Abstract
The genome diversity of RNA viruses allows for rapid adaptation to a wide variety of adverse conditions. Accordingly, viruses can escape inhibition by most antiviral compounds targeting either viral or host factors. Here we exploited the capacity of RNA viruses for rapid adaptation to explore the evolutionary constraints of chaperone-mediated protein folding. We hypothesized that inhibiting a host molecular chaperone required for folding of a viral protein would force the virus to evolve an alternate folding strategy. We identified the chaperone Hsp90 as an essential factor for folding and maturation of picornavirus capsid proteins. Pharmacological inhibition of Hsp90 impaired the replication of poliovirus, rhinovirus, and coxsackievirus in cell culture. Strikingly, anti-Hsp90 treatment did not yield drug-resistant viruses, suggesting that the complexity of capsid folding precludes the emergence of alternate folding pathways. These results reveal tight evolutionary constraints on chaperone-mediated protein folding, which may be exploited for viral inhibition in vivo. Indeed, Hsp90 inhibitors drastically reduced poliovirus replication in infected animals without the emergence of drug-resistant escape mutants. We propose that targeting folding of viral proteins may provide a general antiviral strategy that is refractory to development of drug resistance.
Keywords: Evolution, protein folding, Hsp90, drug resistance, antiviral, picornavirus
RNA viruses possess the greatest capacity for rapid evolution among all organisms. Their ability to adapt stems from having the highest mutation rates in nature, combined with short generation times and very large population sizes (Domingo and Holland 1997; Fields et al. 2001; Elena et al. 2006). In fact, RNA viruses never exist as a single genotype; rather, at any single time, the viral population consists of an ensemble of closely related genotypes termed “quasispecies” (Domingo and Holland 1997; Elena et al. 2006). This property allows RNA viruses to evolve at rates of up to a million times greater than those observed for organisms using DNA to encode their genome (Fields et al. 2001). Such capacity for rapid evolution enables viruses to survive in the face of adverse conditions and successfully replicate in different hosts and changing microenvironments.
The tremendous capacity of viruses for rapid evolution has profound medical consequences, as many antiviral drugs are rendered ineffective by the emergence of drug-resistant viral variants (Sarafianos et al. 2004). The most common antiviral strategy relies on directly inhibiting viral proteins. While leading to specific viral inhibitors, this strategy invariably results in the emergence of drug resistance, as the virus can readily mutate to circumvent inhibition, even under conditions of combinatorial therapy targeting multiple viral proteins (Pillay and Zambon 1998). An alternative strategy is to target host processes required for viral replication, as direct mutation of the drug target is not possible. Strikingly, this approach also results in the emergence of viral drug resistance. For instance, poliovirus replication is strongly inhibited by Brefeldin A (BFA), which targets components of the cellular secretory apparatus required for viral RNA replication (Irurzun et al. 1992; Maynell et al. 1992; Doedens et al. 1994; Cuconati et al. 1998). However, viral variants independent of these factors and resistant to BFA were readily isolated (Crotty et al. 2004). HIV can also rapidly gain resistance to an inhibitor of a cellular prolyl-peptidyl isomerase that is required for infectivity (Aberham et al. 1996). Likewise, HSV can become resistant to an inhibitor of a nuclear export factor, Crm1, involved in export of HSV viral RNAs from the nucleus (Murata et al. 2001). In such cases, the viruses are thought to gain drug resistance by evolving new replication strategies that use alternative cellular factors or dispense with the affected function. The extreme adaptability of viruses observed in these cases provides a unique tool for studying the evolution of new processes and mechanisms.
One of the most complex and poorly understood problems in biology is that of protein folding. The polypeptide sequence must encode the functional and structural information of the native state. In addition, it must contain the blueprint for reaching the native state rapidly and efficiently. As the length and complexity of proteins increased during evolution, so did the challenges of reaching the final folded structure. Accordingly, folding of many cellular proteins is critically dependent on the assistance of a complex set of folding factors termed molecular chaperones (Frydman 2001; Young et al. 2004). Eukaryotic cells contain several different chaperone families, which appear to facilitate folding of different subsets of cellular proteins. However, it is unclear why some proteins require chaperones to fold while others do not and what determines the chaperone specificity of proteins.
Several lines of evidence suggest that viral proteins, like cellular proteins, bind to chaperones or are dependent on chaperone function for folding and assembly (Hu et al. 1997; Braakman and van Anken 2000; Hong et al. 2001; Hung et al. 2002; Li et al. 2004; Basha et al. 2005; Burch and Weller 2005; Kampmueller and Miller 2005; Mayer 2005). However, the role of molecular chaperones in viral protein folding has not been extensively explored. Hsp90 is the only chaperone with specific pharmacological inhibitors; these inhibitors of Hsp90 have been shown to reduce the replication of several viruses in vitro (Hu et al. 1997; Hung et al. 2002; Li et al. 2004; Basha et al. 2005; Burch and Weller 2005; Janin 2005; Kampmueller and Miller 2005; Okamoto et al. 2006). For most of these viruses, it is unclear whether the observed inhibitory effect is directly related to the folding of viral proteins by Hsp90, or due to pleiotropic effects on viral replication arising from inhibition of cellular Hsp90 targets. In the case of herpes and hepatitis B viruses, where the molecular basis of Hsp90 action on a viral protein has been investigated, this chaperone appears to be involved in the regulation of viral polymerase function, rather than polypeptide folding (Hu et al. 1997; Hu and Anselmo 2000; Burch and Weller 2005).
Here, we define the chaperone requirements for capsid assembly of picornaviruses, a large and conserved family of RNA viruses. Our analysis identified a single protein in the picornavirus proteome, the capsid protein P1, which requires the chaperone Hsp90 for folding. Accordingly, Hsp90 inhibitors impaired the replication of three major picornavirus pathogens in tissue culture: poliovirus, the agent of poliomyelitis; rhinovirus, the agent of the common cold; and coxsackievirus, a major agent of viral-induced cardiac disease (Fields et al. 2001). Strikingly, poliovirus was unable to develop escape mutants resistant to an Hsp90 inhibitor, even though its rapid replication rate and high mutation frequency (106 times higher than that of DNA-based genomes) have enabled the isolation of drug-resistant poliovirus variants to virtually all antiviral compounds tested to date (Melnick et al. 1961; Drake 1999; Gitlin et al. 2002; Pfeiffer and Kirkegaard 2003; Crotty et al. 2004; Vignuzzi et al. 2006). These results suggest that stringent constraints prevent proteins from being able to evolve folding pathways that bypass their Hsp90 requirement. Importantly, this finding uncovered a potential target for antiviral therapies that may be refractory to development of drug resistance in vivo. Indeed, we found that administration of Hsp90 inhibitors to infected animals drastically reduced poliovirus replication without eliciting viral drug resistance. Our results highlight the potential of chaperone inhibitors as antiviral agents and may be of immediate relevance to the treatment of a variety of Picornaviridae-related diseases.
