function allele has the opposite effect. Here we showed that Fgd2, a GEF and activator of Rho GTPases, acts in a similar manner. Thus, both Distorters act in parallel, while having opposing effects on their respective target Rho proteins. From these data we conclude that Fgd2 and Tagap1 must regulate different Rho targets. Therefore, there are two signaling cascades exerting opposing effects on Smok1. One pathway, revealed by Tagap1, inhibits Smok1; the other, identified by Fgd2, activates Smok1. The hypermorph Tagap1Tcd1a reduces inhibition of Smok1, while Fgd2Tcd2 most likely enhances activation of Smok1. In this manner both Distorter signaling cascades additively hyperactivate Smok1, followed by impairment of motility parameters in all sperm. Tcr is able to rescue this harmful effect of the Distorters, thus restoring normal flagellar function. Since the effect of Tcr is restricted to t sperm, the latter are able to out-compete the impaired wild-type sperm in the race for oocytes.
Figure 4.
Model of TRD: The t haplotype encodes several Distorters (only two are shown), which are expressed in all sperm cells derived from a t/+ male and act on two opposing Rho signaling pathways regulating Smok1. Smok1 is thought to be involved in sperm motility control. Tagap1Tcd1a and Fgd2Tcd2 represent hypermorphic alleles expressing strongly elevated gene activity as compared with the wild type. Enhanced down-regulation of the inhibitory pathway by Tagap1 and stronger up-regulation of the activating pathway by Fgd2 additively induce hyperactivation of Smok1 in all sperm, resulting in abnormal flagellar function and low fertilization probability. This harmful effect of the Distorters is rescued by the dominant-negative action of Tcr, which is restricted to t sperm, giving the latter an advantage in fertilizing the egg cells. Neither the Rho switch molecules nor their target effector proteins (X, Y) are known. Arrows symbolize activation, bars represent inhibition, green arrow indicates normal signaling, and purple arrow indicates impaired signaling.
Rho signaling has been implicated previously in sperm motility control, though direct functional evidence is still missing (Hinsch et al. 1993). The localization of Ropporin (a binding factor for Rho) to the inner surface of the fibrous sheath (a cytoskeletal structure believed to modulate bending of the flagellum and to influence the shape of the flagellar beat), and the localization of Rhophilin (a binding partner for Ropporin and presumed adaptor protein for Rho to the outer surface of the outer dense fibers that are directly associated with the axonemal microtubule doublets and that face the fibrous sheath) provide additional hints for an important role of Rho signaling in sperm motility, although genetic data are still missing (Eddy et al. 2003). Localization of the Rho modifiers Tagap1 and Fgd2 in the flagellum and biochemical studies might interlink these factors and will further elucidate the role of Rho signaling in the control of sperm motility.
Materials and methods
Mice and genetics
We mapped Fgd2 by Southern blot analysis of PstI-digested genomic DNA derived from various complete and partial t haplotypes obtained by rare recombination between wild-type and t-haplotype chromosomes (see Fig. 1A; Lyon 1984), using a cDNA of Fgd2 as probe (position 170–2576 in accession no. AF017368). We derived an embryonic stem (ES) cell line from the strain BTBR/TF–+tf/+tf and generated the targeted allele, Fgd2tm4Bgh, by standard procedures (Ramirez-Solis et al. 1993). A heterozygous Fgd2tm4Bgh/+ female was mated to a th49/th49 male to generate male littermates of the genotypes +/+; th49/+ and +/Fgd2tm4Bgh; th49/+ to test the effect of the targeted allele on the transmission ratio of the t haplotype (Table 1). We genotyped embryos derived from matings with outbred females (NMRI) for th49 by Southern blot analysis of KpnI-digested (or, alternatively, BamHI-digested) genomic DNA using the 3′ fragment of Tagap1 as probe as described (Bauer et al. 2005).
