Abstract
It is generally accepted that the growth rate of an organism is modulated by the availability of nutrients. One common mechanism to control cellular growth is through the global down-regulation of cap-dependent translation by eIF4E-binding proteins (4E-BPs). Here, we report evidence for a novel mechanism that allows eukaryotes to coordinate and selectively couple transcription and translation of target genes in response to a nutrient and growth signaling cascade. The Drosophila insulin-like receptor (dINR) pathway incorporates 4E-BP resistant cellular internal ribosome entry site (IRES) containing mRNAs, to functionally couple transcriptional activation with differential translational control in a cell that is otherwise translationally repressed by 4E-BP. Although examples of cellular IRESs have been previously reported, their critical role mediating a key physiological response has not been well documented. Our studies reveal an integrated transcriptional and translational response mechanism specifically dependent on a cellular IRES that coordinates an essential physiological signal responsible for monitoring nutrient and cell growth conditions.
Keywords: 4E-BP, Foxo, IRES, insulin receptor
Coupled transcription and protein synthesis is a hallmark of prokaryotic gene expression. The advantages of such a linked system are well recognized as it provides smooth coordination to ensure that cells respond appropriately to signals such as nutrient availability. A rapid response to such environmental signals also allows for multiple points of regulation and a fine-tuning mechanism for controlling gene expression. In eukaryotic organisms, the compartmentalization of the cell nucleus makes the direct coupling of transcription and translation problematic. Nevertheless, like prokaryotes, the metazoan cell must respond to many external as well as internal signals, and a coupled response would be highly advantageous. However, there is currently little evidence for such a direct linkage, either physical or functional, in metazoans. In our attempts to dissect the transcriptional regulatory circuitry of the insulin-like signaling cascade in Drosophila, we have discovered a potentially new mechanism that functionally links transcription and translation.
Metazoan organisms must strictly control both body and organ size during development (Conlon and Raff 1999). Thus, cell size and cell number are tightly controlled to determine the final size of an animal. One of the cues used in determining growth regulation is nutrient availability (Hafen 2004; Puig and Tjian 2006). The insulin receptor (INR) and insulin-like growth factor (IGF) receptor pathways have evolved as key sensors of nutrient availability and play an important role in both cell-autonomous and nonautonomous decisions controlling cellular proliferation, cell size determination, and the response to nutrient availability. In Drosophila, this pathway is critical for determining body and organ size as well as metabolic homeostasis and life span. Perhaps most notably, misregulation of this pathway in humans can lead to type 2 diabetes and all of its associated pathologies, which is becoming a rapidly escalating worldwide epidemic (Saltiel and Kahn 2001).
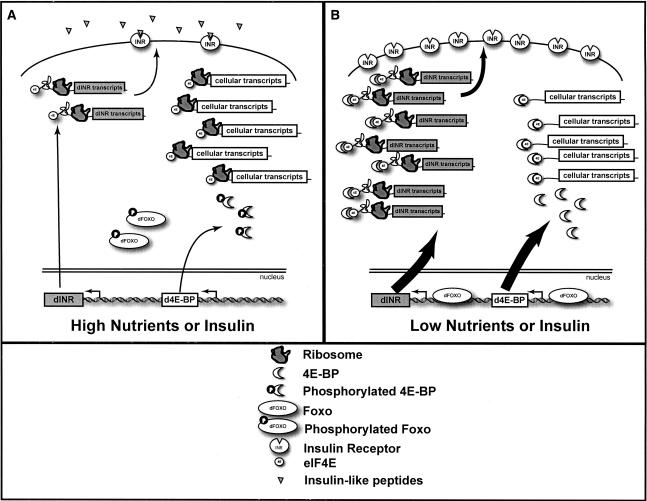
The INR/IGF pathway is highly conserved, with homologs of the key molecular players present in metazoan organisms from flies to humans (Garofalo 2002). The downstream targets of this signaling cascade are thought to separately modulate both transcription and translation to potentiate signals for either growth or stasis. In the presence of insulin or insulin-like peptides, the signaling cascade activates the oncogenic protein kinase Akt. To control RNA synthesis, Akt phosphorylates the Forkhead-box-binding protein (dFOXO) family of transcription factors, sequestering them in the cytoplasm and thus effectively inactivating them. This in turn prevents activated transcription of the dFOXO target genes. In addition, Akt stimulates the modification of the target of rapamycin (TOR) protein, which in turn phosphorylates and inactivates the translation initiation inhibitor eIF4E-binding protein (d4E-BP). In its unphosphorylated and active state, d4E-BP binds to the 7-methyl-guanosine (m7G) cap-binding protein eIF4E. This prevents formation of the translation initiation complex eIF4F, thereby inhibiting cap-dependent translation (Ruggero and Sonenberg 2005). This combination of inactivated dFOXO and inactive d4E-BP efficiently drives the cell toward growth and proliferation (Fig. 1, high nutrients). Conversely, active dFOXO and d4E-BP conspire to arrest cell growth until the cell receives favorable nutrient and physiological signals to continue proliferation (Fig. 1, low nutrients).
Figure 1.