Results
Pharmacological inhibition of Hsp90 impairs viral replication in cultured cells
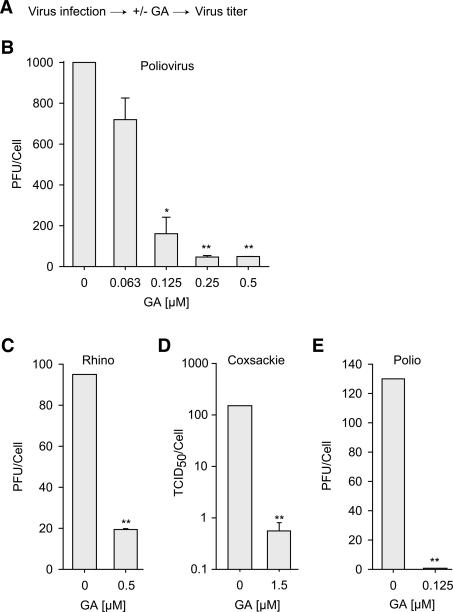
We first tested the effect of pharmacologically inhibiting Hsp90 on the replication of three pathogens of the picornavirus family—poliovirus, rhinovirus, and coxsackievirus (Fig. 1A). Geldanamycin (GA), a specific Hsp90 inhibitor, was used to inhibit Hsp90. HeLa S3 cells were infected with poliovirus in the presence or absence of increasing concentrations of GA, and virus production was measured at 7 h post-infection. GA treatment inhibited poliovirus replication in a dose-dependent manner, with maximal inhibition of 95% and an IC50 of 0.11 μM (±0.026) relative to DMSO-treated controls (Fig. 1B). A similar antiviral effect by GA was observed in TSA201 cells (data not shown). GA treatment also inhibited the replication of rhinovirus (Fig. 1C) and coxsackievirus (Fig. 1D; Li et al. 2004). It has been reported that the Hsp90 machinery of transformed cells is more susceptible to GA than that of untransformed cells (Kamal et al. 2003). To evaluate if GA also possesses antiviral activity in untransformed cells, we examined whether GA can inhibit poliovirus replication in primary human foreskin fibroblasts (Fig. 1E). Strikingly, the inhibitory effect of GA on poliovirus replication was even stronger in these cells than in HeLa S3 or TSA201 cells (Fig. 1E, >99% inhibition at 0.1 μM). These results indicate that Hsp90 function is required for picornavirus replication.
Figure 1.
The Hsp90 inhibitor GA reduces picornavirus replication in cultured cells. (A) Outline of the experiment. (B–E) Effect of GA on poliovirus (B,E), rhinovirus (C), and coxsackievirus (D) production in HeLa S3 cells (B–D) or primary human foreskin fibroblasts (E). Data are represented as the number of plaque-forming units (PFU) or 50% tissue culture infective dose (TCID50) produced per cell and, for comparison reasons, standardized between experiments so as to yield the same number of PFU or TCID50 per cell for DMSO-treated conditions. Results indicate mean and SEM of three independent experiments. (*) p < 0.05, (**) p < 0.001 relative to DMSO-treated condition by t-test.
GA decreases production of mature capsid proteins
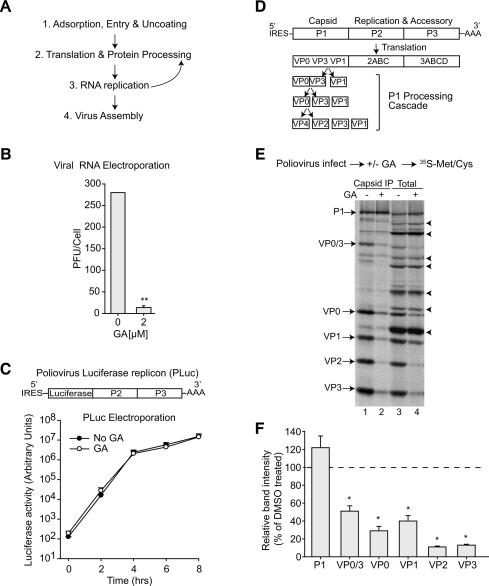
The molecular basis for the anti-picornavirus activity of GA was defined by systematically examining its effect on distinct steps in the viral life cycle (Fig. 2A; Fields et al. 2001). We first tested whether GA affects the early steps of viral replication (Fig. 2B). To bypass the viral entry and uncoating steps, the viral genomic RNA (vRNA) was directly introduced into cells; this allowed us to measure the effect of GA on subsequent steps in virus production (Fig. 2B). GA treatment inhibited virus production in vRNA-transfected cells to the same degree as when cells were infected with intact virus (95% inhibition) (Figs. 2B, 1B), indicating that GA acts downstream from these early steps. Consistent with this conclusion, GA treatment effectively inhibited virus production even when added 2 h post-infection, at a stage subsequent to viral entry and uncoating (Supplementary Fig. S1).
Figure 2.
Inhibition of Hsp90 specifically affects production of mature capsid proteins. (A) Schematic representation of picornavirus life cycle. (B) GA inhibits poliovirus replication from a transfected infectious genomic RNA. Data represent the mean and SEM of three independent experiments. (C) GA does not inhibit translation and replication of a poliovirus luciferase replicon (PLuc), in which the capsid-coding sequence is replaced with luciferase (Herold and Andino 2000). The time course of luciferase activity reports on viral translation and replication. Data show mean and SEM of three independent experiments. (D) Poliovirus-encoded polyprotein, highlighting the processing events for the capsid precursors. (E,F) GA decreases capsid protein production. (E) Steady-state 35S-labeling of poliovirus proteins from infected cells grown in the presence or absence of GA. Total cytoplasmic extracts (lanes 3,4) and immunoprecipitated capsid proteins (lanes 1,2) separated by SDS-PAGE were visualized by autoradiography. P1-derived (labeled arrows) and P2- and P3-derived (arrowheads) proteins are indicated. (F) Relative band intensity of P1 and P1-derived capsid proteins in control and GA-treated cells. Data show means and SEM of four independent experiments performed as in E. (*) p < 0.05, (**) p < 0.001 relative to control-treated cells by t-test.
Following viral entry, the positive-stranded genomic RNA is translated to produce the viral replication machinery, which in turn synthesizes more genomic RNA (Fig. 2A). To examine whether GA treatment targets translation and/or replication of the viral genome, we used a poliovirus replicon, PLuc, which carries the firefly luciferase gene in lieu of the capsid-coding sequence P1 (Herold and Andino 2000). Since PLuc translates and replicates like wild-type virus, luciferase activity provides a quantitative measure of viral translation and RNA replication (Andino et al. 1993). Importantly, GA treatment did not affect luciferase production in PLuc-transfected cells (Fig. 2C). Thus, GA does not inhibit translation or replication of the viral genome.
The poliovirus genome encodes a single ORF that is translated to yield a single polyprotein (Fields et al. 2001). Viral-encoded proteases—such as 2Apro, 3Cpro, or 3Cpro’s precursor, 3CD—liberate three proteins, P1, P2, and P3, that are further processed to generate the mature viral proteins. Because the PLuc replicon encodes all viral proteins except for the capsid precursor P1, these results suggest that Hsp90 is not required for the function of P2- and P3-derived proteins but rather participates in P1 function. P1 maturation involves processing into three capsid proteins: VP0, VP3, and VP1 (see Fig. 2D). VP0 is itself a precursor to VP4 and VP2, but is only cleaved at a late stage of particle assembly, probably following genome encapsidation (Basavappa et al. 1994).
The effect of Hsp90 inhibition on the processing and maturation of viral proteins was further examined using 35S-labeling of poliovirus-infected cells (Fig. 2E,F). Because poliovirus efficiently shuts off cellular translation, only viral proteins are radiolabeled under these conditions (Fields et al. 2001). As expected, both precursors and mature viral proteins were produced in control cells; on the other hand, treatment with GA produced a significant reduction in P1-derived capsid proteins (Fig. 2E,F). However, GA treatment did not affect processing of P2 or P3, in agreement with our findings using the PLuc replicon (Fig. 2C). Of note, GA also impaired P1 processing in rhinovirus-infected cells (Supplementary Fig. S2A,B), suggesting a conserved mode of action for GA within the picornavirus family.
