Abstract
Factor VIIa (EC 3.4.21.21) is a trypsin-like serine protease that plays a key role in the blood coagulation cascade. On injury, factor VIIa forms a complex with its allosteric regulator, tissue factor, and initiates blood clotting. Although the structure of the binary complex has already been determined [Banner, D. W., D’Arcy, A., Chène, C., Winkler, F. K., Guha, A., Konigsberg, W. H., Nemerson, Y. & Kirchhofer, D. (1996) Nature (London) 380, 41–46], the conformational effects of cofactor binding to factor VIIa are not known in detail because of a lack of structural information on free factor VIIa. Here we report the structure of γ-carboxyglutamic acid-domainless human coagulation factor VIIa at a resolution of 2.8 Å. The molecule adopts an extended conformation within the crystal similar to that previously observed for the full-length protein in complex with tissue factor. Detailed comparison of free and tissue factor-bound factor VIIa reveals several structural differences. The binding mode of the active-site inhibitor d-Phe-Phe-Arg methyl ketone differs in the two structures, suggesting a role for the cofactor in substrate recognition. More importantly, a surface-exposed α-helix in the protease domain (residues 307–312), which is located at the cofactor recognition site, is distorted in the free form of factor VIIa. This subtle structural difference sheds light on the mechanism of the dramatic tissue factor-induced enhancement of factor VIIa activity.
Coagulation factor VIIa (fVIIa, EC 3.4.21.21) and other enzymes of the clotting cascade (1), such as factor Xa and thrombin, belong to the family of trypsin-like serine proteases. fVIIa possesses a modular organization with an N-terminal, membrane-binding γ-carboxyglutamic acid (Gla)-containing domain, two epidermal growth factor (EGF)-like domains, and a C-terminal serine protease domain (2). The active form of the enzyme, comprising a light chain (152 residues) and a heavy chain (254 residues) linked by a disulfide bond, is generated by the cleavage of the Arg-152–Ile-153 peptide bond ({15–16}; hereafter, trypsin numbering is given in curly brackets) located between the second EGF-like domain (EGF2) and the protease domain. This internal peptide bond cleavage unleashes the activity of the trypsin-like enzymes by facilitating the maturation of their catalytic machinery (3). fVIIa is unusual, however, in that it remains in a zymogen-like state after cleavage and only becomes an efficient catalyst when associated with its protein cofactor, tissue factor (TF). Thus, a quiescent state of the clotting cascade is maintained as long as TF is physically separated from its ligand, fVIIa. The two proteins form a complex at the site of injury, where extravascular TF becomes exposed to the blood, and fVIIa triggers blood coagulation by autoactivating fVII and activating factors IX and X (1). In addition to acting as an allosteric regulator, TF colocalizes fVIIa and its substrates and places the active site of fVIIa at the proper distance above the surface of the membrane for alignment with the scissile bond (4).
The crystal structure of fVIIa in complex with the extracellular part of TF (sTF) reveals an extensive interface between the two proteins encompassing all four domains in fVIIa, with the light chain contributing the majority of the binding energy (5), and both extracellular domains of TF (6, 7). The intradomain structural changes that accompany the allosteric enhancement of fVIIa’s activity appear to be confined to the protease domain (8), but the precise molecular switch by which TF turns on fVIIa remains elusive. Experimental data demonstrate conformational linkages between the TF binding site, the active site cleft, and the macromolecular substrate exosite in the protease domain of fVIIa (9). An understanding of the mechanism of TF-induced enhancement of fVIIa’s activity is of interest not only in terms of blood coagulation but also given fVIIa’s wider role in signal transduction (10) and metastasis (11). In an attempt to unravel the detailed structural changes that underlie this potentiation, we have determined the structure of Gla-domainless fVIIa (GD-fVIIa) in isolation. Here, we compare the structure of GD-fVIIa with that of fVIIa from a previously published complex with TF (hereafter referred to as fVIIasTF) (6).
MATERIALS AND METHODS
Protein Preparation, Crystallization, and Data Acquisition.