Evolutionary conserved components of the Drosophila insulin-like receptor pathway. In Drosophila, the insulin-like receptor (dINR) is activated by insulin and then phosphorylates insulin receptor substrate (IRS, chico). The pathway continues through phosphatidylinositol-3 kinase (PI3K) and phosphoinositide-dependent kinase 1 (dPDK1) resulting in activation of the Drosophila Akt kinase (dAkt). dAkt phosphorylates the Drosophila TOR (dTOR), which in turn phosphorylates d4E-BP, inactivating it. Akt also directly phosphorylates dFOXO, inactivating it. Thus, in the presence of insulin-like peptides, translation is stimulated by inactivation of d4E-BP, and transcription of dFOXO targets is inhibited. Conversely, in the absence of insulin-like peptides, transcription of dINR and d4E-BP is activated but overall translation is inhibited by an active d4E-BP.
Results
Drosophila melanogaster has proven to be a valuable model organism for working out the molecular details of this conserved pathway (Brogiolo et al. 2001; Miron et al. 2001; Tatar et al. 2001; Junger et al. 2003; Puig et al. 2003; Puig and Tjian 2005). We have recently shown that in the absence of insulin or insulin-like peptides, dFOXO activates the transcription of both the insulin-like receptor (dINR) gene and the gene for Drosophila 4E-BP, establishing a transcriptional signaling loop that sensitizes the cell to receive further nutrient-dependent signals while preventing the cell from proliferating (Puig et al. 2003; Puig and Tjian 2005). In order to investigate this intriguing transcriptional feedback control, we first precisely mapped the start site of transcription for the dINR gene using a modification of the cap-trapping cDNA synthesis method (Carninci et al. 1997). This method, which depends on an intact m7G cap for capture of the mRNA, when combined with rapid amplification of five prime (5′) cDNA ends (5′ RACE) maximizes the yield of full-length 5′ untranslated regions (UTRs). The use of this methodology allowed us to detect critical UTRs associated with the mRNA that had previously gone undocumented. We find that the dINR gene is actually controlled by a complex set of three distinct promoters (P1, P2, and P3) spread over 38 kb of the Drosophila genome (Fig. 2A; Supplemental Material). These combined promoters and associated introns and exons encompass the entire region between the Drosophila E2F gene and the currently annotated dINR gene. This complex control region fills a gap in the genome annotation that contains no other annotated genes or gene predictions.
Figure 2.
Drosophila insulin receptor is regulated by a complex set of promoters. (A) Diagram of the genomic region surrounding the dINR gene on chromosome 3R. The region shown, from left to right, encompasses cytological bands 93E8, 93E7, 93E6, 93E5, and part of 93E4. The boxes indicate exons, and lines indicate introns. The three transcripts are labeled from farthest to nearest the dINR initiator codon (ATG). The transcript labels are shown above their first exons. The exons common to all three transcripts are shaded. The scale bar at the left is equal to 2 kb of genomic DNA. (B) Quantitative PCR (qPCR) analysis of the induction of the dINR promoters in S2 cells. Total RNA from either uninduced cells (white bars) or induced cells (black bars) was used to detect UTR-specific transcripts. The left pair of bars in each graph is the result from cells expressing a wild-type dFOXO. In the presence of insulin, wild-type dFOXO cannot activate transcription even upon induction. In contrast, the right pair shows the results for a constitutively active dFOXO (dFOXO-A3). Upon induction, all three promoters are activated, indicating that they are responsive to dFOXO. The data are plotted as the fraction of the RP49 transcript present in the cell. (C) Total RNA from whole animals was collected at the time after laying indicated below each graph as well as pupae and adult. The amount of specific dINR transcript, relative to the RP49 transcript, is plotted for each specific transcript. (D) Immunoblot detection of dINR protein in S2 cells expressing dFOXO-A3 shows dINR protein level increases. The control on the left is uninduced cells. The tubulin immunoblot below serves as a loading control.
Each of the dINR promoters produces a transcript with a unique and unusually long 5′UTR spliced to a short common exon that is in turn spliced to the first coding exon (Supplemental Material). The UTR originating from P1 is 1118 bases, the UTR originating from P2 is 419 bases, and the UTR originating from P3 is 485 bases. In contrast, the average 5′UTR in Drosophila is only 256 bases (Misra et al. 2002). All three UTRs contain multiple AUG initiator codons upstream of the legitimate INR initiator codon. In the case of the transcript that originates from P1, there are 12 AUGs before the legitimate translational start signal.
The DNA sequences immediately upstream of the mapped transcript start sites contain easily recognizable sequences similar to the computationally and biochemically determined common core promoter elements (Sanders et al. 1986; Burke and Kadonaga 1997; Ohler et al. 2002). P1 contains a TATA box, an Initiator element, and a downstream promoter element (DPE). P2 contains a TATA-like box and a DPE but no recognizable Initiator. P3 contains a recognizable Initiator but no recognizable TATA box or DPE. Importantly, a constitutively active form of dFOXO (dFOXO-A3) (Puig et al. 2003) activates all three promoters in Drosophila Schneider line 2 (S2) cells, and this increased RNA synthesis can produce dINR protein even in the presence of insulin (Fig. 2B,C). The transcript originating at P1 is by far the most abundant transcript under both unactivated and activated conditions. P2 is present at an intermediate level, and P3 is a low-abundance transcript. Interestingly, the level of transcription correlates with the number of recognized core promoter elements, illustrating the important role these different elements play in determining the total level of transcription from a gene in both activated and unactivated states.