The capsid protein P1 is a folding substrate of Hsp90
We next determined whether Hsp90 directly interacts with viral proteins. Coimmunoprecipitation experiments using 35S-labeled poliovirus-infected cells indicated that Hsp90 and its cochaperone p23 both associate with only one viral protein—the capsid precursor P1 (Fig. 3A; Supplementary Fig. S3). This result is consistent with the specific effect of GA on capsid protein production in infected cells. Interestingly, treatment with GA did not disrupt the Hsp90–P1 interaction but abrogated the association of P1 with p23, suggesting that p23 does not interact directly with P1 (Fig. 3A). This result supports previous findings that GA inhibits the p23–Hsp90 interaction, thus blocking progression through the Hsp90 chaperone cycle (Young et al. 2001; Picard 2002; Wegele et al. 2004; Ali et al. 2006). It thus appears that the nucleotide-dependent interaction of p23 with Hsp90 is required for P1 maturation.
Figure 3.
Hsp90 associates with the capsid precursor P1 and is required for its processing to mature capsid proteins. (A) Association of 35S-labeled P1 with Hsp90 and its cochaperone p23 in the presence or absence of GA, measured by immunoprecipitation. (NI) Nonimmune control. (B) Pulse-chase analysis of poliovirus proteins from infected cells grown in the presence or absence of GA. Total cytoplasmic extracts separated by SDS-PAGE were visualized by autoradiography. P1-derived (labeled arrows) and P2- and P3-derived (arrowheads) proteins are indicated. (C) Relative band intensity of P1 and P1-derived capsid proteins in control and GA-treated cells, calculated from B as percent of P1 at 15-min chase time point. (D) GA treatment promotes P1 degradation by the proteasome. The effect of GA on degradation of 35S-labeled P1, expressed in cells by infection with a recombinant vaccinia virus (VV-P1) (Ansardi et al. 1991), was examined in the presence or absence of the proteasome inhibitors LC and ALLN, and the lysosomal protease inhibitor E64. (E) Processing of in vitro translated P1 into capsid proteins by purified 3Cpro is blocked by GA even in the absence of proteasomal function. (CHX) Cycloheximide. (F) Role of Hsp90 in picornavirus capsid maturation. Hsp90 binds newly translated P1, probably in cooperation with Hsp70 (see Discussion; Macejak and Sarnow 1992). Together with ATP and its cofactors, such as p23, Hsp90 folds P1 to a processing-competent conformation (P1*) and protects it from proteasomal degradation. Upon cleavage by 3CPro, the mature capsid proteins no longer interact with Hsp90.
The effect of Hsp90 inhibition on P1 processing was next examined by pulse-chase analysis (Fig. 3B,C). Poliovirus-infected cells were subjected to a brief pulse of 35S-methionine/cysteine to label viral protein precursors and then chased with unlabeled amino acids to examine their processing kinetics. GA treatment did not affect the appearance of some viral proteins, such as 3CD or 2C, consistent with a specific action on P1 (Fig. 3B, right arrowheads). Notably, the kinetics of P1 production also appeared unaffected by the presence of GA (Fig. 3B,C). However, while P1 disappearance was largely unaffected by GA treatment, the appearance of the P1-derived mature capsid proteins was drastically reduced by Hsp90 inhibition (Fig. 3B,C). Together, these results indicate that association of P1 with Hsp90 and its cochaperone p23 are important for generation of mature capsid proteins.
Why does P1 disappear following GA treatment without yielding capsid proteins? We reasoned that if Hsp90 mediates P1 folding, its inhibition by GA could lead to P1 misfolding, which in turn would target P1 for elimination by the cellular quality-control machinery. To directly monitor the effect of Hsp90 inhibition on the fate of P1 in the absence of poliovirus-encoded processing proteases, P1 was expressed using a previously established vaccinia virus expression system (VV-P1) (Ansardi et al. 1991). In the absence of GA, P1 was stable; in contrast, it was readily degraded within 3 h of GA treatment (Fig. 3D). Inhibition of the proteasome pathway with lactacystin (LC) or N-acetyl-L-leucyl-L-leucyl-L-norleucinal (ALLN) protected P1 from degradation (Fig. 3D). On the other hand, addition of the lysosomal protease inhibitor E64 resulted in minimal protection from degradation. Thus, disruption of the Hsp90–p23 complex results in P1 misfolding, which targets it to the proteasome for degradation.
To better define the role of Hsp90 in P1 folding and maturation, a cell-free system was used. 35S-labeled P1 was generated by translation in rabbit reticulocyte lysate, and processing of the capsid precursor into capsid proteins was then monitored following addition of purified 3Cpro (Fig. 3E, lanes 1–5). Inhibition of Hsp90 by GA significantly reduced P1 processing (Fig. 3E, lanes 6–10). Notably, in contrast to our observations in intact cells (Fig. 3B,D), P1 did not disappear upon Hsp90 inhibition. This is consistent with findings that in translating reticulocyte lysates, proteasomal degradation is inhibited by free hemin (Haas and Rose 1981). Thus, even in the absence of proteasomal degradation, interaction with Hsp90 and p23 is still required for capsid protein maturation. These results suggest that Hsp90 does not simply protect P1 from proteasomal degradation but is required to fold it into a processing-competent conformation (Fig. 3F). Thus, inhibition of the Hsp90 chaperone cycle by GA leaves P1 in a misfolded conformation that cannot be recognized by 3Cpro and is instead degraded by the quality-control systems.
The virus cannot bypass the Hsp90 requirement
Having identified Hsp90 as essential for folding of a single protein in the picornavirus proteome, we examined whether the evolutionary capacity of poliovirus can be exploited to drive P1 to fold via an Hsp90-independent pathway. To force the emergence of Hsp90-independent viral variants, poliovirus was subjected to serial passage in the presence of GA (Fig. 4A). This approach was found to yield resistance to a diverse array of antiviral compounds in fewer than six passages (Gitlin et al. 2002; Crotty et al. 2004; Vignuzzi et al. 2006). As a control, we carried out a parallel selection regime to isolate BFA-resistant viruses, which optimally requires the accumulation of two amino acid substitutions in viral proteins (Fig. 4A; Crotty et al. 2004). Importantly, the selection of BFA-resistant variants was carried out at a BFA concentration that initially inhibited viral replication to a similar degree as GA (Fig. 4A, cf. GA and BFA inhibition on the untreated viral population). This ensured a similar selective pressure in both drug selection procedures. Following 10 passages in the presence of the inhibitors, the sensitivity of each virus to BFA and GA was examined. Strikingly, while the virus grown in BFA had become significantly BFA-resistant, no GA resistance was detected for the virus grown in the presence of the Hsp90 inhibitor (Fig. 4A). To extend this result, we next carried out an independent selection for GA-escape mutants for 20 passages in the presence of inhibitor, representing >50 replication cycles (Fig. 4B). Strikingly, no resistance to GA was observed under these conditions (Fig. 4B). Conservative theoretical considerations indicate that each passage in the presence of GA should generate at least 2.7 × 107 potential mutation events in P1 (Drake 1999), and that mutations offering even a 12% fitness advantage to growth in GA would suffice to completely dominate the viral population under our experimental conditions (M.F. Boni, pers. comm.). Given that the Hsp90 requirement of P1 folding cannot be circumvented by compensatory mutations even after so many generations, it appears that the protein folding pathway of P1 is under strong evolutionary constraints that limit its capacity to change its sequence without affecting the viability of the virus.