Recombinant human fVIIa was produced, inhibited with d-Phe-Phe-Arg chloromethyl ketone (FFR), and deprived of the Gla domain (residues 1–44) by digestion with cathepsin G as described (12). The resulting GD-fVIIa was purified by chromatography of the digest on a column of Q-Sepharose Fast Flow (Amersham Pharmacia) equilibrated with 20 mM Tris/2 mM EDTA (pH 9.0) and eluted with a linear NaCl gradient from 0 to 0.4 M. Crystals of GD-fVIIa were grown by using the sitting-drop vapor-diffusion method. Drops composed of a 2:1 ratio of protein (12–20 mg/ml in 50 mM NaCl/10 mM Tris, pH 8) to reservoir solution were equilibrated against reservoir solution (50 mM NaCl/10 mM CaCl2/3.5–3.7 M sodium formate in 100 mM Tris, pH 8.5). Large tetragonal crystals, belonging to space group P43 with unit cell dimensions a = b = 117.96 Å, c = 100.44 Å, grew within 1–2 weeks. To optimally cryoprotect the crystals, the mother liquor was replaced by a solution containing 50 mM NaCl/10 mM CaCl2/5% (wt/vol) xylitol/3.3 M sodium acetate in 100 mM Tris (pH 8.5), and the concentration of xylitol was increased to 35% in 5% steps over 3 hr. Crystals treated in this way belong to space group P4322 (a = b = 115.3 Å, c = 98 Å) and have a single molecule per crystallographic asymmetric unit. Data were collected to a resolution of 2.8 Å at 120 K on a MAR image plate detector mounted on beamline BW7B (λ=0.8342 Å) at the European Molecular Biology Laboratory Hamburg Outstation (DESY/Hamburg, Germany). The data were indexed, reduced, and scaled by using denzo and scalepack (13). Data processing statistics are given in Table 1.
Table 1.
Data collection, refinement, and model validation statistics
| Data Collection | |
| Resolution, Å | 35–2.8 |
| Observations | 238,127 |
| Unique reflections | 16,960 |
| Completeness, % | 95.4 (79.4)* |
| Rmerge†, % | 5.6 (25.3)* |
| Wilson B, Å2 | 70 |
| Refinement | |
| Resolution range, Å | 35–2.8 |
| Reflections used (free reflections) | 15,270 (815) |
| Rcryst(Rfree)‡, % | 21.5 (26.7) |
| Estimated coordinate error§, Å | 0.34 |
| rms deviation from ideal geometry | |
| Bond length, Å | 0.014 |
| Bond angle distance, Å | 0.045 |
| Bonded ΔB, Å2¶ | 2.3 |
| Average B factor, Å2 | 56.7 |
| A, B, L (a, b, l, p), %‖ | 82.8 (17.2) |
Value in parentheses are for highest resolution shell 2.85–2.80 Å.
Rmerge = Σ|/(h) − 〈/(h)〉|/Σ/(h).
Rcryst = 100 × Σ|Fobs − Fcalc|/Σ|Fobs|; Rfree is as Rcryst but calculated over 5% of the data that were excluded from the refinement process.
Based on Rfree from refmac (15).
rms deviation between B factors for all bonded atoms.
Percentage of residues located in most favored (additional) regions of the Ramachandran plot as determined by procheck (17).
Structure Determination.
The position of the protease domain was determined by molecular replacement (14) (amore) using the protease domain from the fVIIa–TF complex (6) as a search model (PDB code: 1dan). The positions of the two remaining EGF-like domains were revealed by the resultant sigmaa-weighted 2|Fo| − |Fc| electron density map calculated after 5 cycles of refinement of the correctly positioned protease domain with refmac (15). The coordinates corresponding to each EGF-like domain (PDB code: 1dan) were manually fitted to the electron density and subjected to rigid-body refinement. This initial model was then refined with refmac by using all data (5% flagged for Rfree) and incorporating a bulk solvent correction as calculated by x-plor (16). During subsequent rounds of refinement and rebuilding, the linker region between the two EGF-like domains, the protease inhibitor, carbohydrate moieties, and solvent molecules were added to the model. Solvent molecules that refined to a B factor >70 Å2 were removed. The final model, comprising 2,928 atoms, has an Rcryst and Rfree of 21.5 and 26.7, respectively, for all data between 35 and 2.8 Å. Refinement and model validation statistics are given in Table 1. All model building was carried out by using quanta (Molecular Simulations, Waltham, MA).