In the animal, all three transcripts are detectable in multiple developmental stages. They are present in whole animal extracts in the same relative order of abundance that is detected in S2 cells (P1 ≫ P2 > P3) (Fig. 2C). When compared with the Rp49 transcript, a common control transcript that changes little over the stages tested, all three transcripts fluctuate in abundance. Notably, all three transcripts diminish significantly in the L3 larva, a time when the animal is voraciously eating. In contrast, these dINR transcripts peak in the pupae, a time when the animal is fasting and expending much of the energy gained during the larval stage. This observation is consistent with our previous finding that dINR expression is linked to nutrient availability (Puig and Tjian 2005).
Strikingly, we find that dINR is not only transcriptionally up-regulated (Puig et al. 2003; Puig and Tjian 2005) but also robustly translated. Growing S2 cells in the absence of serum and insulin causes a marked decrease in the rate of incorporation of radiolabeled cysteine and methionine consistent with a global decrease in the rate of translation (Fig. 3A). Despite this slowing of overall translation, dINR protein accumulates in S2 cells. This is detectable by immunoblot of whole cell extracts with antisera raised against the dINR protein (Fig. 3B). The increase in dINR protein levels is at least partially due to the absence of insulin itself and not another component of serum because the accumulation of dINR protein is inhibited by addition of insulin to media containing insulin-depleted serum (Fig. 3C). In addition, the increased dINR protein level is most likely due to increased synthesis since serum starved cells contain more radiolabeled receptor that binds to insulin-agarose (Fig. 3D). This raises the intriguing question of how translation of dINR can proceed in the presence of a quantitatively dephosphorylated (Pause et al. 1994; Miron et al. 2001, 2003), potently active, and up-regulated inhibitor of protein synthesis, d4E-BP (Fig. 1, low nutrients; Junger et al. 2003; Puig et al. 2003; Teleman et al. 2005; Tettweiler et al. 2005). This paradoxical finding that the dINR pathway transcriptionally up-regulates both dINR and d4E-BP combined with the newly discovered unusually long 5′UTRs of these transcripts suggest that perhaps the INR gene engages the translation machinery in an unconventional manner that bypasses the need for eIF4E. We were intrigued by the potential presence of a d4E-BP resistant internal ribosome entry site (IRES) in these Drosophila genes that contain long UTRs, as has been the precedent from other studies. For example, both the Antennapedia and Ultrabithorax long 5′UTRs contain IRESs, although their physiological role has remained undetermined (Oh et al. 1992; Ye et al. 1997).
Figure 3.
Translational control in S2 cells. (A) S2 cells grown in the presence or absence of serum and insulin were pulse-labeled with radiolabeled methionine and cysteine as described in Materials and Methods. The radiolabel incorporation is normalized to cells grown in serum. (B) Immunoblot detection of dINR protein in S2 cells in response to serum and insulin depletion. The sample on the left is from cells in the presence of serum and insulin. The sample on the right is from cells in the absence of serum and insulin. The tubulin immunoblot below serves as a loading control. (C) Immunoblot detection of dINR protein in S2 cells in response to insulin depletion. The sample on the left is from cells in the presence of insulin. The sample on the right is from cells in the absence of insulin. The tubulin immunoblot below serves as a loading control. (D) Extracts from pulse-labeled S2 cells grown in the presence or absence of insulin and serum were incubated with insulin-agarose to precipitate material that binds to insulin. The precipitated radiolabeled material is plotted as a fraction of the input of radiolabeled proteins.
As a first test of whether the dINR 5′UTRs also contain an IRES activity, we generated a bicistronic construct commonly used to assess IRES activity. The various 5′UTRs of dINR were inserted in both the forward and reverse orientations between the Renilla and firefly luciferase genes (Fig. 4A). The reverse orientation was used as a spacer length control equivalent. The ratio of Renilla luciferase expression to firefly luciferase expression should provide an indication of the cap-independent translational potential of the various 5′UTRs. Since resistance to d4E-BP is most relevant to this pathway, we carried out these experiments in the presence and absence of a constitutively active form of d4E-BP. Because the Renilla luciferase ORF is the first in the mRNA, it should be uniquely sensitive to inhibition of cap-dependent translation, while the firefly gene expression, if any, should be dependent on internal ribosome entry. The data are expressed as a ratio of the activity in the presence of d4E-BP to the activity in the absence of d4E-BP. Therefore, a number close to 1 indicates that there is no resistance to d4E-BP (Fig. 3A). In these cell-based assays, the 5′UTR from both P1 and P2 showed significant resistance to d4E-BP (about fourfold better than the reverse orientation in both cases), but only when inserted in the forward direction. Curiously, the 5′UTR from P3 showed unusual resistance to d4E-BP in either orientation. Indeed, the P3 UTR showed a perplexing increase in expression of the firefly ORF in the presence of d4E-BP compared with no UTR in both orientations. This finding reveals a potential limitation of using the bicistronic assays since we cannot rule out interfering effects from cryptic promoters, cryptic splicing, or secondary effects of expression of d4E-BP with this assay.
Figure 4.
In vivo analysis of dINR UTR function. (A) Above the graph is a schematic diagram of the expected transcript produced by transfection of the bicistronic DNA constructs. The UTR and insertion orientation are indicated below each bar in the graph. The data are plotted as the ratio of the activity in the presence of d4E-BP to the activity in the absence of d4E-BP. (B) Above the graph is a diagram of the expected transcript produced by transfection of the monocistronic DNA constructs. The UTR is indicated below each bar in the graph. The data are plotted as the ratio of the activity in the presence of d4E-BP to the activity in the presence of GFP, a nonspecific control. (C) RNA transfection of UTR-containing RNAs shows significant IRES activity for the dINR UTRs. Luciferase activity is plotted normalized to the activity of a m7G-containing RNA.
