Abstract
Spt6 promotes transcription elongation at many genes and functions as a histone H3 chaperone to alter chromatin structure during transcription. We show here that mammalian Spt6 binds Ser2-phosphorylated (Ser2P) RNA polymerase II (RNAPII) through a primitive SH2 domain, which recognizes phosphoserine rather than phosphotyrosine residues. Surprisingly, a point mutation in the Spt6 SH2 domain (R1358K) blocked binding to RNAPIIo without affecting transcription elongation rates in vitro. However, HIV-1 and c-myc RNAs formed in cells expressing the mutant Spt6 protein were longer than normal and contained splicing defects. Ectopic expression of the wild-type, but not mutant, Spt6 SH2 domain, caused bulk poly(A)+ RNAs to be retained in the nucleus, further suggesting a widespread role for Spt6 in mRNA processing or assembly of export-competent mRNP particles. We cloned the human Spt6-interacting protein, hIws1 (interacts with Spt6), and found that it associates with the nuclear RNA export factor, REF1/Aly. Depletion of endogenous hIws1 resulted in mRNA processing defects, lower levels of REF1/Aly at the c-myc gene, and nuclear retention of bulk HeLa poly(A)+ RNAs in vivo. Thus binding of Spt6 to Ser2-P RNAPII provides a cotranscriptional mechanism to recruit Iws1, REF1/Aly, and associated mRNA processing, surveillance, and export factors to responsive genes.
Keywords: Spt6, SH2 domain, RNAPII CTD, transcription elongation, splicing, nuclear mRNA export
In eukaryotic cells, transcription elongation is tightly coupled to RNA polymerase II (RNAPII) phosphorylation, mRNA processing (5′-end capping, splicing, cleavage, and polyadenylation), surveillance, and assembly of export-competent mRNPs (for recent reviews, see Aguilera 2005; Bentley 2005; Sommer and Nehrbass 2005). To integrate these different steps in gene expression, a subset of protein/histone modification and mRNA processing complexes are loaded directly onto the C-terminal domain (CTD) of the RNAPII Rpb1 subunit during RNA elongation. The nascent mRNA then assembles with RNA-binding proteins to form mRNP particles that can be efficiently exported out of the nucleus. Any event that significantly disrupts the formation of functional mRNP particles leads to nuclear retention of the RNA and can trigger mRNA decay at the nuclear pore or within the exosome, a complex of 3′-to-5′ exonucleases that degrades improperly processed RNAs as part of the mRNA surveillance pathway (Vasudevan and Peltz 2003; Vinciguerra and Stutz 2004; Saguez et al. 2005).
In Saccharomyces cerevisiae, mRNA export is linked to RNAPII transcription elongation through the TREX (transcription export) complex, which is composed of the THO subcomplex proteins (Hpr1, Tho2, Mtf1, Thp2), the RNA export factor, REF1/Aly (Yra1 in yeast), and UAP56 (Sub2 in yeast) (Lei et al. 2001; Strasser and Hurt 2001). REF1/Aly is recruited cotranscriptionally to nascent transcripts by UAP56/Sub2 and the THO complex, and subsequently directs mRNAs to the TAP:NXF1–p15 (Mex67 in yeast) export receptor at the nuclear pore (Lei et al. 2001; Luo et al. 2001; Strasser and Hurt 2001; Lei and Silver 2002; Zenklusen et al. 2002). In mammalian cells, UAP56 and REF1/Aly associate with the exon junction complex (EJC), and can be stably deposited onto spliced RNAs independently of ongoing transcription (Abruzzi et al. 2004; Masuda et al. 2005). However, splicing is not a prerequisite for mRNA export in mammalian cells, and factors like REF1/Aly must be recruited cotranscriptionally to intronless genes. This process may involve eukaryotic THO complex, which has been suggested to link RNAPII elongation with mRNA processing (Rondon et al. 2003; Rehwinkel et al. 2004; Li et al. 2005). Because eukaryotic cells possess multiple mRNA export pathways, it is unclear whether splicing-dependent recruitment is the predominant mechanism(s) by which export factors are loaded onto nascent transcripts (for reviews, see Dimaano and Ullman 2004; Aguilera 2005; Reed and Cheng 2005).
Recent studies have examined the mechanisms that connect RNAPII elongation with later steps in gene expression. Phosphorylation of the RNAPII CTD at the Ser5 position occurs upon promoter clearance, concomitant with the loading of elongation factors like Spt6, FACT/Spt16–Cdc73, the Paf1 complex, and enzymes that mediate histone H3K4 trimethylation (H3K4Me3). Spt6 functions as a H3:H4 chaperone to alter chromatin structure (Bortvin and Winston 1996; DeSilva et al. 1998; Adkins and Tyler 2006) and can act together with FACT, a histone H2A:2B chaperone, to coordinate the local disassembly and reassembly of nucleosomes during transcription (Orphanides et al. 1998; Belotserkovskaya et al. 2003; Saunders et al. 2003; for review, see Reinberg and Sims 2006). Loss of Spt6 histone H3-binding activity leads to increased cryptic intragenic transcription (Kaplan et al. 2003) and decreased levels of H3K36Me3, which is normally associated with elongation. Recruitment of Spt6 to responsive genes requires Paf1 (Adelman et al. 2006; Pavri et al. 2006) and the Kismet chromatin remodeling complex (Srinivasan et al. 2005). Studies in yeast and flies have shown that Spt6 colocalizes extensively with elongating RNAPII (Andrulis et al. 2000; Kaplan et al. 2000) and is essential for transcription elongation at many, but not all, genes (for review, see Sims et al. 2004). In vitro, Spt6 strongly enhances RNAPII elongation rates on nonchromatin templates (Endoh et al. 2004), indicating that it has both chromatin-dependent and -independent roles in transcription. In yeast, the Spt6 and Spt5 elongation factors are also required for pre-mRNA 3′-end processing (Lindstrom et al. 2003; Kaplan et al. 2005). Yeast Spt6 associates with another nuclear SPT family member, designated Iws1 (interacts with Spt6)/SPN1 (Fischbeck et al. 2002; Lindstrom et al. 2003); however, the precise role of Iws1/SPN1 in transcription is unclear.
Many late events in gene expression including splicing, cleavage/polyadenylation, surveillance, and nuclear mRNA export also depend on Ser2 phosphorylation of the RNAPII CTD (Licatalosi et al. 2002; Xiao et al. 2003; Ahn et al. 2004; Bird et al. 2004; Ni et al. 2004), which is mediated by the P-TEFb (CycT1:CDK9) kinase (for review, see Peterlin and Price 2006). P-TEFb/CDK9 is critical for transcription elongation at HIV-1 and many, but not all, regulated genes. The yeast Ser2-specific CTD kinase, Ctk1, is also essential for Set2-mediated H3K36Me3 during elongation and limits the spread of H3K4Me3 (Xiao et al. 2003, 2006; Wood et al. 2006). Both mRNA processing enzymes and the nuclear exosome RNA degradation complex associate with actively transcribing Ser2-P RNAPII (RNAPIIo) complexes in vivo (Andrulis et al. 2002). Although the mechanisms that link mRNA surveillance with elongation are not well defined, nuclear Spt6 has been shown to associate tightly with the nuclear exosome subunit, Rrp6 (Andrulis et al. 2002). Moreover, another exosome subunit, hSki8, is an integral component of the human Paf1 complex (Zhu et al. 2005). In yeast, mutations in the nuclear exosome and mRNA 3′-processing factors cause apparent defects in transcription elongation, which underscores the bidirectional cross-talk between transcription and mRNP biogenesis (Luna et al. 2005).
In this study, we report that mammalian RNAPII transcription elongation can be linked to mRNA processing and export through the Spt6 elongation factor. We show that the conserved SH2 domain of Spt6 mediates direct, phospho-specific binding to Ser2-P RNAPII. A point mutant Spt6 protein that is unable to bind RNAPIIo retains the ability to support transcription elongation; however, the resulting transcripts contain splicing defects and accumulate to high levels within the nucleus. Interestingly, similar mRNA processing and nuclear export defects are observed in cells lacking the human Iws1 protein, a direct binding partner of Spt6. We further show that human Iws1 associates with the RNA export factor, REF1/Aly, and facilitates recruitment of REF1/Aly and the nuclear exosome subunit Rrp6 to Spt6-dependent genes in vivo. These findings suggest important roles for Spt6 in nuclear mRNP biogenesis and surveillance, mediated through binding to the RNAPIIo CTD.
Results
Spt6 regulates HIV-1 transcription elongation
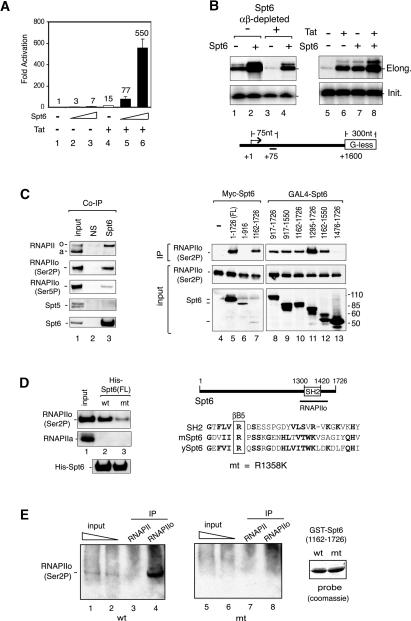
Although Spt6 is a well-defined transcription elongation factor, it was not clear whether it was required for Tat-regulated transactivation of the HIV-1 promoter. Consequently, we used transient expression and small interfering RNA (siRNA) knockdown experiments to determine whether Spt6 promotes HIV-1 Tat-mediated activation of an integrated HIV-1:LacZ reporter gene in HeLa P4 cells. To avoid any unintended effects of Spt6 on HIV-1 Tat protein expression levels, purified recombinant Tat (GST-Tat101) (Fig. 1A, lanes 4–6), or control GST (Fig. 1A, lanes 1–3) proteins were introduced into the cells by chloroquine-mediated protein transduction. Subsequent transient expression of murine Spt6 strongly enhanced Tat activity in these cells and also modestly enhanced basal HIV-1 transcription (Fig. 1A). We next asked whether recombinant Spt6, like Tat, is sufficient to stimulate RNA elongation from the HIV-1 promoter in vitro. Addition of purified recombinant murine Spt6 to a HeLa nuclear extract strongly enhanced HIV-1 transcription from nonchromatin templates in run-off experiments that measure RNA elongation (Fig. 1B, top panel, cf. lanes 1 and 2). In contrast, Spt6 had no effect on transcription initiation from the HIV-1 promoter, as determined by primer extension analysis of the same RNA (Fig. 1B, bottom panel, cf. lanes 1 and 2). Immunodepletion of endogenous Spt6 protein from the HeLa nuclear extract reduced HIV-1 transcription elongation (Fig. 1B, top panel, cf. lanes 3 and 1), without affecting RNA initiation (Fig. 1B, bottom panel, cf. lanes 3 and 1), and RNAPII elongation could be rescued with recombinant Spt6 (Fig. 1B, top panel, cf. lanes 3 and 4). In side-by-side comparisons, either Tat (Fig. 1B, lane 6), Spt6 (Fig. 1B, lane 7), or both proteins (Fig. 1B, lane 8), strongly enhanced HIV-1 RNA elongation without affecting initiation (Fig. 1B, bottom panel, lanes 5–8). Pulse-chase experiments with paused early elongation complexes (EECs) further indicated that Spt6 enhances RNAPII elongation rates on nonchromatin HIV-1 templates in vitro (Supplementary Fig. S1A). Thus, Spt6 cooperates strongly with Tat to induce HIV-1 transcription in vivo and in vitro.