The final model comprises most of the light chain, the entire heavy chain (Ile-153–Pro-406 {16–257}), 5 carbohydrate moieties, the d-Phe-Phe-Arg methylene covalent inhibitor, 1 putative calcium ion, and 84 water molecules. The first four (Ser-45–Asp-48) and last eight (Asn-145–Arg-152) residues of the light chain and residues Arg-315–Lys-316 {170C–170D} of the heavy chain are disordered and have not been included in the final model. The side chains of several surface residues were also not visible in the final map and have been truncated to their Cβ atoms (light chain: Lys-62, Glu-77, Lys-85; heavy chain: Lys-199 {60C}, Arg-202 {62}). Residues Ser-103–Ser-111 of the light chain are disordered in the structure and have been modeled in two alternate conformations.
RESULTS AND DISCUSSION
Light Chain of GD-fVIIa.
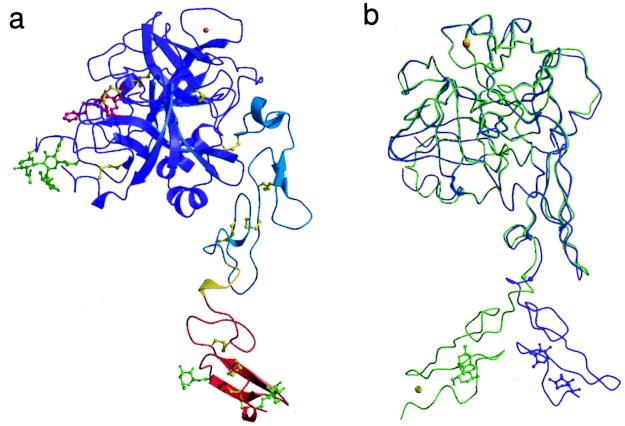
The light chain forms a narrow stalk ≈65 Å in length on which the spherical protease domain sits (Fig. 1a). The EGF2 domain and the protease domain associate into the compact structural unit that is common to factors VIIa (6), IXa (18), and Xa (19–21) and to activated protein C (22). The first EGF-like domain (EGF1) extends away from this compact core at an angle of 120° to yield a molecule with a linear length of ≈90 Å.
Figure 1.
(a) Ribbon representation of Gla-domainless fVIIa. The EGF1, EGF2, and protease domains are colored red, cyan, and purple, respectively. The linker region between the two EGF-like domains is colored yellow. The disulfide bridges (yellow), carbohydrate groups (light green), and FFR inhibitor (magenta) are depicted in ball-and-stick representation. The putative calcium ion (top right) is colored orange. (b) Superposition of free and sTF-complexed fVIIa. GD-fVIIa (purple) and fVIIasTF (light green; PDB code 1dan, ref. 6) are shown after superposition of their EGF2 and protease domains. The carbohydrate groups attached to EGF1 are shown for reference. The Cα position of Gln-88, which acts as the hinge residue in the linker region, is indicated by a small sphere.
The EGF-like modules are stacked approximately “end-on-end” to yield an elongated domain arrangement. There is minimal contact between the two domains with a single stabilizing hydrogen bond between the Nδ2 of Asn-93 and the main-chain carbonyl of Lys-85. The EGF1 domain is, however, rotated ≈180° about the linker hexapeptide (residues 85–90) compared with its position in the fVIIasTF structure (Fig. 1b). The positioning of the EGF1 domain in the GD-fVIIa structure is also different from that adopted by the corresponding region in factor IXa (18) and protein C (22). This different relative orientation reflects both the inherent flexibility of the linker region and the environment of this domain in the two structures and supports the proposal that fVIIa’s association with TF reduces the internal flexibility of the light chain through the stabilization of a particular orientation of the EGF1 and Gla domains (6). Interestingly, the different orientations of EGF1 in the two fVIIa structures arise from a single change in the main-chain torsion angles of Gln-88.