To circumvent some of the inherent idiosyncrasies of the bicistronic constructs, we next used monocistronic constructs that more closely mimic the situation of the endogenous dINR gene. We tested for potential IRES activity in two complementary ways. First, in a DNA-based transient transfection, we expressed either the constitutively active form of d4E-BP or a control protein, green fluorescent protein (GFP), and measured resistance to d4E-BP as the ratio of luciferase activity (provided by a second plasmid) in the presence of d4E-BP to the activity in the presence GFP (Fig. 4B). In this set of experiments, we included the minimal Antennapedia IRES (Oh et al. 1992), a Drosophila 5′UTR known to support cap-independent initiation of translation, as a positive control. Under these cell-based assay conditions, the P1 and P2 UTRs again displayed robust resistance to d4E-BP, while P3 and the common exons showed little resistance. Notably, the P2 5′UTR is as efficient as the minimal Antennapedia IRES, and the P1 5′UTR is actually significantly more efficient than the control IRES. Taken together, these two cell-based assays suggest that the 5′UTRs of at least the P1 and P2 transcripts can direct substantial IRES activity, while the P3 UTR appears to have much less if any such activity in S2 cells. Second, to complement these plasmid-based assays and directly investigate the contribution of the UTRs to translation, we used an RNA-based transfection assay. The RNAs contained either a m7G cap or an ApppG cap mimic. Only the 7mG cap allows cap-dependent translation. The ApppG cap stabilizes the transcript but does not allow cap-dependent translation, so it is a direct measure of the contribution of IRES activity. In this assay, the UTRs again showed significant IRES activity. The P1 UTR confers the same activity with or without a m7G cap, indicating a strong IRES activity. The P2 and P3 UTRs also confer cap-independent translation activity, although the level of activity is not equal to UTR plus cap. In contrast, the common exon or nonspecific UTR retains only ∼20% of their translation potential without the m7G cap (Fig. 4C). Taken together, these cell-based assays provide encouraging evidence for IRES activity of the dINR 5′UTRs.
However, given the well-recognized limitations inherent with using cell-based assays to establish IRES activity, we turned to a Drosophila embryo-derived capdependant in vitro translation system to test more directly the putative IRES activity and more specifically the potential d4E-BP resistance of the INR UTRs (Gebauer et al. 1999; Tuschl et al. 1999). We first treated the translation extracts with micrococcal nuclease to destroy the bulk of competing endogenous transcripts so that translation would be largely dependent on exogenously added RNA (Sanders et al. 1986). As expected, addition of normal capped transcripts results in robust translation from all of the UTR-containing RNAs as well as the common UTR and a short nonspecific UTR control RNA (Fig. 5A). To test the dependence of translation on eIF4E, we added exogenous m7G cap analog as a competitor. This excess free cap efficiently binds and sequesters the available eIF4E, preventing this essential initiation factor from binding capped RNA, thus effectively blocking the nucleation of the eIF4F complex and cap-dependent initiation. Remarkably, only the transcripts containing the P1, P2, and P3 UTRs are resistant to exogenously added competitor cap analog, whereas the common UTR fragment and the short nonspecific leader are effectively inhibited (Fig. 5A). This finding strongly suggests that the various dINR-specific UTRs, indeed, provide a cap-independent mechanism of translation initiation. To directly test the resistance of these transcripts to d4E-BP-mediated translation inhibition, we added recombinant d4E-BP to the reactions. Whereas the common exon and control RNAs are efficiently inhibited by this blocker of eIF4E-mediated translation initiation, the P1, P2, and P3 UTR-containing transcripts are highly resistant to d4E-BP (Fig. 5B). These findings taken together with our previous cell-based assays suggest that, indeed, dINR protein synthesis can proceed via an IRES-mediated eIF4E-independent mechanism of initiation both in vitro and in vivo.
Figure 5.
In vitro analysis of dINR UTR function in the absence of endogenous mRNA. (A) Micrococcal nuclease-treated embryo translation extracts were programmed with monocistronic, capped luciferase transcripts containing the dINR UTR indicated below each set of bars. The extract was challenged with an excess of cap analog, and luciferase activity was measured. The data are plotted as the amount of activity relative to addition of buffer alone. (B) Micrococcal nuclease-treated embryo translation extracts were programmed with monocistronic, capped luciferase transcripts containing the dINR UTR indicated below each set of bars. The extract was challenged with an excess of either a recombinant GST-d4E-BP fusion protein or GST alone. The data are plotted as the amount of activity relative to addition of buffer alone.
What purpose might a cap-independent translation activity serve beyond simple resistance to the active d4E-BP in the absence of insulin? Perhaps by functionally coupling transcription and translation, such a mechanism could serve to amplify the signal received from the insulin receptor pathway. To test this idea, we again turned to the in vitro translation experiments. In the absence of miccrococal nuclease treatment, the endogenous transcripts present in the translation extract should effectively compete with the experimental dINR transcripts for limiting amounts of the translation machinery (Svitkin et al. 2005). We therefore took advantage of this inevitable competition for translation machinery to test the response of the various UTRs in a situation that may more closely reflect the cellular environment, where multiple variable abundant transcripts must compete for a limited supply of the translational apparatus. Under these competitive conditions, addition of either m7G or d4E-BP actually results in an even more robust increase in translation of the dINR UTR-containing RNAs relative to the unchallenged state (Fig. 6A,B). This finding suggests that these RNAs that contain dINR UTRs, and presumably IRES activity, are highly effective at out-competing other transcripts for access to the translational machinery when m7G cap-dependent initiation is inhibited. While we have not unequivocally defined the molecular mechanism of 4E-BP resistance of the dINR transcripts, it is clear that the UTRs allow significant translation in conditions when cap-dependent translation is inhibited.