Figure 1.
Spt6 binds tightly to Ser2-P RNAPII. (A) Spt6 enhances Tat transactivation of an integrated HIV-1:LacZ reporter gene in HeLa P4 cells. HeLa P4 cells were transfected with 5 or 25 ng of Spt6 vector in the presence or absence of recombinant HIV-1 Tat101, which was introduced to the cells by chloroquine-mediated protein transduction. (B) In vitro transcription analysis of an HIV-1 G-less template in untreated (lanes 1,2,5–8) or Spt6-immunodepleted (lanes 3,4) nuclear extracts in the presence of recombinant Spt6 (lanes 2,4,7), HIV-1 Tat (lane 6), or both proteins (lane 8). In vitro synthesized RNA was monitored by run-off (elongation, top panels) or primer extension (initiation, bottom panels) assays. (C) Spt6 associates with Ser2-P RNAPII in nuclear extracts. Immunoblot analysis of HeLa nuclear extract (input, lane 1), or fractions immunoprecipitated with antisera to Spt6 (lane 3) or with nonspecific IgG (NS) antisera (lane 2). Antibodies used for immunoblot are indicated to the left of each panel. (Top, Lanes 4–13, top) To map the RNAPII-interacting domain, transiently expressed Myc-tagged Spt6 or GAL4-Spt6 proteins were analyzed for associated Ser2-P RNAPII by immunoblot. (Lanes 4–13) Input RNAPIIo levels are shown in the middle panels, and the different Spt6 protein domains are shown in the bottom panels. (D, lanes 1–3) Full-length wild-type or R1358K recombinant His-Spt6 proteins (shown in the bottom panel) were coupled to beads, incubated with HeLa nuclear extract, and analyzed for binding to Ser2P RNAPII or RNAPIIa. A schematic shows the sequence of the murine Spt6 SH2 domain and the location of the R1358K mutation. (E) Far-Western analysis of binding of wild-type (wt) or R1358K mutant (mt) GST-Spt6 proteins (amino acids 1162–1726) to immunoprecipitated HeLa Ser2-P RNAPII (H5, lanes 4,8) or total RNAPII (N20, lanes 3,7). Binding of the GST-Spt6 proteins to the immunoprecipitated Ser2-P or total RNAPII complexes was visualized by incubating the membrane with glutathione-horseradish peroxidase (Glut-HRP). The panel at the right shows a Coomassie stain of the wild-type and mutant GST-Spt6 proteins used to probe the blot.
Spt6 and Ser2-P RNAPII associate tightly in nuclear extracts
Although Spt6 has been shown to colocalize extensively with elongating RNAPII transcription complexes in vivo (Andrulis et al. 2000; Kaplan et al. 2000), it is not known whether it binds RNAPII directly. To investigate this question, we generated antisera to human Spt6 and asked whether it associates with endogenous RNAPII in our transcription extracts. Interestingly, human Spt6 coimmunoprecipitated strongly with hyperphosphorylated RNAPIIo complexes (Fig. 1C, top panel, lane 3) that are enriched for Ser2-P RNAPII (Fig. 1C, lane 3). In contrast, Spt6 did not associate with Spt5/DSIF (Fig. 1C) or FACT/Spt16 (data not shown), indicating that these binding conditions are quite specific. To map the region of Spt6 that mediates binding to RNAPIIo, wild-type and mutant Myc- or GAL4-tagged Spt6 proteins were transiently expressed in 293T cells and analyzed for associated RNAPIIo by immunoblot (Fig. 1C, lanes 4–13). We noted that Ser2-P RNAPIIo bound efficiently to several C-terminal fragments of Spt6 that contain the SH2 domain (Fig. 1C, lanes 10–12), but did not recognize an overlapping fragment that lacks the SH2 domain (Fig. 1C, lane 13).
To determine whether the Spt6 SH2 domain directly binds RNAPIIo, we generated a mutant Spt6 protein in which an invariant arginine in the SH2 phospho-amino acid-binding motif (Fig. 1D; Pawson 2004) was replaced with a lysine residue (R1358K). Full-length wild-type and R1358K murine His-Spt6 proteins were then purified, coupled to beads, and analyzed for their ability to bind nuclear RNAPIIo in pulldown experiments. Remarkably, wild-type Spt6, but not the R1358K mutant, bound efficiently to native Ser2-P RNAPII in the extract (Fig. 1D, top panel, cf. lanes 2 and 3). In contrast, neither Spt6 protein associated with hypophosphorylated RNAPII (RNAPIIa). Immunoblots confirmed that equivalent levels of wild-type and mutant His-Spt6 proteins were used (Fig. 1D, bottom panel). To assess whether the interaction is direct, we expressed and purified wild-type and R1358K GST-Spt6 SH2 proteins and analyzed their ability to recognize the RNAPIIo Rpb1 subunit in a Far-Western blot. In these experiments, Ser2-P (H5 antibody) or total (N20 antibody) RNAPII immunoprecipitates were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with recombinant wild-type or R1358K mutant GST-Spt6 SH2 domain proteins. Binding of the GST-Spt6 SH2 domain was visualized by chemiluminescence following incubation of the filter with glutathione-coupled horseradish peroxidase (Glut-HRP). As shown in Figure 1E, the wild-type (lane 4), but not R1358K mutant (lane 8), murine GST-Spt6 SH2 domain protein bound selectively to the endogenous HeLa Ser2-P Rpb1 subunit, as evidenced by Glut-HRP reactivity of the RNAPIIo (H5), but not RNAPII (N20), immunoprecipitates. Taken together, these data indicate that Spt6 selectively binds to RNAPIIo through its conserved SH2 domain.
The SH2 domain of Spt6 interacts directly with the Ser2-P RNAPII CTD in vitro
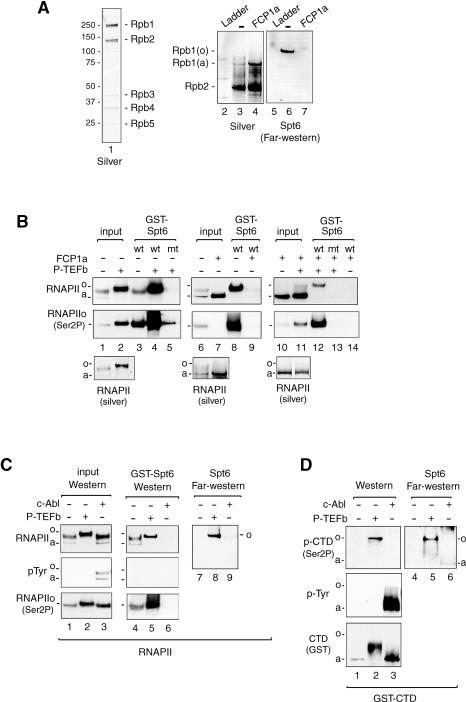
To assess the phospho-specificity of the Spt6:RNAPIIo interaction, both GST-pulldown and Far-Western blot experiments were used to evaluate the binding of recombinant murine Spt6 to affinity-purified HeLa RNAPII or to the recombinant GST-CTD protein. Far-Western blot analysis revealed that the Spt6 SH2 domain fragment binds selectively to the highly phosphorylated Rpb1 subunit (Fig. 2A, lane 6), which is a relatively minor species in the purified HeLa RNAPII fraction (Fig. 2A, lane 3, silver stain). Binding of Spt6 was effectively blocked when the RNAPII fraction was incubated with the CTD phosphatase, FCP1a (Fig. 2A, cf. lanes 6 and 7; Mandal et al. 2002), indicating that Spt6 binds specifically to the hyperphosphorylated RNAPII CTD.
Figure 2.
An SH2 domain mediates binding of Spt6 to Ser2-P RNAPII. (A) Purified HeLa RNAPII (lane 1) was incubated in the presence or absence of the FCP1a CTD phosphatase and analyzed by silver stain (lanes 2–4) or Far-Western blot (lanes 5–7). Binding of the GST-Spt6 SH2 domain (amino acids1162–1726) was detected by staining with Glut-HRP. (B) Binding of the GST-Spt6 SH2 domain (amino acids 1162–1496) to purified HeLa RNAPII was analyzed in the absence or presence of the FCP1a CTD phosphatase or P-TEFb CTD kinase, as indicated above each lane. The RNAPII complexes bound to the GST-Spt6 beads were identified by immunoblot using antisera to total (top) or Ser2-P (middle) RNAPII, and the input RNAPII fractions were also analyzed by silver stain (bottom). (C) Binding of the GST-Spt6 SH2 domain (amino acids 1162–1496) to P-TEFb-phosphorylated (lanes 2,5,8) or c-Abl-phosphorylated (lanes 3,6,9) HeLa RNAPII complexes was examined by means of GST-pulldown (lanes 4–6) or Far-Western blot (lanes 7–9) experiments. Interactions were visualized with antisera to total RNAPII (top), phosphotyrosine (pTyr, middle), or Ser2-P RNAPII (bottom), as indicated next to each panel. The Far-Western blot was visualized with Glut-HRP stain. (D) Far-Western analysis of binding of the GST-Spt6 SH2 domain (amino acids 1162–1726) to purified recombinant murine GST-CTD, either alone (lanes 1,4), or after incubation with recombinant P-TEFb (lanes 2,5) or c-Abl (lanes 3,6) kinases.
To further examine the specificity of this interaction, the purified RNAPII complex was phosphorylated in vitro with the Ser2-P-specific CTD kinase, P-TEFb (CycT1:CDK9), and binding to the GST-Spt6 SH2 domain protein was analyzed using GST-pulldown experiments. Incubation of the purified RNAPII fraction with baculovirus-expressed P-TEFb strongly increased Ser2 phosphorylation in vitro (Fig. 2B, cf. lanes 1 and 2), and significantly enhanced binding to the wild-type (Fig. 2B, cf. lanes 3 and 4), but not R1358K mutant (Fig. 2B, lane 5) GST-Spt6 SH2 proteins. Incubation of purified RNAPII with FCP1a converted most of the Rpb1 subunit to the hypophosphorylated RNAPIIa species (Fig. 2B, cf. lanes 6 and 7), and eliminated binding to GST-Spt6 (Fig. 2B, cf. lanes 8 and 9). Incubation of the FCP1a-treated core RNAPII with recombinant P-TEFb restored Ser2 phosphorylation (Fig. 2B, lanes 10,11) as well as binding of the wild-type (Fig. 2B, cf. lanes 12 and 14), but not the mutant (Fig. 2B, lane 13), GST-Spt6 proteins. Interestingly, the GST-Spt6 SH2 domain protein did not recognize RNAPII complexes phosphorylated in vitro with the c-Abl tyrosine kinase (Baskaran et al. 1997), which presumably targets Tyr1 in the heptad repeats, in either the GST pulldown or Far-Western assays (Fig. 2C, lanes 6,9, respectively). Similarly, Spt6 did not bind to RNAPII complexes phosphorylated by TFIIH/CDK7, a Ser5-specific CTD kinase, in vitro (Supplementary Fig. S1B), indicating that Spt6 recognizes the RNAPII CTD in a Ser2 phospho-specific manner.