Figure 4.
Poliovirus cannot bypass the Hsp90 requirement. (A) Poliovirus can gain resistance to BFA but not GA within 10 passages. For each passage, 106 viruses (MOI of <0.2) were used to inoculate a new dish in the presence of BFA, GA, or no drug. After 10 passages, the sensitivity of each virus to BFA or GA was tested as in Figure 1B. Data represent the mean number of PFU per cell and SEM. (B) Poliovirus remains GA-sensitive following extensive serial passage in the presence of GA. For each passage, an MOI of 0.1–0.002 was used to inoculate a new dish of cells in the presence GA. Data are represented as the number of PFUs produced per cell. To facilitate comparison between experiments, data were standardized to yield the same number of PFUs for DMSO-treated conditions. (**) p < 0.01 relative to the virus passaged untreated by t-test.
Hsp90 inhibitors impair poliovirus replication in infected animals
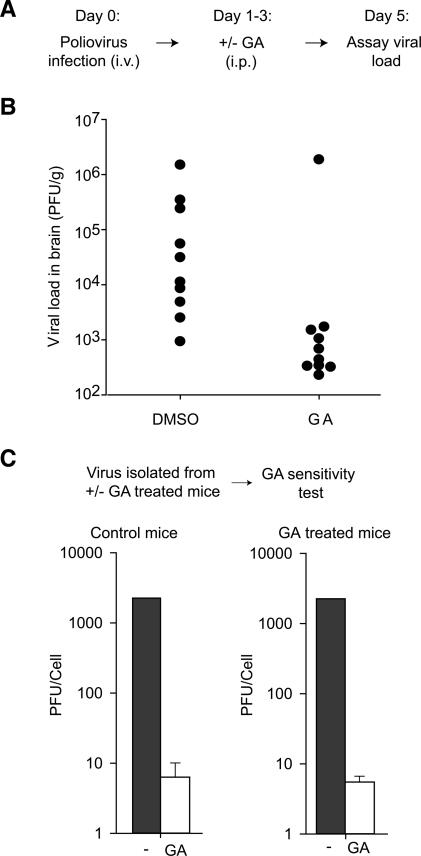
The inability of poliovirus to become Hsp90-independent suggests that protein folding inhibitors may provide an antiviral strategy that can function in vivo without eliciting drug resistance. Despite their use in clinical trials for cancer treatment, the ability of Hsp90 inhibitors to reduce viral replication in infected animals has not been addressed (Dai and Whitesell 2005). We therefore tested whether Hsp90 inhibitors can impair poliovirus replication in infected mice. We initially examined whether GA can inhibit the replication of poliovirus in a transgenic mouse model of poliomyelitis extensively used to study the pathogenesis of poliovirus (Crotty et al. 2002; Vignuzzi et al. 2006). Beginning on the day of infection, the Hsp90 inhibitor GA was administered systemically for 4 d using a dose and formulation previously shown to inhibit an Hsp90-dependent process in mice (Fig. 5A; Bucci et al. 2000). GA treatment significantly reduced the viral load in the central nervous system (CNS) of poliovirus-infected mice compared with vehicle-treated mice in two independent experiments (Fig. 5B; data not shown).
Figure 5.
GA inhibits viral replication in poliovirus-infected animals without eliciting drug resistance. (A) Outline of the experiment. (B) Viral load in the brains of poliovirus-infected cPVR transgenic mice treated with vehicle or GA is expressed as number of PFUs per gram of brain (n = 10 per group, p < 0.01 by Wilcoxon two-sample test). (C) Viral populations recovered from GA-treated animals remain GA sensitive. Poliovirus isolated from the brains of infected animals from B was used to infect HeLa S3 cells at a low MOI (10−4) in the presence or absence of 1 μM GA. Virus production was measured after 48 h by standard plaque assay. Data represent the average number of PFUs produced per cell from all 10 GA-treated animals and four control animals.
Since the infected animals may provide several alternative microenvironments for viral evolution, we examined whether the viral population recovered from GA-treated animals 5 d post-infection had acquired drug resistance (Fig. 5C). Notably, viruses isolated from control and GA-treated animals were equally sensitive to the inhibitor; thus, no drug resistance arose in infected animals during GA treatment (Fig. 5C).
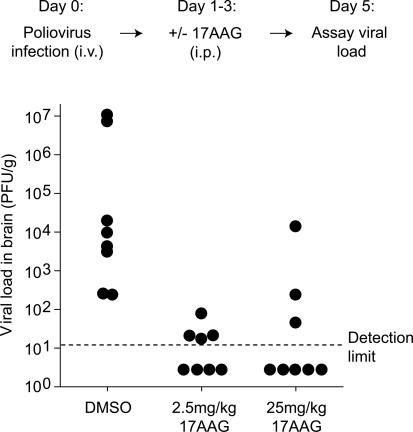
We next examined the antiviral activity of a first-generation GA derivate, 17-(allylamino)-17-demethoxygeldanamycin (17AAG), which is better tolerated than GA, more effective in crossing the blood–brain barrier, and is currently in clinical trials for cancer treatment (Dai and Whitesell 2005; Waza et al. 2005). While poliovirus was readily detected in all vehicle-treated animals, daily 17AAG treatment dramatically reduced the viral load in the CNS (Fig. 6). In fact, the virus decreased to undetectable levels in four of eight mice receiving a lower 17AAG dose and in five of eight mice receiving a higher dose (Fig. 6). Importantly, the short course of 17AAG treatment did not result in any apparent toxicity to the treated animals (data not shown). The improved pharmacological properties of 17AAG over GA may account for its dramatic ability to reduce the viral load in the CNS of infected animals even at the lower dose used here. Together, these results provide a proof-of-principle for the hypothesis that inhibitors of chaperone function can effectively block viral replication in infected animals.
Figure 6.
17AAG inhibits viral replication in poliovirus-infected animals. Viral load in the brains of poliovirus-infected cPVR transgenic mice treated with vehicle or 17AAG expressed as in Figure 5B (n = 8 per group, p < 0.001 for 2.5 mg/kg group and p < 0.005 for 25 mg/kg group by Wilcoxon two-sample test). Animals with no detectable virus (four of eight mice treated with 2.5 mg/kg 17AAG and five of eight mice treated with 25 mg/kg 17AAG) are plotted below the hatched line indicating the detection limit.
Discussion
Here we establish that picornavirus replication is critically dependent on the function of the Hsp90 chaperone machine. Investigation into the molecular basis of the Hsp90 dependence uncovered a requirement for this chaperone in facilitating the maturation of a single viral protein, the large multidomain capsid protein P1. Upon Hsp90 inhibition, P1 misfolds and is targeted for proteasomal degradation, while the other viral proteins remain unaffected.