Figure 6.
In vitro analysis of dINR UTR competition. (A) Untreated embryo translation extracts, still containing endogenous mRNA, were programmed with monocistronic, capped luciferase transcripts containing the dINR UTR indicated below each set of bars. The extract was challenged with an excess of cap analog, and luciferase activity was measured. The data are plotted as the amount of activity relative to addition of buffer alone. (B) Untreated embryo translation extracts, programmed with monocistronic, capped, luciferase transcripts as above. The extract was challenged with an excess of either a recombinant GST-d4E-BP fusion protein or GST alone. The data are plotted as the amount of activity relative to addition of buffer alone.
Discussion
The data we report here, combined with our previous data and those from others, allow us to formulate a new model to explain the effects of nutrients and insulin levels on dINR feedback regulation. In times of high nutrients and therefore high insulin-like peptides, both dFOXO and d4E-BP are phosphorylated and inactive. Under these “rich” conditions, dFOXO is sequestered in the cytoplasm and phosphorylated d4E-BP is unable to interact with eIF4E. This situation allows efficient translation of most cellular transcripts regardless of the mechanism of initiation (cap-dependent vs. cap-independent) (Fig. 7A). In contrast, in low nutrient conditions or in the absence of insulin or insulin-like peptides, both dFOXO and d4E-BP become dephosphorylated and active. Activated dFOXO directs a robust increase in the transcription of both dINR and d4E-BP (among other genes). Additionally, the active and up-regulated d4E-BP effectively inhibits cap-dependent translation, freeing up the protein synthesis machinery to selectively translate IRES-containing transcripts like dINR (Fig. 7B). These two coordinated mechanisms consequently orchestrate the integration of a specific transcriptional response and simultaneously a translational response that greatly amplifies the signal and sensitizes the cell for detection of small changes in nutrient availability as well as, possibly, developmental and environmental cues.
Figure 7.
Model of the role of IRES activity in production of dINR. (A) In conditions of high nutrients or insulin-like peptides, dFOXO and d4E-BP are phosphorylated and inactive. This results in a basal level of expression of dINR and 4E-BP. The translational machinery is divided evenly between dINR and other cellular transcripts. (B) In conditions of low nutrients or insulin-like peptides, dFOXO and d4E-BP are unphosphorylated. dFOXO can activate transcription of both the dINR and d4E-BP genes. The unphosphorylated d4E-BP is able to bind to 4E, preventing the translation of cellular cap-dependent translation. This results in an increase in the dINR transcript and an uneven distribution of the translation machinery amplifying expression of dINR at both the transcription and translation levels. This results in the accumulation of dINR in the cellular membrane, poising the cell for a rapid response to signals when nutrients are again available.
Interestingly, the dFOXO-responsive dINR promoters produce three distinct transcripts. Why such a complex regulatory network? A hint may be that the P3 UTR does not seem to have detectable IRES activity in the S2 cells but shows substantial activity in vitro with extracts derived from whole Drosophila embryos. It is likely that the three transcripts are produced in a tissue- or temporal-specific manner during development, and we speculate that each may depend on cell-specific IRES trans-acting factors (ITAFs) that are required for activity. This would direct tissues to respond differentially to dINR signaling. In tissues lacking specific ITAFs, the IRES activity would be diminished and the tissue may produce only a moderate level of dINR protein.
We also find an interesting parallel between mechanisms for reprogramming the gene expression machinery in a cell to respond to physiological cues and the more commonly observed viral takeover of the cellular macromolecular synthesis machinery (Svitkin et al. 2005). When some viruses, such as polio, infect a cell, they target the translation initiation machinery (either eIF4G or 4E-BP) so that there is a switch from cap-dependent synthesis to IRES-dependent synthesis (Jackson 2005). This leads to a robust and specific stimulation of viral protein synthesis at the expense of most cellular protein synthesis. By the evolution of cellular mechanisms that activate 4E-BP and simultaneously produce transcripts containing cellular IRESs, a critical physiological signaling cascade can evidently adopt a similar mechanism to effectively usurp the macromolecular synthesis machinery to drive cellular physiology in a very specific direction. Indeed, viruses may have merely co-opted the mechanism from cells in the eternal battle between host and virus.
Although our initial characterization of the INR transcriptional feedback loop was carried out in Drosophila, we subsequently found a similar regulatory circuit in vertebrates (Puig et al. 2003; Puig and Tjian 2005). It is interesting to note that the transcripts for human insulin receptor and IGF-2 receptor remain associated with polysomes when cap-dependant translation is inhibited by poliovirus infection (Johannes et al. 1999). We have also observed that although the level of INR mRNA up-regulation by FOXO in mouse muscle cells is only twofold, the levels of INR protein increase much more dramatically (six- to eightfold), consistent with a coupled transcription/translation mechanism of the signal in vertebrates (Puig and Tjian 2005). It seems likely, given the findings we report here, that the same type of coupling between the transcriptional program of FOXO proteins and translational control by IRES activity is also occurring in vertebrate systems. Understanding this novel mechanism that couples transcription and translation may provide new insight into disease states such as insulin-resistant type 2 diabetes.