In the present structure, the precise orientation of EGF1 appears to be largely dictated by crystal-packing forces and therefore probably represents only one of a number of possible positions that are accessible to this domain in solution. It is noteworthy that the same face of the EGF1 domain that interacts with TF also participates in the intermolecular crystal contacts seen here. Despite the difference in relative orientation of the two EGF-like domains, they exhibit essentially identical conformations in the free and sTF-complexed structures having Cα rms differences of 0.49 Å (EGF1; residues 49–84) and 0.54 Å (EGF2; residues 91–142) after superposition. However, in contrast to fVIIasTF, the calcium-binding site located at the N-terminal end of EGF1 is only partially formed and unoccupied in GD-fVIIa despite the inclusion of 10 mM calcium in the crystallization medium. Removal of the Gla domain is known to decrease the calcium affinity of the corresponding site in bovine factor X (23), and poor occupation of this site is also observed in the crystal structure of human Gla-domainless factor Xa (21). Although this truncation does not remove any of the Ca2+-coordinating residues per se, it appears to disrupt their positioning and leads to the destabilization of the binding site. In fact, Asp-46 and Gly-47, two of the equatorial calcium-coordinating groups, are poorly ordered and not visible in the electron density. Residues Asp-63–Ser-67 also adopt a different conformation in the present structure compared with the corresponding region in fVIIasTF. This is presumably also because of the disruption of the calcium-binding site as inferred from the solution structure of the apo form of the EGF1 domain from fVII (24).
Protease Domain (Heavy Chain) of GD-fVIIa.
The differences in the protease domain structure between the free and complexed forms of fVIIa are relatively minor and primarily restricted to the loops flanking the active site cleft. The Cα rms difference between the two structures is 0.66 Å (252 equivalent atoms). Because of the presence of the active site inhibitor, the protease domain exhibits an “active” conformation with the N terminus correctly inserted. This insertion is a prerequisite for protease activation because it facilitates the formation of both the S1 recognition pocket and the oxyanion hole (25). The conformational changes induced by the inhibitor appear to be sufficient to stabilize the critical salt bridge between the α-amino group of Ile-153 {16} and the side chain of Asp-343 {194}. This observation is consistent with the protection of the α-NH2 group against carbamylation induced by either TF binding (26) or chloromethyl ketone inhibitor incorporation (27). A similar induction of a mature catalytic machinery is also observed on the binding of pancreatic trypsin inhibitor to trypsinogen (28). There is clear density at the high-affinity calcium-binding site in GD-fVIIa, but the temperature factor for the modeled calcium ion is very high (73 Å2), suggesting that the site may only be partially occupied.
The most striking structural difference is observed for residues Leu-305–Glu-325 {163–178}. This region in fVIIasTF is composed of a short α-helix (307–312 {165–170}) followed by a loop that defines the outermost face of the active site cleft’s aryl-binding subsite. The N-terminal portion of the helix forms part of the interface between the protease domain and TF and contains a number of residues that are important for proteolytic function and optimal binding to TF (29). For these reasons, this region has been proposed to act as the allosteric control site in the TF-mediated activation of fVIIa (6, 9). Fig. 2 shows a representative view of the electron density for this part of the molecule, which is reasonably well defined apart from the region encompassing residues 315–316. The average temperature factor for the Leu-305–Glu-325 region is 55 Å2, even though residues 317–319 have high individual B values. A similar degree of thermal mobility is also observed for this surface-exposed loop region (315–319) in the fVIIasTF structure (6). In the GD-fVIIa structure, this “transmitter helix” (307–312) is distorted (to 310) and shortened by three residues. The whole region is also shifted by between 0.5 Å and 3 Å toward the active-site cleft relative to its position in the fVIIasTF structure. This movement has a knock-on effect on the positioning of two neighboring loops (Tyr-332–Ser-339 {184–190} and Thr-370–Phe-374 {221–225}), which are shifted away from the active site in the GD-fVIIa structure. It is noteworthy that, in trypsin, these loops form part of the activation domain that is stabilized on conversion of the zymogen to the enzymatically active form (25). However, interpretation of this structural transition is complicated by crystal-packing contacts that may be responsible for the difference between GD-fVIIa and fVIIasTF. Nonetheless, the flexible nature of this portion of fVIIa is clearly highlighted.