To confirm that Spt6 binds directly to the RNAPII CTD, the binding studies were repeated using a recombinant GST-CTD protein. Far-Western blots indicated that the GST-Spt6 SH2 domain binds avidly to P-TEFb-/Ser2-, but not c-Abl-/Tyr1-phosphorylated GST-CTD (Fig. 2D, cf. lanes 5 and 6). As observed with the native RNAPII, Spt6 did not recognize the bulk of intermediate-phosphorylated GST-CTD proteins in the P-TEFb-phosphorylated mixture, but rather selectively bound to a highly phosphorylated form of the protein (Fig. 2D, lane 2, cf. top and bottom panels). Collectively, these data indicate that Spt6 preferentially binds the heavily phosphorylated Ser2-P RNAPII CTD, in vivo and in vitro.
Transcripts formed in the presence of Spt6 R1358K contain splicing defects
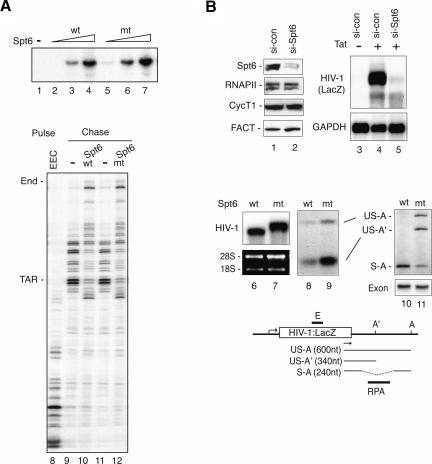
To assess whether binding of Spt6 to the Ser2-P RNAPII CTD is required for transcription, we tested the ability of the recombinant R1358K Spt6 proteins to stimulate transcription elongation of the HIV-1 template in vitro. As shown in Figure 3A, the mutant Spt6 protein was as active as wild-type Spt6, both in run-off transcription experiments (cf. lanes 2–4 and 5–7) as well as in pulse-chase experiments that examine the transcription rate of preassembled EECs (cf. lanes 9,10 and 11,12) on the HIV-1 template.
Figure 3.
Spt6 binds Ser2-P RNAPII to regulate mRNA processing, but not RNAPII transcription elongation. (A, lanes 1–7) Purified recombinant wild-type and R1358K Spt6 proteins enhance RNAPII transcription elongation in vitro. Reactions contained 10 ng (lanes 2,5), 25 ng (lanes 3,6), or 50 ng (lanes 4,7) of wild-type (lanes 2–4) or mutant (lanes 5–7) Spt6 protein. The rate of transcription elongation by recombinant full-length wild-type (lane 10) or R1358K mutant Spt6 (lane 12) was analyzed by pulse-chase analysis of isolated EECs (lane 8). (Lanes 9,11) Where indicated, the HIV-1 EEC was incubated with 100 ng of wild-type Spt6, R1358K Spt6, or GST control, and transcripts were extended following a chase with excess ribonucleotides. (B) siRNA-mediated depletion of endogenous Spt6 blocks transcription of the integrated HIV-1 reporter gene in HeLa P4 cells. Cells treated with control (si-con) or Spt6 (si-Spt6) siRNAs for 84 h were analyzed by immunoblot (lanes 1,2), or by Northern blot for expression of HIV-1:LacZ or GAPDH RNA (lanes 3–5). The bottom panels show Northern blot analyses of HIV-1:LacZ transcripts in HeLa P4 cells expressing siRNA-resistant wild-type (lane 6) or R1358K mutant (lane 7) Spt6 proteins. In this experiment, cells were first treated with an Spt6-siRNA to deplete endogenous Spt6, and HIV-1 transcription was induced by cotransfection with an HIV-1 Tat expression construct. Levels of spliced and unspliced HIV-1:LacZ transcripts were assessed by RPA (lanes 8,9) and RT–PCR (lanes 10,11). (Bottom panel) Quantitative RT–PCR using primers within the exon in HIV-1:LacZ served as an RNA input control. The schematic diagram depicts the integrated HIV-1:LacZ gene in HeLa P4 cells. The HIV-1:LacZ transcripts were amplified using LacZ gene-specific (arrow) and a oligo-dT anchor primers. (S) Spliced transcripts; (US) unspliced transcripts; (A) major polyadenylation sites; (A′) premature polyadenylation sites; (E) Northern probes; (RPA) RPA probes.
To test the activity of the mutant R1358K Spt6 protein in vivo, HeLa P4 cells were first treated with Spt6-siRNA to knockdown the endogenous Spt6 protein. Depletion of the native Spt6 protein caused a dramatic loss of HIV-1:LacZ transcription by Flag-Tat101 (Fig. 3B, cf. lanes 4 and 5). In contrast, loss of Spt6 levels did not affect GAPDH RNA levels (Fig. 3B, bottom panel, cf. lanes 4 and 5), indicating that Spt6 is not essential for the expression of all cellular genes. Neither HIV-1:LacZ nor GAPDH RNA levels were affected in cells expressing a nonsilencing siRNA (si-con) (Fig. 3B, lane 4), indicating that these effects are specific to Spt6. The striking loss of stable HIV-1 RNA was unexpected, given that depletion of Paf1, which recruits Spt6 (Adelman et al. 2006), has a much more modest and transient effect on the expression of the RARβ 2 or fly HSP70 genes in vivo (Adelman et al. 2006; Pavri et al. 2006). Nevertheless, Spt6 strongly contributes to HIV-1 gene transcription, perhaps due to the mechanism of Tat and P-TEFb-regulated transcription elongation. To evaluate the activity of the R1358K mutant Spt6 protein, Spt6-siRNA-treated HeLa P4 cells were cotransfected with vectors that express Flag-Tat101 and the murine (siRNA-resistant) wild-type or R1358K Spt6 proteins, and transcription of the integrated HIV-1:LacZ reporter gene was monitored by Northern blot. Interestingly, HIV-1:LacZ RNA levels were comparable in cells expressing the wild-type or R1358K mutant Spt6 proteins (Fig. 3B, cf. lanes 6 and 7), indicating that the mutant Spt6 protein was not impaired for transcription elongation in vivo. However, the HIV-1:LacZ transcripts produced in the presence of the R1358K Spt6 protein were larger and migrated more slowly than those in normal cells (Fig. 3B, cf. lanes 6 and 7), which suggested that these mRNAs contain processing defects.
As indicated in the schematic diagram, the HIV-1:LacZ gene contains a single 3′-splice site and two possible polyadenylation sites (A′, A). To test for splicing defects, total HIV-1:LacZ RNA was annealed to a probe within the 3′ intron of the HIV-1:LacZ gene and analyzed by means of RNase protection assay (RPA) (Supplemental Material). As shown in Figure 3B, relatively high levels of unspliced transcripts accumulated in cells expressing the R1358K Spt6 protein (cf. lanes 8 and 9). To analyze the HIV-1:LacZ transcripts in greater detail, the 3′ ends of the mRNAs were amplified by RT–PCR using oligo-dT anchor and LacZ-specific primers, and the amplified products were subcloned and sequenced (Supplemental Material). Expression of the mutant Spt6 protein strongly enhanced the use of a premature polyadenylation site (US-A′) (Fig. 3B, lane 9). Consistent with the Northern and RPA data, a strong increase in unspliced HIV-1:LacZ RNA (both US-A and US-A′) was observed in cells expressing the mutant R1358K Spt6 protein (Fig. 3B, cf. lanes 10 and 11), along with lower levels of correctly spliced HIV-1:LacZ RNAs (S-A). Quantitative RT–PCR using primers within the exon (E) ensured that identical amounts of mRNA were analyzed in each reaction (Fig. 3B, exon panel, lanes 10,11). Thus, these data indicate that transcripts with splicing defects accumulate in cells expressing the R1358K mutant Spt6 protein. We cannot exclude that these transcripts may also contain polyadenylation or termination defects, which could contribute to the use of the upstream polyadenylation site.
Characterization of the human Iws1 protein
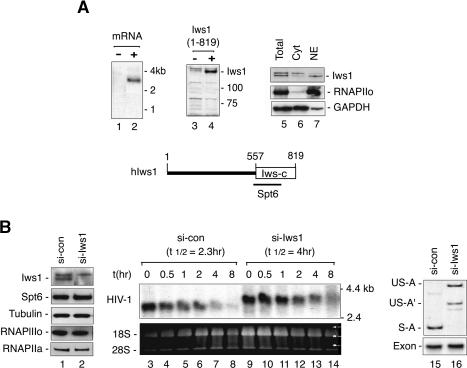
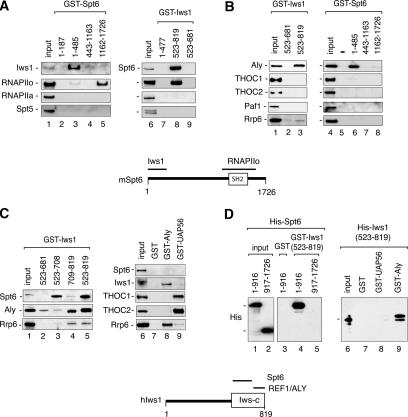
In yeast, Spt6 associates with Iws1/SPN1 (FLJ10006), an essential protein of largely unknown function (Fischbeck et al. 2002; Lindstrom et al. 2003). To characterize the mammalian Iws1 protein, we isolated a full-length human Iws1 cDNA from HeLa total RNA that encodes an 819-amino-acid protein from a 3 kB mRNA (Fig. 4A, lane 2). The predicted human Iws1 is much larger than its yeast ortholog, but contains a homologous 262-amino-acid domain at the C terminus (Fig. 4A, see schematic at the bottom). A fragment of hIws1 spanning the conserved region (amino acids 523–819) was expressed as a bacterial His-tagged protein and used to generate rabbit polyclonal antisera. The anti-Iws1 antisera recognized a 140-kDa protein in immunoblots of HeLa nuclear extracts as well as the ectopically expressed Iws1 protein (Fig. 4A, lanes 3,4), and immunoblots confirmed that hIws1 is a nuclear protein (Fig. 4A, cf. lanes 6 and 7).
Figure 4.