Role of Hsp90 in picornavirus capsid folding and assembly
The formation of viral capsids presents several structural challenges to the capsid proteins that may account for the inability of poliovirus to become Hsp90-independent. The viral capsid must be stable in the harsh extracellular environment yet energetically “spring-loaded” to disassemble upon reinfection (Hogle 2002). Although complete capsid folding must await the final structure, the newly translated capsid precursors must attain a conformation that is competent for proteolytic processing and assembly, and is protected from degradation. Following processing, capsid proteins must assemble into a large, highly regular oligomeric structure and encapsulate the viral genome. Moreover, the capsid assembly process must occur efficiently, as misassembly or incorporation of even a few misfolded subunits into the mature capsid can exert a dominant-negative effect on the whole particle (Crowder and Kirkegaard 2005). It appears that Hsp90 fulfills many of these functions for picornavirus capsid proteins; it maintains P1 in a processing-competent conformation and protects it from proteasomal degradation. Upon cleavage, the capsid proteins no longer interact with Hsp90, suggesting that maturation occludes the chaperone-binding sites. Although the function of Hsp90 in cells remains poorly understood, it is thought to maintain and remodel the active conformation of a unique set of cellular proteins including steroid hormone receptors and protein kinases (Young et al. 2001; Picard 2002; Prodromou and Pearl 2003; Riggs et al. 2004; Wegele et al. 2004). Hsp90 may analogously facilitate the maturation of P1 to a processing-competent conformation. In uninfected cells, Hsp90 receives its cellular clients from Hsp70 in a partially folded state (Pratt and Toft 2003; Riggs et al. 2004; Wegele et al. 2004). Interestingly, P1 has been previously reported to interact with Hsp70 (Macejak and Sarnow 1992). Thus, P1 appears to use the canonical Hsp90-mediated folding pathway described for other client proteins (Fig. 3F).
Evolutionary constraints on the chaperone-mediated P1 folding pathway
The high mutation rate and extreme adaptability of RNA viruses provided a unique system to test the evolutionary constraints of a protein folding pathway. Since P1 is the only poliovirus protein to require Hsp90, we examined whether growth under conditions of Hsp90 inhibition could drive the evolution of viral variants with an altered Hsp90-independent P1 folding pathway. Strikingly, despite the ability of poliovirus to rapidly gain resistance to virtually all antiviral compounds tested to date, the virus was unable to become resistant to Hsp90 inhibitors, either in vitro or in infected animals. We propose that changing the folding pathway of P1 requires such extensive alterations in the polypeptide sequence that sufficient mutations would never arise, even in viruses with a high mutation rate. Alternatively, acquiring mutations that decrease the Hsp90 dependence of capsid maturation may require transiting through several decreased fitness stages, rendering the mutant virus unable to survive, even under selective pressure. In either case, it appears that the folding pathway of a large chaperone-dependent protein such as P1 is highly constrained.
Molecular chaperones as novel antiviral targets
Our findings uncover a potential limitation to the evolutionary capacity of viruses that may be exploited as a therapeutic strategy. Indeed, we demonstrate the ability of Hsp90 inhibitors to act as antiviral compounds in animals without generating drug-resistant variants (Figs. 5, 6). The potential of Hsp90 inhibitors as general anti-picornavirus compounds is of interest, as no antivirals are licensed for the treatment of picornavirus infections. These are extremely common and can result in serious disease, including paralysis, hepatitis, and myocarditis. Importantly, with the scheduled halting of routine poliovirus vaccination using the live attenuated poliovirus vaccine in the near future, an anti-poliovirus drug may become important for treating resurging infections in an increasingly susceptible, immunologically naive population (Committee on Development of a Polio Antiviral and Its Potential Role in Global Poliomyelitis Eradication 2006). Since several Hsp90 inhibitors are presently undergoing clinical trials, this result is of particular interest.
Considering the general complexity of viral capsids and other viral protein complexes, our conclusions may extend beyond the picornavirus family. Indeed, several unrelated viruses are reported to encode proteins that depend on Hsp90 for their activity (Hu et al. 1997; Hung et al. 2002; Li et al. 2004; Basha et al. 2005; Burch and Weller 2005; Janin 2005; Kampmueller and Miller 2005; Okamoto et al. 2006). Accordingly, chaperone inhibitors may hold promise as broad antiviral agents. Since rapidly replicating viruses—particularly lytic viruses—must produce large quantities of viral proteins to successfully propagate, they are likely to impose a heavy load on the specific chaperones required to fold them. Such conditions likely render viruses hypersensitive to inhibitors of these chaperones, disproportionately impairing viral replication with relatively low toxicity to uninfected cells. Given that folding of cellular proteins in uninfected cells is distributed among different chaperones (Frydman 2001; Young et al. 2004), partial inhibition of a single chaperone system, such as Hsp90, does not reduce cell viability under normal growth conditions (Borkovich et al. 1989). Indeed, pharmacological inhibitors of Hsp90 with tolerable toxicity, such as 17AAG, are currently being evaluated in clinical trials as anti-cancer drugs (Dai and Whitesell 2005). Our results suggest that a short course of 17AAG may provide an effective antiviral strategy, without presenting many of the adverse effects associated with the long-term administration of these drugs required for anti-cancer treatments.
Implication for the evolution of chaperone-dependent proteins
Our results have interesting implications for how chaperone dependence influences protein evolution. Why certain proteins, such as P1, require chaperones to fold while others do not is poorly understood. For some proteins, chaperones appear to merely increase the efficiency and/or yield of folding by preventing off-pathway interactions. On the other hand, other proteins exhibit stringent requirements for specific chaperones in order to fold (Kerner et al. 2005).
The information for the native state is encoded in the primary protein sequence. However, to fold spontaneously a protein must also encode in its sequence the information for a smooth, energetically favorable folding pathway. Using chaperone assistance for folding may relieve some proteins from this constraint. As a result, the evolutionary potential of these chaperone-dependent proteins would increase, allowing for a greater degree of functional and structural plasticity. This may be particularly advantageous for protein complexes such as viral capsids, given their complexity. In the particular case of Hsp90, an added advantage lies in its ability to function as a buffer against destabilizing mutations (Whitesell and Lindquist 2005). Such a buffering mechanism would be of particular benefit to viral capsid proteins whose correct folding is essential for assembly but that are under surveillance by the immune system and must constantly mutate surface-exposed epitopes. Furthermore, Hsp90 dependence comes at no cost for the virus, as this chaperone is abundant and ubiquitous. However, an important conclusion from our study is that while chaperones may offer proteins the ability to increase their structural complexity, this strategy may eventually lead to an evolutionary trap, as the obligate chaperone dependence for folding cannot be altered without a dramatic reduction in fitness. These constraints on chaperone-mediated folding may provide an antiviral approach refractory to development of drug resistance.
Materials and methods
Cells, viruses, and reagents
HeLa S3 cells, TSA201, Vero, and human foreskin fibroblasts were cultured under standard procedures. For experiments with human rhinovirus 14 (HRV14), cells were grown at 33°C. GA, 17AAG (LC laboratories), LC (EMD Biosciences), ALLN (Calbiochem), and BFA (LC Laboratories) were dissolved in DMSO and E64 (Boehringer Mannheim) in 70% ethanol. GA was obtained from the National Cancer Institute, Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis. All GA experiments were done under dim light conditions. Poliovirus Mahoney type 1 strain (PV) was generated from plasmid pRib (+)XpA as previously described (Herold and Andino 2000). HRV14 was obtained from American Tissue Culture Collection. The coxsackievirus B3 (CVB3) construct (Klump et al. 1990), a kind gift of Dr. Willem Melchers from the Radboud University Nijmegen, Nijmegen, The Netherlands, was generated as described (Herold and Andino 2000). Vaccinia virus P1 (VV-P1) (Ansardi et al. 1991) was a kind gift of Dr. Casey Morrow from the University of Alabama at Birmingham, Birmingham, AL. Egg phosphatidylcholine was purchased from Avanti Polar Lipids.
Viral infections
PV, CVB3, or HRV14 was allowed to adsorb to cells for 30 min at 37°C (for PV, CVB3) or 33°C (for HRV14), after which cells were washed with PBS and incubated in culture media. VV-P1 infections were carried out for 1.5 h at 37°C. Both VV-P1 and CVB3 infections were carried out in media containing low serum concentrations (2% FCS).