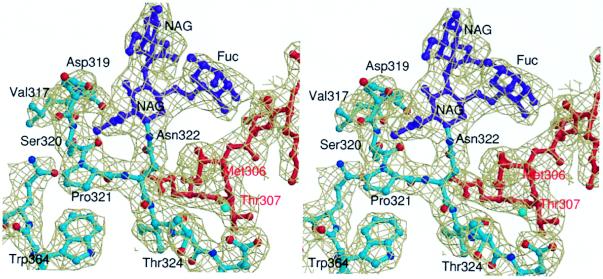
Figure 2.
Stereo view of the 2Fo − Fc electron density around the N-linked carbohydrate attachment site. All atoms are colored according to their type except for the carbohydrate moieties (purple) and the 304–308 {162–166} region of a neighboring molecule in the crystal (red). The map is contoured at 1σ and overlaid on the final refined model.
The conformation of the Leu-305–Glu-325 region of GD-fVIIa is also affected by the N-linked oligosaccharide chain that is attached to Asn-322 {175} in the disulfide-bridged loop (Cys-310–Cys-329 {168–182}) that follows the transmitter helix. Two N-acetylglucosamines and a fucose are well defined in the electron density maps, but the remainder of the oligosaccharide chain is poorly ordered and could not be reliably modeled (Fig. 2). The visible carbohydrate groups are directed away from the body of the protein and do not restrict access to the active-site cleft. This positioning is consistent with the nonessential role of the N-linked carbohydrate in proteolytic function and TF-binding (30) but does not preclude its involvement in substrate recognition. The carbohydrate attachment participates in essential intermolecular contacts. The fucose is wedged in a shallow depression on the surface of an adjacent molecule formed by Phe-275 {129F}, Arg-277 {134}, and Met-306 {164} (Fig. 2). To fulfil these packing contacts, the residues flanking the carbohydrate attachment site (Ser-320–Ile-323 {170H–176}) are shifted 1.6 Å toward the body of the protein resulting in a narrowing of the aryl binding subsite of the active site.
Inhibitor-Binding Mode.
The inhibitor chosen for the present study, FFR, is the same as that used in the fVIIa–sTF complex. It is covalently attached to both the active-site serine (Ser-344 {195}) and histidine (His-193 {57}) and occupies the S1-, S2-, and aryl-binding subsites. The binding mode of the inhibitor, which is well defined in the electron density, is surprisingly different in the structures of GD-fVIIa and fVIIasTF with an all-atom rms difference of 1.5 Å. In both structures, the attachment to the protein and the binding of the arginine side chain in the S1 recognition pocket are identical. However, the orientation of the phenylalanine side chain in S2 and the positioning of the N-terminal d-Phe in the aryl-binding site differ significantly (Fig. 3).
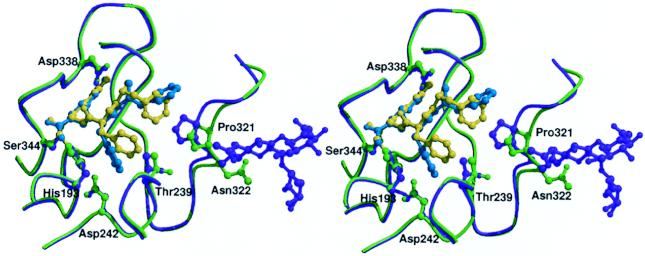
Figure 3.
Comparison of the inhibitor-binding mode in Gla-domainless (purple) and sTF-complexed fVIIa (light green). The FFR inhibitor in the GD-fVIIa and fVIIasTF structures is colored cyan and yellow, respectively. The carbohydrate groups attached to Asn-322 {175} in the GD-fVIIa structure are also shown.
The N-terminal d-Phe is positioned so that the face, rather than the edge of the side chain, packs against Pro-321 {170I}. This change is accomplished by a 1 Å shift in the Cα position of the d-Phe coupled with a χ1 rotation of 120° and adjustments in the main-chain torsion angles of the two phenylalanines. Despite the shift in Cα position, the hydrogen bonds between the main-chain groups of the d-Phe and Gly-365 {216} are maintained. This alteration in the positioning of the d-Phe appears to be a direct result of the narrowing of the cleft induced by the shift in the 320’s loop described above. In the S2 subsite, the benzyl side chain of the l-Phe is thrust deeper into the pocket and sandwiched between the imidazole ring of the active site histidine (His-193 {57}) and Thr-239 {99}. The driving force behind this side-chain rotation is less clear but probably relates to the removal of the potential T stack interaction with the benzyl side chain of the d-Phe present in the fVIIasTF structure. Although crystal contacts clearly affect the way in which the side-chain moieties of the FFR inhibitor are positioned within the active site cleft of GD-fVIIa, its overall binding mode is very similar to that observed for the analogous tripeptidyl d-Phe-Pro-Arg inhibitor complexed with thrombin (31), factor IXa (18), and activated protein C (22). The unusual orientation of the d-Phe in the aryl subsite of the fVIIasTF structure (6) is therefore rather intriguing and may reflect a role for TF in substrate binding.