Materials and methods
Cell culture
Drosophila S2 cells were maintained at 25°C in M3BPYE as described by the Drosophila Genomics Resource Center (http://dgrc.cgb.indiana.edu) supplemented with 10% fetal bovine serum (FBS) and 5 μg/mL bovine insulin pen per strep, and 2 mM glutamine (complete media). S2 cells expressing dFOXO or dFOXO-A3 have been described (Puig et al. 2003). For dFOXO induction in Figure 2D, cells were treated with 0.5 mM CuSO4 for 12 h at 25°C. For insulin-dependent experiments, FBS was treated with charcoal-dextran to remove insulin (CDFBS) (Herbert et al. 1965). Briefly, charcoal-dextran charcoal was prepared by adding 2.5 g of activated charcoal into a solution that contains 0.25 g of dextran and 1000 mL of Tris-HCL (0.1 M at pH 7.9). The suspension was stirred overnight at 4°C. The dextran-coated charcoal pellets were mixed with 200 mL heat-inactivated sera and stirred overnight at 4°C. Charcoal was removed, and the sera were sterilized by filtering. The CDFBS was used at 10% in M3BPYE. For insulin-containing samples, insulin was added to 5 μg/mL. The cells were incubated for 36 h at 25°C and lysed in modified RIPA Buffer (PBS containing 10 mM EDTA, 10 mM β-glycerophosphate, 10 mM pyrophosphate, 10 mM NaF, 1 mM sodium vanadate, 1% Triton X-100, 1% SDS, 1% deoxycholate, aprotinin, leupeptin, 1× complete protease inhibitor [Roche], 10% glycerol). Total protein concentration was determined by BCA assay (Pierce), and equal amounts of total protein were loaded on a 7.5% SDS–polyacrylamide gel. For pulse-labeling experiments, cells were grown in complete M3BPYE media or M3BPYE lacking serum and insulin for 24 h. The cells were then labeled with 143 μCi (10 μL) of Pro-mix (GE Healthcare) for 1 h. Cells were harvested and washed one time in PBS, and incorporation was measured by glass filter binding of TCA-precipitated material and scintillation counting. For insulin-agarose-binding experiments, pulse-labeled cells were incubated in PBS containing 3% Triton X-100 and 1× EDTA-free complete protease inhibitors (Roche) for 30 min on ice. Insoluble material was removed by centrifugation in a microfuge for 30 min at 16,000 rpm. The supernatant was diluted 1:10 with PBS containing 0.1% Triton X-100 and combined with 50 μL of insulin-agarose (Sigma) prewashed with PBS and 0.1% Triton X-100. The samples were incubated overnight at 4°C. The samples were poured into an empty 2-mL column and washed with 80 bead volumes (4 mL) PBS containing 0.1% Triton X-100. Proteins were eluted with 10 column volumes of 5 M urea, 50 mM sodium acetate (pH 4.5), and 0.1% Triton X-100. The eluted material was precipitated with 10% TCA and assayed by binding to glass filters and scintillation counting.
Transient transfection
For expression in S2 cells, the dINR UTR-containing constructs and the d4E-BP constructs were driven by a version of the Actin 5C promoter truncated to remove negative elements (Chung and Keller 1990). The expression plasmid in Figure 3A expresses a d4E-BP mutant that binds eIF4E more tightly than wild-type d4E-BP [pACd4E-BP (LL)] (Miron et al. 2001). The control was an empty version of the expression plasmid. The expression plasmid used in Figure 3B expresses a d4E-BP cDNA (pACd4E-BP; AALL) that contains the eIF4E-binding enhancing mutations as well as phosphorylation site mutations to prevent insulin-dependent phosphorylation (Miron et al. 2003). The control plasmid in Figure 3B expresses GFP under the same promoter. S2 cells (0.25 × 106 per well) were plated in 0.4 mL of M3BPYE 10% FBS and allowed to adhere. A total of 1 μg of plasmid DNA (13:1 expression plasmid to reporter plasmid) was transfected per well using Effectene Transfection Reagent (Qiagen) following the manufacturer’s instructions as modified for S2 cells.
For RNA transfections, RNA was transcribed in vitro in the presence of m7G(5′)ppp(5′) or G(5′)ppp(5′)A cap analog (New England Biolabs) using Megascript T7 kits (Ambion). RNA was purified on a G-50 spin column and by LiCl precipitation. S2 cells were transfected in 24-well plates with 100 ng of firefly reporter and 50 ng of capped Renilla RNA as a transfection control using Effectene Reagent (Qiagen). Luciferase activity was measured 24 h post-transfection.
Cloning of the dINR 5′UTRs
The dINR UTRs were mapped using the cap-trapper methodology as previously described with the following modifications (Carninci et al. 1997). A gene-specific primer that hybridized to the dINR ORF was used for first-strand synthesis using poly(A)-purified RNA. RNase protection was performed with RNase A and RNase T1. The RNA/DNA hybrid was purified by biotinylated cap as previously described. The single-stranded cDNA was purified by a combination of RNase H and NaOH treatment followed by spin column purification (Qiagen). The first strand was dC tailed using terminal deoxytransferase. The 5′UTRs were amplified by two rounds of nested PCR and cloned into pCR4 (Invitrogen) and sequenced. Detailed protocol is available upon request. Intron/exon boundaries were deduced by mapping the UTR sequences onto the Drosophila genomic sequence.