Effect of GA on viral replication in cultured cells
HeLa S3, TSA201, or HFF cells were infected at a multiplicity of infection (MOI) of 1–5 and plated in the presence or absence of GA. A 45-min preincubation step with GA was included in Figure 1, B and C. Virus production was measured by standard plaque assay (PV, HRV14) or end-point titration on Vero cells (CVB3).
In vitro transcription and RNA electroporation
Poliovirus genomic or replicon RNA was transcribed from pRib (+)XpA or pRib (+)RLuc plasmids, respectively, as previously described (Herold and Andino 2000). For in vitro translation, P1 was amplified from pRib (+)XpA by PCR and cloned into pCDNA3.1 (+) using HindIII and XhoI restriction sites. Capped RNA was generated using the MegaScript T7 kit (Ambion) after linearization with XhoI following the manufacturer’s protocol. For electroporations, HeLa S3 cells (4 × 106) were pulsed with 10 μg of RNA in 0.8 mL of Ca2+/Mg2+-free PBS using a BTX electroporator set to 950 μF, 128 Ω, and 300 V in a 0.4-cm cuvette. When indicated, GA was used at 2 μM concentrations.
Radiolabeling and immunoprecipitation
Cells were infected at an MOI of >25. GA (0.5 μM) was added 2 h post-infection and maintained for the remainder of the experiment. For steady-state pulse experiments, cells were incubated with 30 μCi/mL 35S-methionine/cysteine for 2 h in media lacking these amino acids. Cells were then washed in PBS, lysed in lysis buffer (25 mM Tris at pH 7.5, 150 mM NaCl, 1% NP-40, protease inhibitor cocktail [Sigma]), and analyzed by 12% SDS-PAGE and autoradiography on a Typhoon PhosphorImager (Amersham Biosciences). For pulse-chase experiments, cells were starved for 15 min at 3 h post-infection, incubated in media containing 35S-methionine/cysteine for 5 min, and chased in media containing the amino acids for the indicated time prior to analysis as above. For experiments with VV-P1, cells were infected at an MOI of 10, radiolabeled as above for 1.5 h, and washed, and media containing LC (20 μM), ALLN (10 μM), or E64 (25 μM) was added. GA (0.5 μM) was added 3 h later, and cells were incubated for an additional 3 h prior to lysis in RIPA buffer and immunoprecipitation with polyclonal αVirion-N1 antibodies. All quantifications were performed using ImageQuant software (Amersham Biosciences).
Chaperone immunoprecipitations
Confluent 10-cm dishes were infected with poliovirus at an MOI of 50. DMSO or GA (1 μM) was added 2 h post-infection and maintained for the remainder of the experiment. Four hours post-infection, cells were starved and radiolabeled for 30 min as above prior to lysis in Hsp90 Lysis Buffer (20 mM HEPES at pH 7.5, 100 mM NaCl, 20 mM sodium molybdate, 5 mM EDTA, 10% glycerol, 0.01% NaN3, protease inhibitor cocktail [Sigma]) containing 10 mg/mL BSA. Nuclei were removed by centrifugation, and supernatants were incubated with antibodies to Hsp90 (SPA840; Stressgen), p23 (JJ3), or a control antibody, for 1 h on ice. Lysates were then incubated with protein G-Sepharose (Amersham Biosciences) for 45 min, washed four times in Hsp90 buffer, and analyzed by 12% SDS-PAGE and autoradiography.
In vitro translation and 3Cpro-HA purification
In vitro transcribed P1 RNA was translated in Flexi Rabbit Reticulocyte Lysate (Promega) following the manufacturer’s instructions. Reactions were stopped by incubation with cycloheximide (0.1 mg/mL) and RNase A (80 μg/mL) for 5 min followed by addition of DMSO, GA (0.5 mM), or EDTA (15 mM) for 10 min at 30°C. Bacterially purified 3Cpro was then added (0.6 mg/mL) for the indicated time, and processing was analyzed by SDS-PAGE and autoradiography as above. 3Cpro was purified via a C-terminal HIS6 tag as described below.
Protein purification
To purify the protease, 3Cpro was PCR-amplified from pRib (+)XpA with NcoI and XhoI restriction sites, cloned into a pET28-a (+) vector (Novagen) in frame with a C-terminal HIS6 tag, and used to transform BL21 (DE3) bacteria. At 0.6 OD600, a 1.2-L culture was induced with 100 μM IPTG for 3 h at 37°C. Purification was performed using TALON metal affinity resin (BD Biosciences) following standard protocols. Purified protein was dialyzed against dialysis buffer (20 mM HEPES, 100 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM EDTA at pH 7.6) and frozen at −80°C.
Animal experiments
On day 0, 6- to 10-wk-old male and female cPVR transgenic mice (Crotty et al. 2002) were injected with the Hsp90 inhibitors or vehicle (i.p.) and infected 4–6 h later with 107 plaque-forming units (PFU) of poliovirus by tail vein injection. Hsp90 inhibitors or vehicle alone were administered daily for the subsequent 3 d. Virus production was determined on day 5 as previously described (Crotty et al. 2002). For GA experiments, each injection contained 2.5 μg (0.1 mg/kg) of GA in 5 μL of DMSO and 45 μL of PEG 400:H2O (1:1). For experiments with 17AAG, each injection contained either 0.05 mg (2.5 mg/kg) or 0.5 mg (25 mg/kg) of 17AAG in 5 μL of DMSO and 45 μL of 2% egg phosphatidylcholine and 5% dextrose (NSC 704,057). All animal experiments were in accordance with institutional guidelines.
Statistical analysis
All data for in vitro and tissue culture experiments are represented as the mean of the indicated number of experiments. Error bars indicate SEM. Significance was tested with a two-tail t-test. For in vivo experiments, a two-tail Wilcoxon two-sample test was employed using the NPAR1WAY procedure on SAS software.