The human Iws1 protein is necessary for splicing of HIV-1:LacZ mRNA in vivo. (A) Analysis of human Iws1 gene expression by Northern blot of HeLa mRNA (lanes 1,2) and immunoblot (lanes 3,4) of a lysate from cells transfected with the hIws1 cDNA (lane 4). (Lanes 5–7) Nuclear and cytoplasmic HeLa extracts were analyzed for hIws1 by immunoblot. At the bottom is a schematic of the hIws1 protein, indicating a C-terminal region (Iws-C, boxed) that is homologous to the S. cerevisiae Iws1/SPN1 protein. (B) Northern blot analysis of HIV-1:LacZ RNA in cells transfected with control- or Iws1-specific siRNAs. (Lanes 1,2) Specific depletion of endogenous hIws1 was assessed by immunoblot. (Lanes 3–14) Cells were treated with ActD (5 μg/mL) for the various times indicated above each lane to block ongoing transcription, and HIV-1:LacZ transcripts were analyzed by Northern blot. (Lanes 15,16) RT–PCR analysis of HIV-1:LacZ RNA in the absence of ActD (0 h) in cells treated with control or Iws1-siRNAs.
To examine whether hIws1 is required for the accumulation or processing of HIV-1:LacZ transcripts, HeLa P4 cells were treated with Iws1-siRNAs or control siRNAs, and HIV-1 RNAs were analyzed by Northern blot. The specificity and efficiency of the knockdown was confirmed by immunoblot (Fig. 4B, cf. lanes 1 and 2). Ongoing transcription was blocked by treating the cells with ActD, which allowed us to examine the stability of HIV-1:LacZ transcripts in the presence or absence of hIws1 (Fig. 4B). Unlike Spt6, Iws1 was not required for the accumulation of stable HIV-1:LacZ RNAs. However, the viral transcripts produced in the absence of hIws1 (Fig. 4B, lanes 9–14) were larger and migrated more slowly than RNAs synthesized in control siRNA-treated cells (Fig. 4B, lanes 3–8). The aberrant HIV-1:LacZ transcripts were somewhat more stable and persisted longer in the presence of ActD (Fig. 4B). RT–PCR analysis indicated that unspliced polyadenylated (US-A) and prematurely polyadenylated (US-A′) HIV-1:LacZ transcripts predominate in cells depleted of hIws1 (Fig. 4B, cf. lanes 15 and 16). Thus, these data indicate that hIws1 is not required for elongation of HIV-1 transcription in vivo, and we also found that recombinant hIws1 has no effect on basal or HIV-1 Tat-induced transcription of nonchromatin HIV-1 templates in vitro (data not shown). Thus, we were unable to detect an elongation defect in cells lacking hIws1; however, transcription of Spt6-dependent genes yielded mRNA processing defects similar to those seen in cells expressing the R1358K mutant Spt6 protein.
Human Iws1 interacts directly with Spt6 and the REF1/Aly mRNA export factor
The yeast Iws1/SPN1 shows strong genetic and biochemical interactions with Spt6, but the interacting regions have not been identified. Consequently, the interacting domains of mammalian Iws1 and Spt6 were mapped using GST pulldown experiments with HeLa nuclear extract and purified recombinant factors. Immunoblot analysis of the GST-pulldown reactions revealed that the N-terminal region of murine Spt6 (GST-Spt6, amino acids 1–485) (Fig. 5A, lane 3) interacts with Iws1, whereas RNAPIIo interacts with the C-terminal SH2 domain fragment (amino acids 1162–1726) (Fig. 5A, lane 5). No binding was observed to RNAPIIa or Spt5, or the Rrp6 in this experiment, indicating that the binding conditions are specific. In reciprocal pulldown experiments using different GST-Iws1 proteins, endogenous Spt6 and RNAPIIo bound avidly to the conserved CTD of hIws1 (amino acids 523–819) (Fig. 5A, lane 8). As expected, the GST-Iws1 (amino acids 523–819) protein did not associate with RNAPIIa or Spt5. Competition experiments further suggested that the binding of hIws1 to RNAPII is indirect and mediated through Spt6 (Supplementary Fig. S1C).
Figure 5.
Human Iws1 interacts directly with Spt6 and REF1/Aly in vitro. (A) GST-Spt6 (lanes 1–5) or GST-Iws1 (lanes 6–9) proteins were coupled to beads, incubated with HeLa nuclear extract, and analyzed for binding to RNAPII and other factors by immunoblot using the antisera indicated to the left of each panel. (B) Immunoblot analysis of the REF1/Aly and other factors to GST-Iws (lanes 1–3) or GST-Spt6 (lanes 4–8) protein-coupled beads in GST-pulldown experiments with HeLa nuclear extract. (C) Analysis of the ability of different transcription factors to bind GST-hIws1 domains (lanes 1–5), GST (lane 7), GST-Aly (lane 8), or GST-UAP56 (lane 9) in GST-pulldown experiments carried out with HeLa nuclear extract. Interacting proteins were visualized by immunoblot using the antisera indicated to the left of each panel. (D) Direct binding of mSpt6 to hIws1 and of hIws1 to hREF1/Aly was analyzed in GST-pulldown experiments carried out with purified recombinant factors. Recombinant His-Spt6 (amino acids 1–916 or 917–1726) proteins were incubated with GST (lane 3) or GST-Iws1 (amino acids 523–819; lanes 4,5) beads, and binding was assessed by immunoblot. For the right panel, His-Iws1 (amino acids 523–819) protein was incubated with GST (lane 7), GST-UAP56 (lane 8), and GST-Aly (lane 9) beads, and binding was examined by immunoblot.
The observation that depletion of hIws1, or mutation of Spt6, gives rise to mRNA processing defects suggested to us that these factors may associate with THO or Paf1 complexes, which have been implicated to couple RNAPII elongation to mRNA processing and export. Although neither Spt6 nor hIws1 associated with Paf1 or with the human THO/TREX subunits, Thoc1 or Thoc2, we noted that both GST-Spt6 (amino acids 1–485) and GST-Iws1 (amino acids 523–819) interacted strongly with the RNA export adaptor protein, REF1/Aly (Fig. 5B, lanes 3,6). Moreover, both hIws1 and REF1/Aly recognize the same region of Spt6 (amino acids 1–485), suggesting that Spt6 recognizes REF1/Aly indirectly, through Iws1. In contrast, hIws1 appears to contain distinct binding sites for Spt6 (Fig. 5C, lane 3) and REF1/Aly (Fig. 5C, lane 4). Reciprocal GST-pulldown experiments showed that GST-Aly strongly associates with hIws1, but not with Spt6 (Fig. 5C, lane 8), in HeLa nuclear extracts. Interestingly, GST-REF1/Aly did not interact with nuclear Thoc1 or Thoc2 under the stringent conditions used in these assays, whereas both of these THO complex subunits bound avidly to the GST-UAP56 beads (Fig. 5C, lane 9). Although Spt6 colocalizes in vivo with the nuclear exosome subunit Rrp6 (Andrulis et al. 2002), Spt6 did not bind Rrp6 in our pulldown experiments (Fig. 5B, lanes 4–8), indicating that this association is not direct. However, low levels of nuclear Rrp6 were recovered in pulldown experiments with GST-Iws1 beads (Fig. 5C, bottom panel, lanes 4,5) and significantly higher levels were recovered with GST-REF1/Aly beads (Fig. 5C, lane 8), indicating that REF1/Aly may bridge the association between Spt6 and Rrp6 in vivo.
To determine whether hIws1 interacts directly with Spt6 and REF1/Aly, the recombinant proteins and putative interacting domains were purified and tested in GST pulldown experiments in vitro. These studies showed that the N-terminal half of mSpt6 (His-Spt6, amino acids 1–916) interacts with hIws1 (amino acids 523–819) (Fig. 5D, lane 4), but not with the GST control (Fig. 5D, lane 3). In contrast, the C-terminal half of Spt6, which includes the SH2 domain, did not recognize Iws1 (Fig. 5D, lane 5), indicating that Spt6 has distinct binding sites for RNAPIIo and Iws1. Moreover, recombinant hIws1 (His-Iws1, amino acids 523–819) bound tightly to GST-Aly (Fig. 5D, lane 9), but did not recognize the GST (Fig. 5D, lane 7) or GST-UAP56 (Fig. 5D, lane 8) proteins. We conclude that Iws1 interacts directly with Spt6 and REF1/Aly, both in vivo and in vitro.
Nuclear retention of bulk poly(A)+ RNAs in cells depleted of hIws1
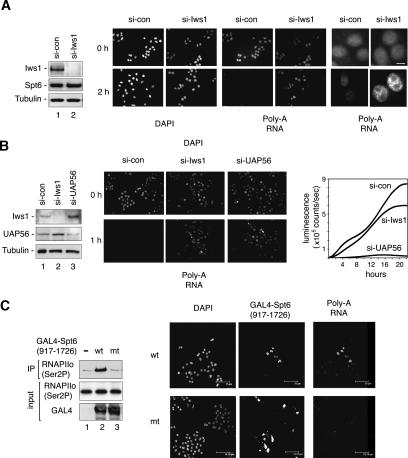
The observation that Iws1 affects mRNA processing of HIV-1 transcripts, and interacts with the REF1/Aly nuclear mRNA export factor led us to ask whether global polyadenylated RNAs accumulate in the nucleus in cells lacking endogenous Iws1 protein. To address this question, HeLa cells were transfected with hIws1-siRNA to deplete endogenous hIws1, as assessed by immunoblot (Fig. 6A), and cellular RNA trafficking was analyzed by fluorescent in situ hybridization (RNA-FISH), using an oligo(dT) probe to detect endogenous polyadenylated RNA. To reduce the RNA signal from ongoing transcription, cells were treated with ActD for 2 h prior to analysis by RNA-FISH. Interestingly, higher levels of poly(A)+ RNA were detected in the nuclei of Iws1-siRNA-expressing cells as compared with the nuclei of control siRNA-treated cells [Fig. 6A, poly(A) RNA panels]. The panels at the far right in Figure 6A show the accumulation of poly(A)+ RNA in Iws1-siRNA-expressing cells at higher resolution. Immunoblot analysis confirmed that ActD induces a similar decline in global Ser2-phosphorylated (Ser2P) RNAPII levels in cells expressing either the control- or the hIws1-specific siRNAs (data not shown). Thus, these data indicate that hIws1 is required for efficient mRNP assembly or export.
Figure 6.
Depletion of endogenous hIws1 impairs export of bulk polyadenylated mRNAs. (A) HeLa cells were transfected with control or hIws1-siRNAs and analyzed by immunoblot (lanes 1,2), immunofluorescence microscopy (DAPI stain), or RNA-FISH to detect poly(A)+ RNAs, as indicated. Cells were either untreated (top panels) or treated with ActD (5 μg/mL) for 2 h (bottom panels) prior to RNA-FISH. The panels on the far right show the fixed nuclei at higher magnification. Bar, 10 μm. (B) HeLa cells were transfected with si-control, si-Iws1, or si-UAP56, and analyzed by immunoblot (lanes 1–3) or by RNA-FISH for poly(A)+ RNAs (bottom panels). Cells were treated with ActD for 1 h prior to RNA-FISH. Graph at the far right compares the rate of expression of an HIV-1:luciferase reporter gene in Tat-expressing HeLa cells treated with control-, Iws1-, or UAP56-specific siRNAs. Luciferase levels were measured by luminescence in counts per second over a 22-h time period. (C) Overexpression of the GAL4-Spt6 SH2 domain blocks the nuclear export of poly(A)+ RNAs in HeLa cells. (Lanes 1–3) Transiently expressed wild-type or mutant GAL4-Spt6 SH2 domain proteins were immunoprecipitated with an anti-GAL4 antibody, and binding to RNAPIIo was determined by immunoblot. The panels show RNA-FISH analysis of HeLa cells transfected with wild-type (wt) or R1358K mutant (mt) GAL4-Spt6-SH2 (amino acids 917–1726) domain proteins. Cells were treated with ActD for 1 h and analyzed by DAPI stain, immunostaining with GAL4-specific antisera, or in situ hybridization for poly(A)+ RNA, as indicated above each panel. Bar, 75 μm.