Implications for the Triggering of Blood Coagulation.
One absolute requirement for optimal macromolecular substrate conversion by the vitamin K-dependent coagulation factors is a mature active site cleft. fVIIa is unique among these factors in that its zymogen-to-enzyme transition is labile, i.e., its active conformation and consequent amidolytic activity are cofactor-dependent. The observation that the 307–312 {165–170} helix is distorted in free GD-fVIIa sheds light on the mode by which TF enhances fVIIa’s activity and hence, triggers blood clotting. However, it should be kept in mind that the elucidation of the complete structural impact of TF on fVIIa is masked by the presence of the FFR inhibitor in GD-fVIIa.
The transmitter helix (residues 307–312) may be destabilized in GD-fVIIa because of the lack of a traditional N-capping residue at position 306 {164}. Serine proteases that express constitutive, cofactor-independent activity, e.g., factor Xa (19–21), thrombin (31), and trypsin (25), have an Asp or Glu residue at this position and exhibit a helical conformation. Asp is the most commonly found residue in the capping position immediately prior to a helix (32). In contrast, this position in fVIIa is occupied by a conserved Met residue and, instead, fVIIa appears to rely on TF to provide the N-capping function for this α-helix (Fig. 4). In the complex between fVIIa and TF (6), Met-306 {164} is sandwiched in a hydrophobic groove on the surface of TF with the side chains of Glu-91 and Tyr-94 capping the 307–312 helix (Fig. 4b). The importance of the lock-and-key interaction of TF with Met-306 {164} for the allosteric activation of fVIIa can be inferred from mutagenesis studies. The functional deficit of the fVIIa–TF complex introduced by mutation of Tyr-94 in TF is more dramatic than anticipated (33), considering the small energetic contribution of this residue to fVIIa binding (34). In addition, Met-306 and adjacent residues in fVIIa were found to influence the proteolytic activity of the complex but barely decrease the affinity of fVIIa for TF (29).
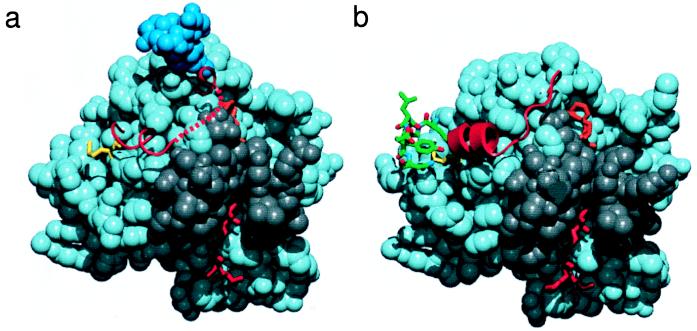
Figure 4.
The protease domain of Gla-domainless fVIIa (a) and fVIIasTF (b, PDB code 1dan). The majority of the domain is shown in space-filling representation. The d-Phe-Phe-Arg inhibitor (orange), the N-terminal region Ile-153–Gly-156 {16–19} (red), and Met-306 {164} (yellow) are drawn in stick representation. The N-linked carbohydrate moiety observed in the GD-fVIIa structure is colored blue, and the serine protease activation domain (Gly-285–Ala-294 {142–152}, Gly-331–Gly-342 {184–193}, and Gly-365–Gly-372 {216–223}; ref. 25) is colored gray. The region encompassing Thr-307–Pro-321 {165–170I} is depicted as a red string and schematically shows the local secondary structure. A region of TF (Glu-91–Asn-96) that interacts with the protease domain around Met-306 {164} is shown in green (b). The red dotted line schematically represents the unmodeled region (Arg-315–Lys-316) of the GD-fVIIa structure. This figure was generated by using quanta (Molecular Simulations, Waltham, MA).