Acknowledgments
We thank Drs. John Boothroyd, Casey Morrow, and Willem Melchers for their generous gift of cells and reagents. We also thank Dr. Lisbeth A. Guethlein for help with statistical analysis, Dr. Maciej F. Boni for help with theoretical probability of emergence of viral resistance, and members of the Andino and Frydman laboratories for useful discussions and comments on the manuscript. This work was supported by NIH grants GM56433 (to J.F.) and AI40085 (to R.A.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1505307
References
- Aberham C., Weber S., Phares W., Weber S., Phares W., Phares W. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. J. Virol. 1996;70:3536–3544. doi: 10.1128/jvi.70.6.3536-3544.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M.M., Roe S.M., Vaughan C.K., Meyer P., Panaretou B., Piper P.W., Prodromou C., Pearl L.H., Roe S.M., Vaughan C.K., Meyer P., Panaretou B., Piper P.W., Prodromou C., Pearl L.H., Vaughan C.K., Meyer P., Panaretou B., Piper P.W., Prodromou C., Pearl L.H., Meyer P., Panaretou B., Piper P.W., Prodromou C., Pearl L.H., Panaretou B., Piper P.W., Prodromou C., Pearl L.H., Piper P.W., Prodromou C., Pearl L.H., Prodromou C., Pearl L.H., Pearl L.H. Crystal structure of an Hsp90–nucleotide–p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R., Rieckhof G.E., Achacoso P.L., Baltimore D., Rieckhof G.E., Achacoso P.L., Baltimore D., Achacoso P.L., Baltimore D., Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansardi D.C., Porter D.C., Morrow C.D., Porter D.C., Morrow C.D., Morrow C.D. Coinfection with recombinant vaccinia viruses expressing poliovirus P1 and P3 proteins results in polyprotein processing and formation of empty capsid structures. J. Virol. 1991;65:2088–2092. doi: 10.1128/jvi.65.4.2088-2092.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavappa R., Syed R., Flore O., Icenogle J.P., Filman D.J., Hogle J.M., Syed R., Flore O., Icenogle J.P., Filman D.J., Hogle J.M., Flore O., Icenogle J.P., Filman D.J., Hogle J.M., Icenogle J.P., Filman D.J., Hogle J.M., Filman D.J., Hogle J.M., Hogle J.M. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: Structure of the empty capsid assembly intermediate at 2.9 Å resolution. Protein Sci. 1994;3:1651–1669. doi: 10.1002/pro.5560031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha W., Kitagawa R., Uhara M., Imazu H., Uechi K., Tanaka J., Kitagawa R., Uhara M., Imazu H., Uechi K., Tanaka J., Uhara M., Imazu H., Uechi K., Tanaka J., Imazu H., Uechi K., Tanaka J., Uechi K., Tanaka J., Tanaka J. Geldanamycin, a potent and specific inhibitor of Hsp90, inhibits gene expression and replication of human cytomegalovirus. Antivir. Chem. Chemother. 2005;16:135–146. doi: 10.1177/095632020501600206. [DOI] [PubMed] [Google Scholar]
- Borkovich K.A., Farrelly F.W., Finkelstein D.B., Taulien J., Lindquist S., Farrelly F.W., Finkelstein D.B., Taulien J., Lindquist S., Finkelstein D.B., Taulien J., Lindquist S., Taulien J., Lindquist S., Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I., van Anken E., van Anken E. Folding of viral envelope glycoproteins in the endoplasmic reticulum. Traffic. 2000;1:533–539. doi: 10.1034/j.1600-0854.2000.010702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci M., Roviezzo F., Cicala C., Sessa W.C., Cirino G., Roviezzo F., Cicala C., Sessa W.C., Cirino G., Cicala C., Sessa W.C., Cirino G., Sessa W.C., Cirino G., Cirino G. Geldanamycin, an inhibitor of heat shock protein 90 (Hsp90) mediated signal transduction has anti-inflammatory effects and interacts with glucocorticoid receptor in vivo. Br. J. Pharmacol. 2000;131:13–16. doi: 10.1038/sj.bjp.0703549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch A.D., Weller S.K., Weller S.K. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone hsp90 for proper localization to the nucleus. J. Virol. 2005;79:10740–10749. doi: 10.1128/JVI.79.16.10740-10749.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on Development of a Polio Antiviral Its Potential Role in Global Poliomyelitis Eradication . Exploring the Role of Antiviral Drugs in the Eradication of Polio. The National Academies Press; Washington, DC.: 2006. [Google Scholar]
- Crotty S., Hix L., Sigal L.J., Andino R., Hix L., Sigal L.J., Andino R., Sigal L.J., Andino R., Andino R. Poliovirus pathogenesis in a new poliovirus receptor transgenic mouse model: Age-dependent paralysis and a mucosal route of infection. J. Gen. Virol. 2002;83:1707–1720. doi: 10.1099/0022-1317-83-7-1707. [DOI] [PubMed] [Google Scholar]
- Crotty S., Saleh M.C., Gitlin L., Beske O., Andino R., Saleh M.C., Gitlin L., Beske O., Andino R., Gitlin L., Beske O., Andino R., Beske O., Andino R., Andino R. The poliovirus replication machinery can escape inhibition by an antiviral drug that targets a host cell protein. J. Virol. 2004;78:3378–3386. doi: 10.1128/JVI.78.7.3378-3386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder S., Kirkegaard K., Kirkegaard K. Trans-dominant inhibition of RNA viral replication can slow growth of drug-resistant viruses. Nat. Genet. 2005;37:701–709. doi: 10.1038/ng1583. [DOI] [PubMed] [Google Scholar]
- Cuconati A., Molla A., Wimmer E., Molla A., Wimmer E., Wimmer E. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J. Virol. 1998;72:6456–6464. doi: 10.1128/jvi.72.8.6456-6464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Whitesell L., Whitesell L. HSP90: A rising star on the horizon of anticancer targets. Future Oncol. 2005;1:529–540. doi: 10.2217/14796694.1.4.529. [DOI] [PubMed] [Google Scholar]
- Doedens J., Maynell L.A., Klymkowsky M.W., Kirkegaard K., Maynell L.A., Klymkowsky M.W., Kirkegaard K., Klymkowsky M.W., Kirkegaard K., Kirkegaard K. Secretory pathway function, but not cytoskeletal integrity, is required in poliovirus infection. Arch. Virol. Suppl. 1994;9:159–172. doi: 10.1007/978-3-7091-9326-6_16. [DOI] [PubMed] [Google Scholar]
- Domingo E., Holland J.J., Holland J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- Drake J.W. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann. N. Y. Acad. Sci. 1999;870:100–107. doi: 10.1111/j.1749-6632.1999.tb08870.x. [DOI] [PubMed] [Google Scholar]
- Elena S.F., Carrasco P., Daros J.A., Sanjuan R., Carrasco P., Daros J.A., Sanjuan R., Daros J.A., Sanjuan R., Sanjuan R. Mechanisms of genetic robustness in RNA viruses. EMBO Rep. 2006;7:168–173. doi: 10.1038/sj.embor.7400636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B.N., Knipe D.M., Howley P.M., Griffin D.E., Knipe D.M., Howley P.M., Griffin D.E., Howley P.M., Griffin D.E., Griffin D.E. Fields’ virology. Lippincott Williams & Wilkins; Philadelphia: 2001. [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu. Rev. Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Gitlin L., Karelsky S., Andino R., Karelsky S., Andino R., Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- Haas A.L., Rose I.A., Rose I.A. Hemin inhibits ATP-dependent ubiquitin-dependent proteolysis: Role of hemin in regulating ubiquitin conjugate degradation. Proc. Natl. Acad. Sci. 1981;78:6845–6848. doi: 10.1073/pnas.78.11.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Andino R., Andino R. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle J.M. Poliovirus cell entry: Common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Choi G., Park S., Chung A.S., Hunter E., Rhee S.S., Choi G., Park S., Chung A.S., Hunter E., Rhee S.S., Park S., Chung A.S., Hunter E., Rhee S.S., Chung A.S., Hunter E., Rhee S.S., Hunter E., Rhee S.S., Rhee S.S. Type D retrovirus Gag polyprotein interacts with the cytosolic chaperonin TRiC. J. Virol. 2001;75:2526–2534. doi: 10.1128/JVI.75.6.2526-2534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Anselmo D., Anselmo D. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: Posttranslational activation by Hsp90. J. Virol. 2000;74:11447–11455. doi: 10.1128/jvi.74.24.11447-11455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Toft D.O., Seeger C., Toft D.O., Seeger C., Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung J.J., Chung C.S., Chang W., Chung C.S., Chang W., Chang W. Molecular chaperone Hsp90 is important for vaccinia virus growth in cells. J. Virol. 2002;76:1379–1390. doi: 10.1128/JVI.76.3.1379-1390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irurzun A., Perez L., Carrasco L., Perez L., Carrasco L., Carrasco L. Involvement of membrane traffic in the replication of poliovirus genomes: Effects of brefeldin A. Virology. 1992;191:166–175. doi: 10.1016/0042-6822(92)90178-r. [DOI] [PubMed] [Google Scholar]