For comparison, the magnitude of the nuclear mRNA accumulation defect was analyzed in cells lacking UAP56, a putative RNA helicase required for global mRNA export. As shown in Figure 6B, siRNA-mediated depletion of either hIws1 or UAP56 in HeLa cells induced poly(A)+ RNAs to accumulate in the nucleus, as visualized by RNA-FISH. However, loss of Iws1 did not significantly impair the activity of an intronless HIV-1:luciferase reporter gene at 48-h post-transfection (data not shown), indicating that the HIV-1 transcripts produced in these cells are eventually exported and translated. To examine the kinetics of expression of the Tat-dependent HIV-1:luciferase gene reporter, luminescence was monitored in real time at 10-min intervals over a 22-h period in HeLa cells expressing either hIws1siRNAs or UAP56-siRNAs. As shown in the graph in Figure 6B, the maximum rate of luciferase gene expression was significantly reduced in cells lacking hIws1, as compared with cells expressing a control siRNA. In contrast, very little luciferase activity was detected in cells depleted of UAP56 during this time interval. Although hIws1 is not essential for mRNA export and translation, as is UAP56, these data indicate that it is nevertheless broadly required for efficient mRNA processing or nuclear export of many polyadenylated RNAs. In addition, transcripts formed in cells lacking Iws1 may be more stable and accumulate to higher levels in the nucleus as a result of impaired transport to the cytoplasm.
To assess whether binding of Spt6 to the Ser2-P RNAPII is similarly required for optimal export of polyadenylated RNAs, we examined the levels of nuclear poly(A)+ RNA in cells expressing high levels of the GAL4-Spt6 SH2 domain (amino acids 917–1726). As expected, endogenous RNAPIIo efficiently coimmunoprecipitated with the wild-type, but not R1358K mutant, GAL4-Spt6 proteins (Fig. 6C, cf. lanes 2 and 3). To determine whether the GAL4-Spt6 SH2 domain affects nuclear RNA levels, cells were treated with ActD to block ongoing transcription, and nuclear poly(A)+ transcripts were analyzed by RNA-FISH. As shown in Figure 6C, poly(A)+ RNAs accumulated to high levels in HeLa nuclei expressing the wild-type GAL4-Spt6-SH2 protein as determined by costaining with anti-GAL4 antibody, as compared with neighboring untransfected cells (Fig. 6C, top, right panel). In contrast, the distribution of poly(A)+ RNAs in cells expressing the GAL4-Spt6 R1358K mutant (amino acids 917–1726) protein, was indistinguishable from that observed in untransfected cells (Fig. 6C, bottom right panel). We conclude that the GAL4-Spt6 SH2 domain protein effectively competes with the endogenous Spt6 protein for binding to RNAPIIo, and disrupts the timely nuclear export of bulk poly(A)+ RNAs in vivo.
hIws1 facilitates recruitment of REF1/Aly and Rrp6 to Spt6-responsive genes in vivo
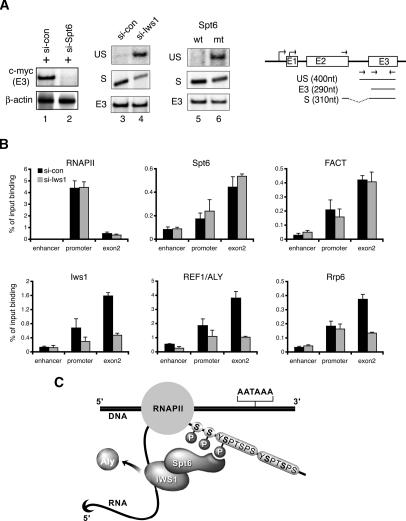
Based on these findings, we speculated that hIws1 might help recruit REF1/Aly to Spt6-responsive genes. Although Spt6 is critical for Tat-regulated HIV-1 transcription, it is not essential for expression of all RNAPII genes, and may be most important for genes regulated at the level of RNAPII elongation. As shown in Figure 7A, we determined that transcription of the endogenous human c-myc gene is highly impaired in 293T cells treated with Spt6-siRNA (lane 2), indicating that c-myc transcription requires Spt6. Moreover, RT–PCR analysis revealed that unspliced c-myc mRNAs accumulate in cells depleted of endogenous hIws1 protein (Fig. 7A, cf. lanes 3 and 4) by RNA interference (RNAi), or in cells that overexpress the R1358K Spt6 mutant protein (Fig. 7A, cf. lanes 5 and 6). Immunoblots confirmed specific depletion of hIws1 protein in cells treated with Iws1-siRNA (see Fig. 6A). Chromatin immunoprecipitation (ChIP) experiments at the endogenous active c-myc gene in 293T cells revealed a strong decline in hIws1 levels, both at the promoter and within exon 2, in cells treated with the Iws1-siRNA, whereas levels of RNAPII, Spt6, or FACT were unaffected (Fig. 7B). Interestingly, cells treated with the Iws1-siRNA contained significantly reduced levels of REF1/Aly within the c-myc exon2 region, and an apparent decline in levels of exon-associated Rrp6 as well. Immunoblots confirmed that neither REF1/Aly nor Rrp6 protein levels were reduced in lysates from cells expressing the Iws1-siRNA (data not shown). Thus, these data indicate that binding of Spt6 to the RNAPIIo CTD may facilitate the recruitment of hIws1, REF1/Aly, and associated mRNA splicing, surveillance, or export factors to active transcription complexes, as illustrated in the schematic shown in Figure 7C.
Figure 7.
The hIws1 protein recruits REF1/Aly to the c-myc gene in vivo. (A) RT–PCR analysis of c-myc mRNA levels in 293T cells expressing control siRNA (lanes 1,3), Spt6-siRNA (lane 2), hIws1-siRNA (lane 4), or the mutant R1358K Spt6 protein (lane 6). The schematic shows RT–PCR products of spliced (S), unspliced (US), and total (E3) c-myc mRNA. (B) ChIP analysis of the c-myc gene in 293T cells treated with control or hIws1-siRNAs. Chromatin was immunoprecipitated with the indicated antisera and analyzed by Q-PCR using primers specific for the enhancer, promoter, or second exon of the human c-myc gene. (C) Working model depicting the association of hIws1 with Spt6 bound to the Ser2-P RNAPII CTD. Our data indicate that this complex is required to facilitate steps required for proper mRNA processing and export, and may also serve to recruit REF1/Aly to Spt6-dependent genes.
Discussion
In this study, we show that the Spt6 elongation factor (Kaplan et al. 2000) binds with high affinity and specificity to the Ser2-P RNAPII CTD in vivo and in vitro. Spt6 promotes transcription elongation at many, but not all, RNAPII genes, and interacts with histone H3 (Bortvin and Winston 1996) to facilitate nucleosome disassembly in conjunction with the FACT complex (Kaplan et al. 2003; Saunders et al. 2003; for review, see Reinberg and Sims 2006). Mutations that prevent binding of Spt6 to histone H3 severely disrupt the repressive chromatin structure at promoters and bypass the need for transcriptional activators (Adkins and Tyler 2006), resulting in high levels of spurious intragenic transcription (Kaplan et al. 2003). The role of Spt6 as a histone H3:H4 chaperone also likely contributes to its role in elongation-associated trimethylation of histone H3K36 (Xiao et al. 2003; Kizer et al. 2005). In addition, Spt6 connects RNAPII elongation with mRNA surveillance through its association with Rrp6 (Andrulis et al. 2002), a subunit of the nuclear exosome. Interestingly, we find that Spt6 is essential for Tat-regulated HIV-1 and c-myc gene transcription in vivo, but is not required for expression of other genes (e.g., β-actin, GAPDH). Similarly, Spt6 strongly enhances HIV-1 transcription elongation in vitro, but has only a minimal effect on the adenovirus major late promoter (data not shown). Thus, the role of Spt6 in transcription elongation is gene specific. Our findings indicate that after recruitment, Spt6 is eventually transferred to the Ser2P RNAPII CTD and functions there to coordinate elongation with later steps in gene expression, including mRNA processing, surveillance, and assembly of mRNP export complexes.
Spt6 is a Ser2-specific RNAPII CTD-binding protein
In vitro, Spt6 binds with strict specificity to Ser2-P (P-TEFb-phosphorylated) RNAPII, and does not recognize RNAPII complexes phosphorylated by the CDK7/TFIIH (Ser5-specific) or c-Abl (Tyr1-specific) CTD kinases (Fig. 2). Surprisingly, this Ser2-P-specific binding to the CTD is mediated by the highly conserved SH2 domain of Spt6, and a conservative point mutation (R1358K) within the Spt6 SH2 phospho-amino acid-binding pocket eliminates binding to RNAPIIo in vivo and in vitro. Interestingly, the Spt6 SH2 domain of the protein is the only SH2 domain in yeast (Maclennan and Shaw 1993), indicating that it originally evolved to recognize phosphoserine, rather than phosphotyrosine, protein substrates. Indeed, several examples of non-phosphotyrosine-binding mammalian SH2 domains have been reported, which may turn out to bind phosphoserine substrates as well. Thus, it will be important to identify the structural features of SH2 domains that specify binding to Ser-phosphorylated rather than Tyr-phosphorylated targets. Moreover, it would be interesting to learn whether the Spt6 SH2 domain could be engineered to bind phosphotyrosine CTD substrates, which could provide a useful reagent for monitoring P-Tyr RNAPII complexes in vivo.
In Far-Western binding experiments, Spt6 recognizes a highly phosphorylated form of the RNAPII CTD, despite the fact that this is a minor species in the in vitro P-TEFb phosphorylation reaction. Thus Spt6 may bind preferentially to the extended conformation of the highly phosphorylated RNAPII CTD (Meinhart et al. 2005), or it may need to recognize multiple phosphoheptad repeats. Our preliminary data indicate that Spt6 cannot bind a single CTD repeat (S. Yoh, unpubl.), and further studies are needed to determine whether it binds multiple repeats through an induced fit mechanism, as observed for certain CTD-interacting proteins, like Set2 (Meinhart and Cramer 2004).
The Spt6 SH2 domain is unique among the various types of protein interaction motifs that have been shown to interact directly with the RNAPII CTD. For example, the Pcf11 3′-processing/transcription termination factor specifically binds Ser2-P RNAPII through a CTD interaction domain (CID) (Licatalosi et al. 2002; Meinhart and Cramer 2004), whereas the Set2 histone methyltransferase, which mediates H3K36 trimethylation during elongation, contains a novel SRI domain that binds to tandem heptad repeats that are doubly phosphorylated at the Ser2 and Ser5 positions (Kizer et al. 2005). Given that the CTD phosphorylation pattern can vary widely among the different imperfect heptad repeats, and that this pattern changes significantly as transcription progresses (for review, see Phatnani and Greenleaf 2006), it will be interesting to learn how the Spt6 is packaged onto the CTD relative to other mRNA processing and histone-modifying complexes, and how the actions of these different complexes are integrated during transcription.