Transcript analysis
We constructed a plasmid cDNA library from testis RNA derived from a t6/tw5 male using the SuperScript plasmid cDNA cloning system (Life Technologies) and screened it by colony filter hybridization using Fgd2-derived cDNAs as probes. We also obtained cDNAs encompassing the full coding sequence from t haplotypes and wild type by RT–PCR. We sequenced clones from both sources and analyzed the results using the Lasergene DNA Star package. We isolated total RNA using Trizol (Invitrogen). For quantitative real-time PCR we used an ABI PRISM 7900 HT SDS (Applied Biosystems). As a reference gene, we analyzed Gapdh expression with the mouse GAPDH assay (Applied Biosystems). We isolated poly(A)+ RNA using the Fast Track system (Invitrogen) and performed Northern blot analysis using the Ambion GlyMAX Northern kit. In situ hybridization on 10-μm cryostat sections was essentially performed as described (Brent et al. 2003). We produced riboprobes complementary to both, the large and the small Fgd2 transcripts by in vitro transcription from the TOPO pCRII vector containing a Fgd2 cDNA fragment obtained by PCR amplification. Digoxygenin-labeled probes were detected by phosphatase reaction of the substrate NBT/BCIP (Sigma).
Gene targeting
We isolated the left (3.9 kb) and right (2.6 kb) homology regions by PCR amplification of genomic DNA derived from the strain BTBR/TF–+tf/+tf. Using restriction sites included in the oligonucleotides we ligated the homology regions to either side of a PGK-promoter/neo resistance gene/triple-pA cassette inserted in a pBluescript vector containing the diphtheria toxin-A chain gene (kindly provided by Achim Gossler), which thereby flanked the left homology region. An EcoRV restriction site creating a restriction fragment length polymorphism (RFLP) for genotyping of the targeted allele was introduced by ligation of the right homology region to the selection cassette. We linearized the resulting targeting vector with SalI. We established a BTBR/TF ES cell line, electroporated the targeting construct, selected and isolated ES cell clones, prepared DNA in 96-well plates, and performed Southern blot analysis according to standard procedures, using 5′ and 3′ probes obtained by PCR amplification of genomic DNA. The targeting event replaced exons 3–6 and part of exon 7 by the neo selection cassette, which also removed a genomic NdeI site, creating another RFLP for genotyping. We verified correct targeting of the locus and genotyped mice using EcoRV-digested DNA hybridized with the 3′ probe, which detects an 11-kb fragment derived from the wild-type allele and a 4.4-kb fragment derived from the targeted Fgd2 allele. The 5′ probe detects a 7.5-kb fragment in NdeI-digested DNA derived from the wild-type allele and a 14-kb fragment derived from the mutant Fgd2 allele. Out of 132 clones analyzed, one displayed the expected mutant fragments with both probes, demonstrating successful targeting of the Fgd2 locus by homologous recombination.
Oligonucleotides and PCR conditions are listed in Supplementary Table 1.
Acknowledgments
We are grateful to Mary Lyon for mouse strains, Sigrid Schaper for protein work and discussions, Birgit Koschorz for excellent technical assistance during the initial phase of the project, Philip Grote for critical comments on the manuscript, Moises Mallo for providing the vector pPGKneoflox, Achim Gossler for the vector pDT Bluescript, Ingo Voigt for chimera production, Ludger Hartmann for supervision of the animal facility, and Rebecca Kalsow for expert animal caretaking. This project was supported by a grant from the Deutsche Forschungsgemeinschaft to B.G.H.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.414807
References
- Bauer H., Willert J., Koschorz B., Herrmann B.G., Willert J., Koschorz B., Herrmann B.G., Koschorz B., Herrmann B.G., Herrmann B.G. The t complex-encoded GTPase-activating protein Tagap1 acts as a transmission ratio distorter in mice. Nat. Genet. 2005;37:969–973. doi: 10.1038/ng1617. [DOI] [PubMed] [Google Scholar]
- Brent A.E., Schweitzer R., Tabin C.J., Schweitzer R., Tabin C.J., Tabin C.J. A somitic compartment of tendon progenitors. Cell. 2003;113:235–248. doi: 10.1016/s0092-8674(03)00268-x. [DOI] [PubMed] [Google Scholar]
- Eddy E.M., Toshimori K., O’Brien D.A., Toshimori K., O’Brien D.A., O’Brien D.A. Fibrous sheath of mammalian spermatozoa. Microsc. Res. Tech. 2003;61:103–115. doi: 10.1002/jemt.10320. [DOI] [PubMed] [Google Scholar]