- Janin Y.L. Heat shock protein 90 inhibitors. A text book example of medicinal chemistry? J. Med. Chem. 2005;48:7503–7512. doi: 10.1021/jm050759r. [DOI] [PubMed] [Google Scholar]
- Kamal A., Thao L., Sensintaffar J., Zhang L., Boehm M.F., Fritz L.C., Burrows F.J., Thao L., Sensintaffar J., Zhang L., Boehm M.F., Fritz L.C., Burrows F.J., Sensintaffar J., Zhang L., Boehm M.F., Fritz L.C., Burrows F.J., Zhang L., Boehm M.F., Fritz L.C., Burrows F.J., Boehm M.F., Fritz L.C., Burrows F.J., Fritz L.C., Burrows F.J., Burrows F.J. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- Kampmueller K.M., Miller D.J., Miller D.J. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J. Virol. 2005;79:6827–6837. doi: 10.1128/JVI.79.11.6827-6837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner M.J., Naylor D.J., Ishihama Y., Maier T., Chang H.C., Stines A.P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Naylor D.J., Ishihama Y., Maier T., Chang H.C., Stines A.P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Ishihama Y., Maier T., Chang H.C., Stines A.P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Maier T., Chang H.C., Stines A.P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Chang H.C., Stines A.P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Stines A.P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Frishman D., Hayer-Hartl M., Mann M., Hayer-Hartl M., Mann M., Mann M., et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Klump W.M., Bergmann I., Muller B.C., Ameis D., Kandolf R., Bergmann I., Muller B.C., Ameis D., Kandolf R., Muller B.C., Ameis D., Kandolf R., Ameis D., Kandolf R., Kandolf R. Complete nucleotide sequence of infectious Coxsackievirus B3 cDNA: Two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J. Virol. 1990;64:1573–1583. doi: 10.1128/jvi.64.4.1573-1583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.H., Tao P.Z., Liu Y.Z., Jiang J.D., Tao P.Z., Liu Y.Z., Jiang J.D., Liu Y.Z., Jiang J.D., Jiang J.D. Geldanamycin, a ligand of heat shock protein 90, inhibits the replication of herpes simplex virus type 1 in vitro. Antimicrob. Agents Chemother. 2004;48:867–872. doi: 10.1128/AAC.48.3.867-872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macejak D.G., Sarnow P., Sarnow P. Association of heat shock protein 70 with enterovirus capsid precursor P1 in infected human cells. J. Virol. 1992;66:1520–1527. doi: 10.1128/jvi.66.3.1520-1527.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.P. Recruitment of Hsp70 chaperones: A crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 2005;153:1–46. doi: 10.1007/s10254-004-0025-5. [DOI] [PubMed] [Google Scholar]
- Maynell L.A., Kirkegaard K., Klymkowsky M.W., Kirkegaard K., Klymkowsky M.W., Klymkowsky M.W. Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 1992;66:1985–1994. doi: 10.1128/jvi.66.4.1985-1994.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick J.L., Crowther D., Barrera-Oro J., Crowther D., Barrera-Oro J., Barrera-Oro J. Rapid development of drug-resistant mutants of poliovirus. Science. 1961;134:557. doi: 10.1126/science.134.3478.557. [DOI] [PubMed] [Google Scholar]
- Murata T., Goshima F., Koshizuka T., Takakuwa H., Nishiyama Y., Goshima F., Koshizuka T., Takakuwa H., Nishiyama Y., Koshizuka T., Takakuwa H., Nishiyama Y., Takakuwa H., Nishiyama Y., Nishiyama Y. A single amino acid substitution in the ICP27 protein of herpes simplex virus type 1 is responsible for its resistance to leptomycin B. J. Virol. 2001;75:1039–1043. doi: 10.1128/JVI.75.2.1039-1043.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T., Nishimura Y., Ichimura T., Suzuki K., Miyamura T., Suzuki T., Moriishi K., Matsuura Y., Nishimura Y., Ichimura T., Suzuki K., Miyamura T., Suzuki T., Moriishi K., Matsuura Y., Ichimura T., Suzuki K., Miyamura T., Suzuki T., Moriishi K., Matsuura Y., Suzuki K., Miyamura T., Suzuki T., Moriishi K., Matsuura Y., Miyamura T., Suzuki T., Moriishi K., Matsuura Y., Suzuki T., Moriishi K., Matsuura Y., Moriishi K., Matsuura Y., Matsuura Y. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 2006;25:5015–5025. doi: 10.1038/sj.emboj.7601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer J.K., Kirkegaard K., Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. 2003;100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay D., Zambon M., Zambon M. Antiviral drug resistance. BMJ. 1998;317:660–662. doi: 10.1136/bmj.317.7159.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt W.B., Toft D.O., Toft D.O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Prodromou C., Pearl L.H., Pearl L.H. Structure and functional relationships of Hsp90. Curr. Cancer Drug Targets. 2003;3:301–323. doi: 10.2174/1568009033481877. [DOI] [PubMed] [Google Scholar]
- Riggs D.L., Cox M.B., Cheung-Flynn J., Prapapanich V., Carrigan P.E., Smith D.F., Cox M.B., Cheung-Flynn J., Prapapanich V., Carrigan P.E., Smith D.F., Cheung-Flynn J., Prapapanich V., Carrigan P.E., Smith D.F., Prapapanich V., Carrigan P.E., Smith D.F., Carrigan P.E., Smith D.F., Smith D.F. Functional specificity of co-chaperone interactions with Hsp90 client proteins. Crit. Rev. Biochem. Mol. Biol. 2004;39:279–295. doi: 10.1080/10409230490892513. [DOI] [PubMed] [Google Scholar]
- Sarafianos S.G., Das K., Hughes S.H., Arnold E., Das K., Hughes S.H., Arnold E., Hughes S.H., Arnold E., Arnold E. Taking aim at a moving target: Designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr. Opin. Struct. Biol. 2004;14:716–730. doi: 10.1016/j.sbi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M., Stone J.K., Arnold J.J., Cameron C.E., Andino R., Stone J.K., Arnold J.J., Cameron C.E., Andino R., Arnold J.J., Cameron C.E., Andino R., Cameron C.E., Andino R., Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waza M., Adachi H., Katsuno M., Minamiyama M., Sang C., Tanaka F., Inukai A., Doyu M., Sobue G., Adachi H., Katsuno M., Minamiyama M., Sang C., Tanaka F., Inukai A., Doyu M., Sobue G., Katsuno M., Minamiyama M., Sang C., Tanaka F., Inukai A., Doyu M., Sobue G., Minamiyama M., Sang C., Tanaka F., Inukai A., Doyu M., Sobue G., Sang C., Tanaka F., Inukai A., Doyu M., Sobue G., Tanaka F., Inukai A., Doyu M., Sobue G., Inukai A., Doyu M., Sobue G., Doyu M., Sobue G., Sobue G. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat. Med. 2005;11:1088–1095. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- Wegele H., Muller L., Buchner J., Muller L., Buchner J., Buchner J. Hsp70 and Hsp90—A relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- Whitesell L., Lindquist S.L., Lindquist S.L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Young J.C., Moarefi I., Hartl F.U., Moarefi I., Hartl F.U., Hartl F.U. Hsp90: A specialized but essential protein-folding tool. J. Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.C., Agashe V.R., Siegers K., Hartl F.U., Agashe V.R., Siegers K., Hartl F.U., Siegers K., Hartl F.U., Hartl F.U. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]