Binding of Spt6 to the RNAPII CTD is required for mRNA processing and exit from the nucleus
Two lines of evidence indicate that the binding of Spt6 to the RNAPII CTD is not required for its ability to promote transcription elongation. First, we found that the R1358K mutant Spt6 protein, which is unable to bind RNAPIIo, retains the ability to stimulate HIV-1 transcription elongation rates in vitro. Second, the mutant Spt6 protein is able to rescue production of HIV-1 and c-myc gene transcription in vivo, in cells that have been depleted of endogenous Spt6 by RNAi. However, the RNAs produced by the R1358K Spt6 protein were longer than normal transcripts and contained splicing defects. Moreover, overexpression in HeLa cells of the isolated Spt6 WT-SH2 domain induced bulk polyadenylated mRNAs to be retained in the nucleus, as determined by RNA-FISH, whereas RNA trafficking was unaffected in cells expressing the Spt6 R1358K SH2 domain. Thus, competitive displacement of the endogenous wild-type Spt6 protein from the RNAPIIo CTD leads to a widespread disruption in mRNA processing or assembly of functional, export-competent mRNP particles, and separates the function of Spt6 in transcription elongation from subsequent downstream events.
Iws1 connects Spt6 to REF1/Aly
These new roles for Spt6 are further supported by the observation that the Spt6-interacting protein, Iws1, appears to play a dedicated role in mRNA processing or formation of functional mRNP particles. In yeast, Iws1/SPN1 was identified as an essential member of the SPT family that interacts with Spt6, but affects transcription of only a small subset of genes (Fischbeck et al. 2002; Lindstrom et al. 2003). We find that the human Iws1 protein is required for mRNA processing, but not transcription, of the HIV-1 and c-myc genes in vivo, and that recombinant hIws1 does not stimulate HIV-1 transcription elongation in vitro. As expected for a broad-acting mRNA processing activity, bulk poly(A)+ mRNAs were found to be retained in the nucleus in cells lacking Iws1, and mRNA export and translation were delayed, but not blocked, in these cells. In comparison, depletion of the UAP56 RNA helicase causes a more profound block to mRNA expression than does loss of either Iws1 or the REF1/Aly RNA adaptor protein (Gatfield and Izaurralde 2002).
In yeast, UAP56 (Sub2) and REF1/Aly (Yra1) are recruited to genes through interactions with THO elongation complex proteins, forming the TREX complex that connects elongation with mRNA splicing and export. The mechanism is complicated by the fact that REF1/Aly is recruited to both DNA (Abruzzi et al. 2004) and RNA (Masuda et al. 2005), potentially through different mechanisms. REF1/Aly and UAP56 associate with the splicing EJC, and UAP56 is thought to recruit REF1/Aly to spliced mRNAs independently of transcription. However, intronless genes also require REF1/Aly for export, and thus far it has been unclear how it is recruited to DNA.
Biochemical interaction experiments shown here indicate that Spt6 can bind simultaneously to Iws1 and RNAPIIo, and we find that Iws1 interacts directly with REF1/Aly, but not with THO subcomplex proteins (Thoc1, Thoc2) or Paf1. Reciprocal pulldown experiments in nuclear extracts confirmed that REF1/Aly interacts strongly with Iws1, but not Spt6, Thoc1, or Thoc2, whereas UAP56 preferentially binds Thoc1, Thoc2, and REF1/Aly, but not Iws1. These findings suggested that Iws1 may connect Spt6 to REF1/Aly (Fig. 7C). Indeed, ChIP experiments revealed that depletion of Iws1 impairs recruitment of REF1/Aly to the c-myc gene in 293T cells without affecting the binding of RNAPII, Spt6, or FACT (Fig. 7B). Thus, the failure to recruit REF1/Aly to Spt6-dependent genes, or its failure to transfer to the growing mRNP, may underlie the widespread accumulation of nuclear mRNAs observed in cells lacking Iws1.
Spt6 integrates transcription elongation with downstream mRNA processing events
Recent studies have shown that export of nuclear mRNAs is directly coupled to the mRNA surveillance pathway (Vasudevan and Peltz 2003; Vinciguerra and Stutz 2004; Saguez et al. 2005). The nuclear exosome subunit, Rrp6, is required for release of nascent mRNAs from sites of active transcription and for mRNA export, and studies in Drosophila show that Rrp6 colocalizes with Spt6 and active RNAPII transcription complexes in vivo (Andrulis et al. 2002). Our data indicate that Rrp6 does not interact directly with nuclear or recombinant Spt6, but rather that Rrp6 associates strongly with REF1/Aly in HeLa nuclear extracts. These observations support a previous report that the yeast Yra1 (REF1/Aly) protein associates with Rrp6 and Rrp45 to link exosome-mediated degradation of incomplete mRNP particles with nuclear mRNA export (Zenklusen et al. 2002). We speculate that Iws1 could also play a role in this process by bridging REF1/Aly and associated exosome components to the DNA-bound Spt6:RNAPIIo transcription complex. Paf1 may also contribute to this regulation, given its role in recruiting Spt6 to promoters (Adelman et al. 2006), influence on poly(A) site utilization and post-transcriptional events (Mueller et al. 2004; Penheiter et al. 2005; Zhu et al. 2005) and ability to traffic with nascent mRNAs as they exit from the nucleus (Porter et al. 2005).
Taken together, our data suggest a mechanism for cotranscriptional loading of mRNA processing and export factors through Spt6 during RNAPII transcription elongation in mammalian cells. This process would provide an alternative route to loading of mRNA export factors through the EJC splicing complex, with potential parallels to yeast, where export factors are loaded through the TREX elongation complex, or the promoter-bound SAGA histone acetylation complex (Rodriguez-Navarro et al. 2004). Additional evidence for cotranscriptional recruitment of export factors in mammalian cells comes from the observation that the herpes virus pUL69 mRNA export protein interacts directly with both Spt6 (Winkler et al. 2000) and UAP56 (Lischka et al. 2006) to control the export of unspliced viral mRNAs. Thus, the model described here provides a framework from which to investigate whether the Spt6–Iws1 interaction has a direct role in REF1/Aly-mediated mRNP export, and whether binding to the RNAPII CTD plays a role in the other important functions of Spt6, such as nucleosome disassembly with the FACT complex or trimethylation of H3K36 by the CTD-bound Set2 complex during transcription elongation.
Materials and methods
Recombinant proteins and antibodies
Wild-type and mutant murine Spt6 proteins and human Cyclin T1 were affinity-purified from recombinant baculovirus-infected Sf9 cells using Talon metal resin (BD Bioscience) chromatography. CDK9 was affinity-purified from recombinant baculovirus-infected Sf9 cell extracts using anti-Flag M2 agarose (Sigma). The recombinant GST-CTD protein (from murine Rpb1) was expressed in bacteria and purified using glutathione-S-agarose chromatography. RNAPII and TFIIH/CDK7 were purified from HeLa S3 nuclear pellet or extracts, respectively (Maldonado et al. 1996). Recombinant FCP1a was purified from baculovirus-infected Sf9 cells as described (Cho et al. 1999). Rabbit polyclonal antibodies were raised against murine Spt6 (amino acids 1508–1726), human Spt5 (amino acids 1–215), and human Iws1 (amino acids 523–819) at Pocono Rabbit Farm and Laboratory, Inc. The following commercial antisera were used: H5 (Ser2-P RNAPII), H14 (Ser5P RNAPII), and 8WG16 (RNAPIIa; Covance); N20 (RNAPII) and GAL4-DBD (Santa Cruz Biotechnology); THOC2 (CIM); REF1/Aly and THOC1 (Gene Tex); FACT/Spt16 (Upstate Biotechnology); Myc-tag and γ-tubulin (Sigma).
In vitro transcription experiments
Primer extension and run-off transcription assays with the HIV-1 G-less template were carried out as described by Bres et al. (2005). For pulse-chase experiments, the immobilized HIV-1 linear template (−340 to +400) was synthesized using a biotinylated forward primer and coupled to Streptavidin-coated paramagnetic beads (Dynabeads M-280). Approximately 500 ng of linear immobilized HIV-1 template was incubated with 20 μL of HeLa nuclear extract in an 85-μL reaction (final volume) containing transcription buffer (20 mM HEPES-Hcl at pH 7.9, 50 mM KCl, 6.25 mM MgCl2, 0.1 mM EDTA, 8.5% glycerol, 20 mM ZnCl2, 3 mM DTT) for 10 min at room temperature. The preinitiation mixture was then pulsed for 30 sec with 600 μM each rATP, rUTP, and rGTP and 1 μM [α-32P]rCTP and washed with Sarkosyl buffer (50 mM Tris-Hcl at pH 7.9, 120 mM NaCl, 0.5% NP-40, 0.1% Sarkosyl, 4 mM DTT). One-tenth of the reaction was incubated with 100 ng of either BSA or recombinant Spt6 for 5 min at room temperature and chased with an rNTP mixture (10 μM rUTP and 0.6 mM each rATP, rCTP, and rGTP).
Protein–protein interaction experiments
The Far-Western procedure was as described by Nollau and Mayer (2001). Briefly, aliquots of 10–50 μg of HeLa nuclear extract, 5 ng of the purified RNAPII fraction, or 1 μg of GST-CTD, which had been irreversibly cross-linked to glutathione, were separated on SDS-PAGE and transferred to a nitrocellulose membrane. GST-Spt6 (amino acids 1162–1726) protein was labeled with glutathione-coupled horseradish peroxidase and binding to the membrane was visualized by conventional chemiluminescence (Marligen). For the GST pulldown experiments, 5–10 μg of GST fusion proteins were coupled to glutathione-S-agarose beads and incubated with either 100 μL of HeLa nuclear extract or 50 ng of purified RNAPII fraction in a final volume of 500 μL of buffer (10 mM HEPES-Hcl at pH 8.0, 300 mM KCl, 0.2 mM EDTA, 0.5% NP-40, 10% glycerol, 1 mM DTT, 1 mM sodium orthovanadate, 1 mM sodium fluoride, 20 mM β-glycerophosphate, 1× protease inhibitor [Roche], 0.2 mM PMSF, supplemented with 10 mg/mL of BSA). The beads were washed extensively in buffer lacking BSA and analyzed by SDS-PAGE and immunoblot.
RNAi
Human siRNAs for Spt6 (5′-CUGCUCGGGAGUAAGCCCAdTdT-3′ and 5′-UGGGCUUACUCCCGAGCAGdTdT-3′), Iws1 (5′-GGAUGAUGUAAAUGAGCAAdTdT-3′ and 5′-UU GCUCAUUUACAUCAUCCdTdC-3′), or UAP56 (5′-GUCAC ACUCGGGAGUUGGCdTdT-3′ and 5′-GCCAACUCCCGAG UGUGACdTdT-3′) were transfected into HeLa P4 cells. Vectors expressing HIV-1 Tat, murine Spt6, or both were introduced 36–60 h post-transfection, and total RNA was isolated 24 h after the second transfection.