Stabilization of the 307–312 {165–170} helix appears to be a prerequisite for the proper formation of the activation domain. The helix lies in a region adjacent to the activation domain of fVIIa and is attached to the body of the protein via the Cys-310–Cys-329 {168–182} disulfide bridge. The TF-induced helix stabilization appears to promote the formation of the hydrogen bond between the amide of Arg-315 {170C} and the carbonyl of Gly-372 {223} (6). This interaction links the helix to the activation domain and positions the side chains of Arg-315 {170C} and Lys-316 {170D} properly for C capping of the helix. Because of structural disorder, residues Arg-315 and Lys-316 are not resolved in GD-fVIIa. When bound to TF, the activation region of fVIIa contains a well defined hydrogen-bonded network of water molecules that is stabilized by both the 307–312 {165–170} helix and the amino group of the N-terminal Ile-153 {16} (6). In addition, active site occupancy adds to the stabilization of the activation domain. Consequently, the structure of the activation region is less defined in GD-fVIIa. The flexibility of this region in free fVIIa is supported by the susceptibility of the Cys-310–Cys-329 and Cys-340–Cys-368 disulfide bonds to reduction (35).
It therefore appears that fVIIa has evolved to contain a Met residue in position 306, whereas constitutively active serine proteases have a residue suitable for N-terminal capping of an α-helix in the corresponding position. The resulting unstable 307–312 {165–170} helix, and the consequently disordered activation domain, prevents the expression of fVIIa activity in the absence of TF and ensures an optimal temporal and spatial procoagulant response to injury.
Acknowledgments
We thank Helen Burrell for help in crystallization and Anette Østergaard, Annette Danielsen, and Lone Odborg for protein preparation. This work was funded by Novo Nordisk A/S. We also thank the European Union for support of the work at European Molecular Biology Laboratory DESY/Hamburg through the TMR/LSF program to the European Molecular Biology Laboratory Hamburg Outstation. The infrastructure of the York Structural Biology Laboratory is supported by the Biotechnology and Biological Sciences Research Council.
ABBREVIATIONS
- fVIIa
coagulation factor VIIa
- GD-fVIIa
Gla-domainless factor VIIa
- TF
tissue factor
- sTF
the extracellular domains of tissue factor
- fVIIasTF
tissue factor-bound factor VIIa
- Gla
γ-carboxyglutamic acid
- EGF
epidermal growth factor
- FFR
d-Phe-Phe-Arg chloromethyl ketone
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID 1QFK).
References
- 1.Davie E W, Fujikawa K, Kisiel W. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 2.Furie B, Furie B C. Cell. 1988;53:505–518. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 3.Khan A R, James M N G. Protein Sci. 1998;7:815–836. doi: 10.1002/pro.5560070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCallum C D, Hapak R C, Neuenschwander P F, Morrissey J H, Johnson A E. J Biol Chem. 1996;271:28168–28175. doi: 10.1074/jbc.271.45.28168. [DOI] [PubMed] [Google Scholar]
- 5.Persson E. FEBS Lett. 1997;413:359–363. doi: 10.1016/s0014-5793(97)00941-1. [DOI] [PubMed] [Google Scholar]
- 6.Banner D W, D’Arcy A, Chène C, Winkler F K, Guha A, Konigsberg W H, Nemerson Y, Kirchhofer D. Nature (London) 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 7.Zhang E, Charles R S, Tulinsky A. J Mol Biol. 1999;285:2089–2104. doi: 10.1006/jmbi.1998.2452. [DOI] [PubMed] [Google Scholar]