In vitro translation
Embryo translation extracts were prepared as described from 0- to 12-h embryos (Gebauer et al. 1999; Tuschl et al. 1999). In vitro transcription templates were linearized with XhoI and purified on Qiagen spin columns. Capped transcripts were generated with mMessage mMachine T7 Ultra kits (Ambion), desalted on a G-50 spin column, and precipitated with LiCl. Transcripts were resuspended in 10 mM Tris-HCl (pH 8.5). Transcription products were checked on denaturing agarose gels. In vitro translation reactions contained 6 μL of Drosophila embryo extract, 0.1 mM spermidine, 60 μm Amino Acids total (Roche), 16.8 mM creatine phosphate, 800 ng of creatine kinase, 24 mM HEPES (pH 7.4), 0.4 mM Mg acetate, 30 mM K acetate, 1 μg of calf liver tRNA, and 12 ng of template RNA in a 10-μL reaction. 3′-0-Me-7meG(ppp)G RNA Cap Structure Analog (New England Biolabs), GST-d4E-BP, or GST was added to a final concentration of 500 nm. Reactions were preincubated for 10 min at 27°C before the addition of RNA templates. Translation was conducted for 40 min at 27°C, and luciferase activity was measured using 8 μL of the reaction in 100 μL of luciferase substrate (Promega).
Acknowledgments
We thank Jennifer Doudna and Wendy Gilbert for critical reading of the manuscript and for sharing data prior to publication. We also thank Nahum Sonenberg for comments on the manuscript. We thank Yoh Isogai, Kevin Wright, Yick Fong, and Francisco Herrera for comments and suggestions on the manuscript; and Mallory Haggart for sequencing and oligonucleotide synthesis. We thank Fatima Gebauer for the Antennapedia construct and for advice on making the Drosophila translation extracts. M.T.M. was supported by a fellowship from the Damon Runyon Cancer Research Foundation (DRG #1684) for part of this work. R.T. is supported by grants from the NIH and Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1506407
References
- Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., Hafen E., Stocker H., Ikeya T., Rintelen F., Fernandez R., Hafen E., Ikeya T., Rintelen F., Fernandez R., Hafen E., Rintelen F., Fernandez R., Hafen E., Fernandez R., Hafen E., Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Burke T.W., Kadonaga J.T., Kadonaga J.T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes & Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Westover A., Nishiyama Y., Ohsumi T., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Westover A., Nishiyama Y., Ohsumi T., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Nishiyama Y., Ohsumi T., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Ohsumi T., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Okazaki Y., Muramatsu M., Schneider C., Muramatsu M., Schneider C., Schneider C., et al. High efficiency selection of full-length cDNA by improved biotinylated cap trapper. DNA Res. 1997;4:61–66. doi: 10.1093/dnares/4.1.61. [DOI] [PubMed] [Google Scholar]
- Chung Y.T., Keller E.B., Keller E.B. Positive and negative regulatory elements mediating transcription from the Drosophila melanogaster actin 5C distal promoter. Mol. Cell. Biol. 1990;10:6172–6180. doi: 10.1128/mcb.10.12.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon I., Raff M., Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Garofalo R.S. Genetic analysis of insulin signaling in Drosophila. Trends Endocrinol. Metab. 2002;13:156–162. doi: 10.1016/s1043-2760(01)00548-3. [DOI] [PubMed] [Google Scholar]
- Gebauer F., Corona D.F., Preiss T., Becker P.B., Hentze M.W., Corona D.F., Preiss T., Becker P.B., Hentze M.W., Preiss T., Becker P.B., Hentze M.W., Becker P.B., Hentze M.W., Hentze M.W. Translational control of dosage compensation in Drosophila by Sex-lethal: Cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 1999;18:6146–6154. doi: 10.1093/emboj/18.21.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafen E. Cancer, type 2 diabetes, and ageing: News from flies and worms. Swiss Med. Wkly. 2004;134:711–719. doi: 10.4414/smw.2004.09885. [DOI] [PubMed] [Google Scholar]
- Herbert V., Lau K.S., Gottlieb C.W., Bleicher S.J., Lau K.S., Gottlieb C.W., Bleicher S.J., Gottlieb C.W., Bleicher S.J., Bleicher S.J. Coated charcoal immunoassay of insulin. J. Clin. Endocrinol. Metab. 1965;25:1375–1384. doi: 10.1210/jcem-25-10-1375. [DOI] [PubMed] [Google Scholar]
- Jackson R.J. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- Johannes G., Carter M.S., Eisen M.B., Brown P.O., Sarnow P., Carter M.S., Eisen M.B., Brown P.O., Sarnow P., Eisen M.B., Brown P.O., Sarnow P., Brown P.O., Sarnow P., Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junger M.A., Rintelen F., Stocker H., Wasserman J.D., Vegh M., Radimerski T., Greenberg M.E., Hafen E., Rintelen F., Stocker H., Wasserman J.D., Vegh M., Radimerski T., Greenberg M.E., Hafen E., Stocker H., Wasserman J.D., Vegh M., Radimerski T., Greenberg M.E., Hafen E., Wasserman J.D., Vegh M., Radimerski T., Greenberg M.E., Hafen E., Vegh M., Radimerski T., Greenberg M.E., Hafen E., Radimerski T., Greenberg M.E., Hafen E., Greenberg M.E., Hafen E., Hafen E. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron M., Verdu J., Lachance P.E., Birnbaum M.J., Lasko P.F., Sonenberg N., Verdu J., Lachance P.E., Birnbaum M.J., Lasko P.F., Sonenberg N., Lachance P.E., Birnbaum M.J., Lasko P.F., Sonenberg N., Birnbaum M.J., Lasko P.F., Sonenberg N., Lasko P.F., Sonenberg N., Sonenberg N. The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat. Cell Biol. 2001;3:596–601. doi: 10.1038/35078571. [DOI] [PubMed] [Google Scholar]