RNA-FISH analysis of polyadenylated RNAs
For RNA-FISH and immunofluorescence staining, cells were permeabilized for 10 min in PBS with 0.1% Triton X-100, washed twice in PBS to remove cytoplasmic proteins, and fixed with PBS containing 4% paraformaldehyde. Hybridization was carried out overnight at room temperature in a buffer containing 4× SSC, 5× Denhart’s solution, 10% dexatransulfate, 50% formamide, 1 mg/mL tRNA, 0.5 mg/mL heparin-sulfate, 10 U/mL RNase inhibitors (Invitrogen), and 1 μg/mL Texas red-oligo-dT (18-mer). Images were collected either with an Axioplan2 fluorescence-imaging microscope outfitted with an Axiophot 2 camera, or with a Leica TCS SP2 AOB confocal microscope, and were processed with Openlab software. To analyze the rate of reporter gene expression, HeLa cells expressing the siRNAs were cotransfected with HIV-1 Tat and HIV-1 LTR:Luc vectors. Four hours post-transfection, the cells were replaced with culture medium supplemented with 2 mM luciferin, and luminescence was measured as counts per second for 30 sec at 10-min intervals over a 22-h period using a Lumicycle (Actimatrics). Excess luciferin was added to the culture medium to rapidly quench luciferase activity and ensure that only newly synthesized luciferase proteins were detected.
ChIP experiments
ChIP experiments were used to analyze the endogenous c-myc gene in 293T cells by real-time quantitative PCR with SYBR (Applied Biosystems). The following primer pairs were used: enhancer (5′-CCCAAAAAAAGGCACGGAA-3′ and 5′-TATTGGAAATGCGGTCATGC-3′), promoter (5′-TCCTCTCTCGCTAATCTCCGC-3′ and 5′-CGTCCAGACCCTCGCATTA TA-3′), and exon2 (5′-CCCTCAACGTTAGCTTCACCA-3′ and 5′-ACCGAGTCGTAGTCGAGGTCA-3′).
Acknowledgments
We thank D. Reinberg (University of Medicine and Dentistry of New Jersey) for FCP1a reagents, M. Green (Worchester, MA) for UAP56 and REF1/Aly reagents, G. Pruijn (Nijmegen Centre for Molecular Life Sciences) for anti-hRrp6 antibodies, J. Noel (The Salk Institute) for structural assessment of the Spt6 SH2 domain, and R.E. Verdun and J. Karlseder (The Salk Institute) for assistance with RNA-FISH. This study was funded by grants from the NIH to K.A.J. (AI044615-07) and R.M.E., who is also an HHMI Investigator and March of Dimes Chair in Molecular and Developmental Biology. S.M.Y was supported by a fellowship from the University-wide AIDS Research Program, and H.C. was an HHMI Research Associate.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1503107
References
- Abruzzi K.C., Lacadie S., Rosbash M., Lacadie S., Rosbash M., Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K., Wei W., Ardehali M.B., Werner J., Zhu B., Reinberg D., Lis J.T., Wei W., Ardehali M.B., Werner J., Zhu B., Reinberg D., Lis J.T., Ardehali M.B., Werner J., Zhu B., Reinberg D., Lis J.T., Werner J., Zhu B., Reinberg D., Lis J.T., Zhu B., Reinberg D., Lis J.T., Reinberg D., Lis J.T., Lis J.T. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol. Cell. Biol. 2006;26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins M.W., Tyler J.K., Tyler J.K. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell. 2006;21:405–416. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Aguilera A. Cotranscriptional mRNP assembly: From the DNA to the nuclear pore. Curr. Opin. Cell Biol. 2005;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Ahn S.H., Kim M., Buratowski S., Kim M., Buratowski S., Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Andrulis E.D., Guzman E., Doring P., Werner J., Lis J.T., Guzman E., Doring P., Werner J., Lis J.T., Doring P., Werner J., Lis J.T., Werner J., Lis J.T., Lis J.T. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes & Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E.D., Werner J., Nazarian A., Erdjument-Bromage H., Tempst P., Lis J.T., Werner J., Nazarian A., Erdjument-Bromage H., Tempst P., Lis J.T., Nazarian A., Erdjument-Bromage H., Tempst P., Lis J.T., Erdjument-Bromage H., Tempst P., Lis J.T., Tempst P., Lis J.T., Lis J.T. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Baskaran R., Chiang G.G., Mysliwiec T., Kruh G.D., Wang J.Y., Chiang G.G., Mysliwiec T., Kruh G.D., Wang J.Y., Mysliwiec T., Kruh G.D., Wang J.Y., Kruh G.D., Wang J.Y., Wang J.Y. Tyrosine phosphorylation of RNA polymerase II carboxyl-terminal domain by the Abl-related gene product. J. Biol. Chem. 1997;272:18905–18909. doi: 10.1074/jbc.272.30.18905. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D., Orphanides G., Studitsky V.M., Reinberg D., Studitsky V.M., Reinberg D., Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Bentley D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bird G., Zorio D.A., Bentley D.L., Zorio D.A., Bentley D.L., Bentley D.L. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 2004;24:8963–8969. doi: 10.1128/MCB.24.20.8963-8969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A., Winston F., Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- Bres V., Gomes N., Pickle L., Jones K.A., Gomes N., Pickle L., Jones K.A., Pickle L., Jones K.A., Jones K.A. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes & Dev. 2005;19:1211–1226. doi: 10.1101/gad.1291705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Kim T.K., Mancebo H., Lane W.S., Flores O., Reinberg D., Kim T.K., Mancebo H., Lane W.S., Flores O., Reinberg D., Mancebo H., Lane W.S., Flores O., Reinberg D., Lane W.S., Flores O., Reinberg D., Flores O., Reinberg D., Reinberg D. A protein phosphatase functions to recycle RNA polymerase II. Genes & Dev. 1999;13:1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSilva H., Lee K., Osley M.A., Lee K., Osley M.A., Osley M.A. Functional dissection of yeast Hir1p, a WD repeat-containing transcriptional corepressor. Genetics. 1998;148:657–667. doi: 10.1093/genetics/148.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano C., Ullman K.S., Ullman K.S. Nucleocytoplasmic transport: Integrating mRNA production and turnover with export through the nuclear pore. Mol. Cell. Biol. 2004;24:3069–3076. doi: 10.1128/MCB.24.8.3069-3076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M., Zhu W., Hasegawa J., Watanabe H., Kim D.K., Aida M., Inukai N., Narita T., Yamada T., Furuya A., Zhu W., Hasegawa J., Watanabe H., Kim D.K., Aida M., Inukai N., Narita T., Yamada T., Furuya A., Hasegawa J., Watanabe H., Kim D.K., Aida M., Inukai N., Narita T., Yamada T., Furuya A., Watanabe H., Kim D.K., Aida M., Inukai N., Narita T., Yamada T., Furuya A., Kim D.K., Aida M., Inukai N., Narita T., Yamada T., Furuya A., Aida M., Inukai N., Narita T., Yamada T., Furuya A., Inukai N., Narita T., Yamada T., Furuya A., Narita T., Yamada T., Furuya A., Yamada T., Furuya A., Furuya A., et al. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 2004;24:3324–3336. doi: 10.1128/MCB.24.8.3324-3336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbeck J.A., Kraemer S.M., Stargell L.A., Kraemer S.M., Stargell L.A., Stargell L.A. SPN1, a conserved gene identified by suppression of a postrecruitment-defective yeast TATA-binding protein mutant. Genetics. 2002;162:1605–1616. doi: 10.1093/genetics/162.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D., Izaurralde E., Izaurralde E. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 2002;159:579–588. doi: 10.1083/jcb.200207128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C.D., Morris J.R., Wu C., Winston F., Morris J.R., Wu C., Winston F., Wu C., Winston F., Winston F. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes & Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C.D., Laprade L., Winston F., Laprade L., Winston F., Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Kaplan C.D., Holland M.J., Winston F., Holland M.J., Winston F., Winston F. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10–GAL7 locus. J. Biol. Chem. 2005;280:913–922. doi: 10.1074/jbc.M411108200. [DOI] [PubMed] [Google Scholar]
- Kizer K.O., Phatnani H.P., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D., Phatnani H.P., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D., Hall H., Greenleaf A.L., Strahl B.D., Greenleaf A.L., Strahl B.D., Strahl B.D. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P., Silver P.A., Silver P.A. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes & Dev. 2002;16:2761–2766. doi: 10.1101/gad.1032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P., Krebber H., Silver P.A., Krebber H., Silver P.A., Silver P.A. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang X., Zhang X., Goodrich D.W., Wang X., Zhang X., Goodrich D.W., Zhang X., Goodrich D.W., Goodrich D.W. Human hHpr1/p84/Thoc1 regulates transcriptional elongation and physically links RNA polymerase II and RNA processing factors. Mol. Cell. Biol. 2005;25:4023–4033. doi: 10.1128/MCB.25.10.4023-4033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D.D., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J.B., Bentley D.L., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J.B., Bentley D.L., Minet M., Schroeder S., Cilli K., McNeil J.B., Bentley D.L., Schroeder S., Cilli K., McNeil J.B., Bentley D.L., Cilli K., McNeil J.B., Bentley D.L., McNeil J.B., Bentley D.L., Bentley D.L. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Lindstrom D.L., Squazzo S.L., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Squazzo S.L., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Yates J.R., III, Hartzog G.A., Hartzog G.A. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischka P., Toth Z., Thomas M., Mueller R., Stamminger T., Toth Z., Thomas M., Mueller R., Stamminger T., Thomas M., Mueller R., Stamminger T., Mueller R., Stamminger T., Stamminger T. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell. Biol. 2006;26:1631–1643. doi: 10.1128/MCB.26.5.1631-1643.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R., Jimeno S., Marin M., Huertas P., Garcia-Rubio M., Aguilera A., Jimeno S., Marin M., Huertas P., Garcia-Rubio M., Aguilera A., Marin M., Huertas P., Garcia-Rubio M., Aguilera A., Huertas P., Garcia-Rubio M., Aguilera A., Garcia-Rubio M., Aguilera A., Aguilera A. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell. 2005;18:711–722. doi: 10.1016/j.molcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Luo M.L., Zhou Z., Magni K., Christoforides C., Rappsilber J., Mann M., Reed R., Zhou Z., Magni K., Christoforides C., Rappsilber J., Mann M., Reed R., Magni K., Christoforides C., Rappsilber J., Mann M., Reed R., Christoforides C., Rappsilber J., Mann M., Reed R., Rappsilber J., Mann M., Reed R., Mann M., Reed R., Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- Maclennan A.J., Shaw G., Shaw G. A yeast SH2 domain. Trends Biochem. Sci. 1993;18:464–465. doi: 10.1016/0968-0004(93)90006-9. [DOI] [PubMed] [Google Scholar]