- 8.Freskgård P-O, Olsen O H, Persson E. Protein Sci. 1996;5:1531–1540. doi: 10.1002/pro.5560050809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruf W, Dickinson C D. Trends Cardiovasc Med. 1998;8:350–356. doi: 10.1016/s1050-1738(98)00031-0. [DOI] [PubMed] [Google Scholar]
- 10.Poulsen L K, Jacobsen N, Sørensen B B, Bergenhem N C H, Kelly J D, Foster D C, Thastrup O, Ezban M, Petersen L C. J Biol Chem. 1998;273:6228–6232. doi: 10.1074/jbc.273.11.6228. [DOI] [PubMed] [Google Scholar]
- 11.Mueller B M, Ruf W. J Clin Invest. 1998;101:1372–1378. doi: 10.1172/JCI930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sørensen B B, Persson E, Freskgård P-O, Kjalke M, Ezban M, Williams T, Rao L V M. J Biol Chem. 1997;272:11863–11868. doi: 10.1074/jbc.272.18.11863. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 14.Collaborative Computational Project No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 15.Murshudov G N, Vagin A A, Dodson E J. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 16.Brünger A. x-plor, A System for x-ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1993. , Version 3.1. [Google Scholar]
- 17.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 18.Brandstetter H, Bauer M, Huber R, Lollar P, Bode W. Proc Natl Acad Sci USA. 1995;92:9796–9800. doi: 10.1073/pnas.92.21.9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padmanabhan K, Padmanabhan K P, Tulinsky A, Park C H, Bode W, Huber R, Blankenship D T, Cardin A D, Kisiel W. J Mol Biol. 1993;232:947–966. doi: 10.1006/jmbi.1993.1441. [DOI] [PubMed] [Google Scholar]
- 20.Brandstetter H, Kühne A, Bode W, Huber R, von der Saal W, Wirthensohn K, Engh R A. J Biol Chem. 1996;271:29988–29992. doi: 10.1074/jbc.271.47.29988. [DOI] [PubMed] [Google Scholar]
- 21.Kamata K, Kawamoto H, Honma T, Iwama T, Kim S-H. Proc Natl Acad Sci USA. 1998;95:6630–6635. doi: 10.1073/pnas.95.12.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mather T, Oganessyan V, Hof P, Hubert R, Foundling S, Esmon C, Bode W. EMBO J. 1996;15:6822–6831. [PMC free article] [PubMed] [Google Scholar]
- 23.Persson E, Hogg P J, Stenflo J. J Biol Chem. 1993;268:22531–22539. [PubMed] [Google Scholar]
- 24.Muranyi A, Finn B E, Gippert G P, Forsén S, Stenflo J, Drakenberg T. Biochemistry. 1998;37:10605–10615. doi: 10.1021/bi980522f. [DOI] [PubMed] [Google Scholar]
- 25.Huber R, Bode W. Acc Chem Res. 1978;11:114–122. [Google Scholar]
- 26.Higashi S, Nishimura H, Aita K, Iwanaga S. J Biol Chem. 1994;269:18891–18898. [PubMed] [Google Scholar]
- 27.Higashi S, Matsumoto N, Iwanaga S. J Biol Chem. 1996;271:26569–26574. doi: 10.1074/jbc.271.43.26569. [DOI] [PubMed] [Google Scholar]
- 28.Bode W, Schwager P, Huber R. J Mol Biol. 1978;118:99–112. doi: 10.1016/0022-2836(78)90246-2. [DOI] [PubMed] [Google Scholar]
- 29.Dickinson C D, Kelly C R, Ruf W. Proc Natl Acad Sci USA. 1996;93:14379–14384. doi: 10.1073/pnas.93.25.14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickinson C D, Ruf W. J Biol Chem. 1997;272:19875–19879. doi: 10.1074/jbc.272.32.19875. [DOI] [PubMed] [Google Scholar]
- 31.Bode W, Turk D, Karshikov A. Protein Sci. 1992;1:426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aurora R, Rose G D. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley R F, Costas K E, O’Connell M P, Lazarus R A. Biochemistry. 1995;34:10383–10392. doi: 10.1021/bi00033a009. [DOI] [PubMed] [Google Scholar]
- 34.Ruf W, Kelly C R, Schullek J R, Martin D M A, Polikarpov I, Boys C W G, Tuddenham E G D, Edgington T S. Biochemistry. 1995;34:6310–6315. doi: 10.1021/bi00019a008. [DOI] [PubMed] [Google Scholar]
- 35.Higashi S, Matsumoto N, Iwanaga S. J Biol Chem. 1997;272:25724–25730. doi: 10.1074/jbc.272.41.25724. [DOI] [PubMed] [Google Scholar]