- Miron M., Lasko P., Sonenberg N., Lasko P., Sonenberg N., Sonenberg N. Signaling from Akt to FRAP/TOR targets both 4E-BP and S6K in Drosophila melanogaster. Mol. Cell. Biol. 2003;23:9117–9126. doi: 10.1128/MCB.23.24.9117-9126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S., Crosby M.A., Mungall C.J., Matthews B.B., Campbell K.S., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Crosby M.A., Mungall C.J., Matthews B.B., Campbell K.S., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Mungall C.J., Matthews B.B., Campbell K.S., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Matthews B.B., Campbell K.S., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Campbell K.S., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Kaminker J.S., Millburn G.H., Prochnik S.E., Millburn G.H., Prochnik S.E., Prochnik S.E., et al. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0083. RESEARCH0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.K., Scott M.P., Sarnow P., Scott M.P., Sarnow P., Sarnow P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes & Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- Ohler U., Liao G.C., Niemann H., Rubin G.M., Liao G.C., Niemann H., Rubin G.M., Niemann H., Rubin G.M., Rubin G.M. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0087. RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A., Belsham G.J., Gingras A.C., Donze O., Lin T.A., Lawrence J.C., Jr., Sonenberg N., Belsham G.J., Gingras A.C., Donze O., Lin T.A., Lawrence J.C., Jr., Sonenberg N., Gingras A.C., Donze O., Lin T.A., Lawrence J.C., Jr., Sonenberg N., Donze O., Lin T.A., Lawrence J.C., Jr., Sonenberg N., Lin T.A., Lawrence J.C., Jr., Sonenberg N., Lawrence J.C., Jr., Sonenberg N., Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Puig O., Tjian R., Tjian R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes & Dev. 2005;19:2435–2446. doi: 10.1101/gad.1340505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Tjian R., Tjian R. Nutrient availability and growth: Regulation of insulin signaling by dFOXO/FOXO1. Cell Cycle. 2006;5:503–505. doi: 10.4161/cc.5.5.2501. [DOI] [PubMed] [Google Scholar]
- Puig O., Marr M.T., Ruhf M.L., Tjian R., Marr M.T., Ruhf M.L., Tjian R., Ruhf M.L., Tjian R., Tjian R. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes & Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D., Sonenberg N., Sonenberg N. The Akt of translational control. Oncogene. 2005;24:7426–7434. doi: 10.1038/sj.onc.1209098. [DOI] [PubMed] [Google Scholar]
- Saltiel A.R., Kahn C.R., Kahn C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Sanders M.M., Triemer D.F., Olsen A.S., Triemer D.F., Olsen A.S., Olsen A.S. Regulation of protein synthesis in heat-shocked Drosophila cells. Soluble factors control translation in vitro. J. Biol. Chem. 1986;261:2189–2196. [PubMed] [Google Scholar]
- Svitkin Y.V., Herdy B., Costa-Mattioli M., Gingras A.C., Raught B., Sonenberg N., Herdy B., Costa-Mattioli M., Gingras A.C., Raught B., Sonenberg N., Costa-Mattioli M., Gingras A.C., Raught B., Sonenberg N., Gingras A.C., Raught B., Sonenberg N., Raught B., Sonenberg N., Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M., Kopelman A., Epstein D., Tu M.P., Yin C.M., Garofalo R.S., Kopelman A., Epstein D., Tu M.P., Yin C.M., Garofalo R.S., Epstein D., Tu M.P., Yin C.M., Garofalo R.S., Tu M.P., Yin C.M., Garofalo R.S., Yin C.M., Garofalo R.S., Garofalo R.S. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Teleman A.A., Chen Y.W., Cohen S.M., Chen Y.W., Cohen S.M., Cohen S.M. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes & Dev. 2005;19:1844–1848. doi: 10.1101/gad.341505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettweiler G., Miron M., Jenkins M., Sonenberg N., Lasko P.F., Miron M., Jenkins M., Sonenberg N., Lasko P.F., Jenkins M., Sonenberg N., Lasko P.F., Sonenberg N., Lasko P.F., Lasko P.F. Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes & Dev. 2005;19:1840–1843. doi: 10.1101/gad.1311805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl T., Zamore P.D., Lehmann R., Bartel D.P., Sharp P.A., Zamore P.D., Lehmann R., Bartel D.P., Sharp P.A., Lehmann R., Bartel D.P., Sharp P.A., Bartel D.P., Sharp P.A., Sharp P.A. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Fong P., Iizuka N., Choate D., Cavener D.R., Fong P., Iizuka N., Choate D., Cavener D.R., Iizuka N., Choate D., Cavener D.R., Choate D., Cavener D.R., Cavener D.R. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol. Cell. Biol. 1997;17:1714–1721. doi: 10.1128/mcb.17.3.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]