- Gummere G.R., McCormick P.J., Bennett D., McCormick P.J., Bennett D., Bennett D. The influence of genetic background and the homologous chromosome 17 on t-haplotype transmission ratio distortion in mice. Genetics. 1986;114:235–245. doi: 10.1093/genetics/114.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann B.G., Koschorz B., Wertz K., McLaughlin K.J., Kispert A., Koschorz B., Wertz K., McLaughlin K.J., Kispert A., Wertz K., McLaughlin K.J., Kispert A., McLaughlin K.J., Kispert A., Kispert A. A protein kinase encoded by the t complex responder gene causes non-Mendelian inheritance. Nature. 1999;402:141–146. doi: 10.1038/45970. [DOI] [PubMed] [Google Scholar]
- Hinsch K.D., Habermann B., Just I., Hinsch E., Pfisterer S., Schill W.B., Aktories K., Habermann B., Just I., Hinsch E., Pfisterer S., Schill W.B., Aktories K., Just I., Hinsch E., Pfisterer S., Schill W.B., Aktories K., Hinsch E., Pfisterer S., Schill W.B., Aktories K., Pfisterer S., Schill W.B., Aktories K., Schill W.B., Aktories K., Aktories K. ADP-ribosylation of Rho proteins inhibits sperm motility. FEBS Lett. 1993;334:32–36. doi: 10.1016/0014-5793(93)81674-o. [DOI] [PubMed] [Google Scholar]
- Katz D.F., Erickson R.P., Nathanson M., Erickson R.P., Nathanson M., Nathanson M. Beat frequency is bimodally distributed in spermatozoa from T/t12 mice. J. Exp. Zool. 1979;210:529–535. doi: 10.1002/jez.1402100316. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. Transmission ratio distortion in mouse t-haplotypes is due to multiple distorter genes acting on a responder locus. Cell. 1984;37:621–628. doi: 10.1016/0092-8674(84)90393-3. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. Male sterility of the mouse t-complex is due to homozygosity of the distorter genes. Cell. 1986;44:357–363. doi: 10.1016/0092-8674(86)90770-1. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. Deletion of mouse t-complex distorter-1 produces an effect like that of the t-form of the distorter. Genet. Res. 1992;59:27–33. doi: 10.1017/s0016672300030147. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. Transmission ratio distortion in mice. Annu. Rev. Genet. 2003;37:393–408. doi: 10.1146/annurev.genet.37.110801.143030. [DOI] [PubMed] [Google Scholar]
- Olds-Clarke P., Johnson L.R., Johnson L.R. t haplotypes in the mouse compromise sperm flagellar function. Dev. Biol. 1993;155:14–25. doi: 10.1006/dbio.1993.1002. [DOI] [PubMed] [Google Scholar]
- Olson M.F., Pasteris N.G., Gorski J.L., Hall A., Pasteris N.G., Gorski J.L., Hall A., Gorski J.L., Hall A., Hall A. Faciogenital dysplasia protein (FGD1) and Vav, two related proteins required for normal embryonic development, are upstream regulators of Rho GTPases. Curr. Biol. 1996;6:1628–1633. doi: 10.1016/s0960-9822(02)70786-0. [DOI] [PubMed] [Google Scholar]
- Pasteris N.G., Gorski J.L., Gorski J.L. Isolation, characterization, and mapping of the mouse and human Fgd2 genes, faciogenital dysplasia (FGD1; Aarskog syndrome) gene homologues. Genomics. 1999;60:57–66. doi: 10.1006/geno.1999.5903. [DOI] [PubMed] [Google Scholar]
- Ramirez-Solis R., Davis A.C., Bradley A., Davis A.C., Bradley A., Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1993;225:855–878. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- Rossman K.L., Der C.J., Sondek J., Der C.J., Sondek J., Sondek J. GEF means go: Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Schimenti J. Segregation distortion of mouse t haplotypes the molecular basis emerges. Trends Genet. 2000;16:240–243. doi: 10.1016/s0168-9525(00)02020-5. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Hall A., Hall A. Guanine nucleotide exchange factors for Rho GTPases: Turning on the switch. Genes & Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Silver L.M., Remis D., Remis D. Five of the nine genetically defined regions of mouse t haplotypes are involved in transmission ratio distortion. Genet. Res. 1987;49:51–56. doi: 10.1017/s0016672300026720. [DOI] [PubMed] [Google Scholar]
- Whitehead I.P., Abe K., Gorski J.L., Der C.J., Abe K., Gorski J.L., Der C.J., Gorski J.L., Der C.J., Der C.J. CDC42 and FGD1 cause distinct signaling and transforming activities. Mol. Cell. Biol. 1998;18:4689–4697. doi: 10.1128/mcb.18.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willison K., Ashworth A., Ashworth A. Mammalian spermatogenic gene expression. Trends Genet. 1987;3:351–355. [Google Scholar]