- Maldonado E., Drapkin R., Reinberg D., Drapkin R., Reinberg D., Reinberg D. Purification of human RNA polymerase II and general transcription factors. Methods Enzymol. 1996;274:72–100. doi: 10.1016/s0076-6879(96)74009-0. [DOI] [PubMed] [Google Scholar]
- Mandal S.S., Cho H., Kim S., Cabane K., Reinberg D., Cho H., Kim S., Cabane K., Reinberg D., Kim S., Cabane K., Reinberg D., Cabane K., Reinberg D., Reinberg D. FCP1, a phosphatase specific for the heptapeptide repeat of the largest subunit of RNA polymerase II, stimulates transcription elongation. Mol. Cell. Biol. 2002;22:7543–7552. doi: 10.1128/MCB.22.21.7543-7552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R., Das R., Cheng H., Hurt E., Dorman N., Reed R., Cheng H., Hurt E., Dorman N., Reed R., Hurt E., Dorman N., Reed R., Dorman N., Reed R., Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes & Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A., Cramer P., Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- Meinhart A., Kamenski T., Hoeppner S., Baumli S., Cramer P., Kamenski T., Hoeppner S., Baumli S., Cramer P., Hoeppner S., Baumli S., Cramer P., Baumli S., Cramer P., Cramer P. A structural perspective of CTD function. Genes & Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- Mueller C.L., Porter S.E., Hoffman M.G., Jaehning J.A., Porter S.E., Hoffman M.G., Jaehning J.A., Hoffman M.G., Jaehning J.A., Jaehning J.A. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- Ni Z., Schwartz B.E., Werner J., Suarez J.R., Lis J.T., Schwartz B.E., Werner J., Suarez J.R., Lis J.T., Werner J., Suarez J.R., Lis J.T., Suarez J.R., Lis J.T., Lis J.T. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- Nollau P., Mayer B.J., Mayer B.J. Profiling the global tyrosine phosphorylation state by Src homology 2 domain binding. Proc. Natl. Acad. Sci. 2001;98:13531–13536. doi: 10.1073/pnas.241215998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., LeRoy G., Chang C.H., Luse D.S., Reinberg D., LeRoy G., Chang C.H., Luse D.S., Reinberg D., Chang C.H., Luse D.S., Reinberg D., Luse D.S., Reinberg D., Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D., Trojer P., Mandal S., Shilatifard A., Reinberg D., Mandal S., Shilatifard A., Reinberg D., Shilatifard A., Reinberg D., Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pawson T. Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- Penheiter K.L., Washburn T.M., Porter S.E., Hoffman M.G., Jaehning J.A., Washburn T.M., Porter S.E., Hoffman M.G., Jaehning J.A., Porter S.E., Hoffman M.G., Jaehning J.A., Hoffman M.G., Jaehning J.A., Jaehning J.A. A posttranscriptional role for the yeast Paf1–RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Peterlin B.M., Price D.H., Price D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Phatnani H.P., Greenleaf A.L., Greenleaf A.L. Phosphorylation and functions of the RNA polymerase II CTD. Genes & Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Porter S.E., Penheiter K.L., Jaehning J.A., Penheiter K.L., Jaehning J.A., Jaehning J.A. Separation of the Saccharomyces cerevisae Paf1 complex from RNA polymerase II results in changes in its subnuclear localization. Eukaryot. Cell. 2005;4:209–220. doi: 10.1128/EC.4.1.209-220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Cheng H., Cheng H. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 2005;17:269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Herold A., Gari K., Kocher T., Rode M., Ciccarelli F.L., Wilm M., Izaurralde E., Herold A., Gari K., Kocher T., Rode M., Ciccarelli F.L., Wilm M., Izaurralde E., Gari K., Kocher T., Rode M., Ciccarelli F.L., Wilm M., Izaurralde E., Kocher T., Rode M., Ciccarelli F.L., Wilm M., Izaurralde E., Rode M., Ciccarelli F.L., Wilm M., Izaurralde E., Ciccarelli F.L., Wilm M., Izaurralde E., Wilm M., Izaurralde E., Izaurralde E. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2004;11:558–566. doi: 10.1038/nsmb759. [DOI] [PubMed] [Google Scholar]
- Reinberg D., Sims R.J., III, Sims R.J., III de FACTo nucleosome dynamics. J. Biol. Chem. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S., Fischer T., Luo M.J., Antunez O., Brettschneider S., Lechner J., Perez-Ortin J.E., Reed R., Hurt E., Fischer T., Luo M.J., Antunez O., Brettschneider S., Lechner J., Perez-Ortin J.E., Reed R., Hurt E., Luo M.J., Antunez O., Brettschneider S., Lechner J., Perez-Ortin J.E., Reed R., Hurt E., Antunez O., Brettschneider S., Lechner J., Perez-Ortin J.E., Reed R., Hurt E., Brettschneider S., Lechner J., Perez-Ortin J.E., Reed R., Hurt E., Lechner J., Perez-Ortin J.E., Reed R., Hurt E., Perez-Ortin J.E., Reed R., Hurt E., Reed R., Hurt E., Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- Rondon A.G., Jimeno S., Garcia-Rubio M., Aguilera A., Jimeno S., Garcia-Rubio M., Aguilera A., Garcia-Rubio M., Aguilera A., Aguilera A. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 2003;278:39037–39043. doi: 10.1074/jbc.M305718200. [DOI] [PubMed] [Google Scholar]
- Saguez C., Olesen J.R., Jensen T.H., Olesen J.R., Jensen T.H., Jensen T.H. Formation of export-competent mRNP: Escaping nuclear destruction. Curr. Opin. Cell Biol. 2005;17:287–293. doi: 10.1016/j.ceb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Saunders A., Werner J., Andrulis E.D., Nakayama T., Hirose S., Reinberg D., Lis J.T., Werner J., Andrulis E.D., Nakayama T., Hirose S., Reinberg D., Lis J.T., Andrulis E.D., Nakayama T., Hirose S., Reinberg D., Lis J.T., Nakayama T., Hirose S., Reinberg D., Lis J.T., Hirose S., Reinberg D., Lis J.T., Reinberg D., Lis J.T., Lis J.T. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- Sims R.J., III, Belotserkovskaya R., Reinberg D., Belotserkovskaya R., Reinberg D., Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes & Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Sommer P., Nehrbass U., Nehrbass U. Quality control of messenger ribonucleoprotein particles in the nucleus and at the pore. Curr. Opin. Cell Biol. 2005;17:294–301. doi: 10.1016/j.ceb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Armstrong J.A., Deuring R., Dahlsveen I.K., McNeill H., Tamkun J.W., Armstrong J.A., Deuring R., Dahlsveen I.K., McNeill H., Tamkun J.W., Deuring R., Dahlsveen I.K., McNeill H., Tamkun J.W., Dahlsveen I.K., McNeill H., Tamkun J.W., McNeill H., Tamkun J.W., Tamkun J.W. The Drosophila trithorax group protein Kismet facilitates an early step in transcription elongation by RNA polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- Strasser K., Hurt E., Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- Vasudevan S., Peltz S.W., Peltz S.W. Nuclear mRNA surveillance. Curr. Opin. Cell Biol. 2003;15:332–337. doi: 10.1016/s0955-0674(03)00051-6. [DOI] [PubMed] [Google Scholar]
- Vinciguerra P., Stutz F., Stutz F. mRNA export: An assembly line from genes to nuclear pores. Curr. Opin. Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Winkler M., Aus Dem Siepen T., Stamminger T., Aus Dem Siepen T., Stamminger T., Stamminger T. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 2000;74:8053–8064. doi: 10.1128/jvi.74.17.8053-8064.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A., Shukla A., Schneider J., Lees J.S., Stanton J.D., Dzuiba T., Swanson S.K., Florens L., Washburn M.P., Wyrick J., Shukla A., Schneider J., Lees J.S., Stanton J.D., Dzuiba T., Swanson S.K., Florens L., Washburn M.P., Wyrick J., Schneider J., Lees J.S., Stanton J.D., Dzuiba T., Swanson S.K., Florens L., Washburn M.P., Wyrick J., Lees J.S., Stanton J.D., Dzuiba T., Swanson S.K., Florens L., Washburn M.P., Wyrick J., Stanton J.D., Dzuiba T., Swanson S.K., Florens L., Washburn M.P., Wyrick J., Dzuiba T., Swanson S.K., Florens L., Washburn M.P., Wyrick J., Swanson S.K., Florens L., Washburn M.P., Wyrick J., Florens L., Washburn M.P., Wyrick J., Washburn M.P., Wyrick J., Wyrick J., et al. Ctk complex mediated regulation of histone methylation by COMPASS. Mol. Cell. Biol. 2006 doi: 10.1128/MCB.01627-06. doi: 10.1128/MCB.01627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T., Hall H., Kizer K.O., Shibata Y., Hall M.C., Borchers C.H., Strahl B.D., Hall H., Kizer K.O., Shibata Y., Hall M.C., Borchers C.H., Strahl B.D., Kizer K.O., Shibata Y., Hall M.C., Borchers C.H., Strahl B.D., Shibata Y., Hall M.C., Borchers C.H., Strahl B.D., Hall M.C., Borchers C.H., Strahl B.D., Borchers C.H., Strahl B.D., Strahl B.D. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes & Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T., Shibata Y., Rao B., Laribee R.N., O’Rourke R., Buck M.J., Greenblatt J.F., Krogan N.J., Lieb J.D., Strahl B.D., Shibata Y., Rao B., Laribee R.N., O’Rourke R., Buck M.J., Greenblatt J.F., Krogan N.J., Lieb J.D., Strahl B.D., Rao B., Laribee R.N., O’Rourke R., Buck M.J., Greenblatt J.F., Krogan N.J., Lieb J.D., Strahl B.D., Laribee R.N., O’Rourke R., Buck M.J., Greenblatt J.F., Krogan N.J., Lieb J.D., Strahl B.D., O’Rourke R., Buck M.J., Greenblatt J.F., Krogan N.J., Lieb J.D., Strahl B.D., Buck M.J., Greenblatt J.F., Krogan N.J., Lieb J.D., Strahl B.D., Greenblatt J.F., Krogan N.J., Lieb J.D., Strahl B.D., Krogan N.J., Lieb J.D., Strahl B.D., Lieb J.D., Strahl B.D., Strahl B.D. The RNA Pol II kinase Ctk1 regulates positioning of a 5xy: unknown character box histone methylation boundary along genes. Mol. Cell. Biol. 2006 doi: 10.1128/MCB.01628-06. doi: 10.1128/MCB.01628-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra P., Wyss J.C., Stutz F., Vinciguerra P., Wyss J.C., Stutz F., Wyss J.C., Stutz F., Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Mandal S.S., Pham A.D., Zheng Y., Erdjument-Bromage H., Batra S.K., Tempst P., Reinberg D., Mandal S.S., Pham A.D., Zheng Y., Erdjument-Bromage H., Batra S.K., Tempst P., Reinberg D., Pham A.D., Zheng Y., Erdjument-Bromage H., Batra S.K., Tempst P., Reinberg D., Zheng Y., Erdjument-Bromage H., Batra S.K., Tempst P., Reinberg D., Erdjument-Bromage H., Batra S.K., Tempst P., Reinberg D., Batra S.K., Tempst P., Reinberg D., Tempst P., Reinberg D., Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes & Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]