Abstract
TRF2 is a telomere-binding protein with roles in telomere protection and telomere-length regulation. The fact that TRF2 is up-regulated in some human tumors suggests a role of TRF2 in cancer. Mice that overexpress TRF2 in the skin, K5TRF2 mice, show critically short telomeres and are susceptible to UV-induced carcinogenesis as a result of deregulated XPF/ERCC1 activity, a nuclease involved in UV damage repair. Here we demonstrate that, when in combination with telomerase deficiency, TRF2 acts as a very potent oncogene in vivo. In particular, we show that telomerase deficiency dramatically accelerates TRF2-induced epithelial carcinogenesis in K5TRF2/Terc−/− mice, coinciding with increased chromosomal instability and DNA damage. Telomere recombination is also increased in these mice, suggesting that TRF2 favors the activation of alternative telomere maintenance mechanisms. Together, these results demonstrate that TRF2 increased expression is a potent oncogenic event that along with telomerase deficiency accelerates carcinogenesis, coincidental with a derepression of telomere recombination. These results are of particular relevance given that TRF2 is up-regulated in some human cancers. Furthermore, these data suggest that telomerase inhibition might not be effective to cease the growth of TRF2-overexpressing tumors.
Keywords: TRF2, cancer, mouse models, recombination, telomerase, telomeres
Telomeres are heterochromatic regions at the ends of chromosomes that consist of tandem TTAGGG repeats and of associated proteins (Chan and Blackburn, 2002; de Lange 2002; Garcia-Cao et al. 2004). The telomeric chromatin protects the single-stranded 3′ chromosome end (G-strand overhang) from degradation and repair activities, and it is essential for chromosome stability (de Lange 2005). Critical telomere shortening and loss of function of telomere-binding proteins result in loss of telomere protection, end-to-end chromosome fusions, and cell cycle arrest or apoptosis (van Steensel et al. 1998; Goytisolo and Blasco 2002; de Lange 2005). Telomere shortening is envisioned as a potent tumor suppressor mechanism (González-Suárez et al. 2000; Blasco and Hahn 2003; Blasco 2005). Telomerase activity, in turn, is able to elongate telomeres in those cells where it is highly expressed, such as the vast majority of human cancers, and it is associated with cell immortalization (Shay and Wright 2006). Mice that lack telomerase activity show premature loss of organismal viability when their telomeres become critically short and are resistant to cancer (Lee et al. 1998; Greenberg et al. 1999; Herrera et al. 1999; González-Suárez et al. 2000), with the only exception of p53-deficient genetic backgrounds (Chin et al. 1999; Artandi et al. 2000).
The possibility that telomere-binding proteins could play a role in cancer and aging is largely unexplored despite their well-known roles in telomere regulation. In particular, the telomere-binding protein TRF2 regulates both telomere protection and telomere length (van Steensel et al. 1998; Smogorzewska et al. 2000; Wang et al. 2004; Celli and de Lange 2005; Muñoz et al. 2005). TRF2 also interacts with several DNA repair proteins that are involved in human chromosomal instability syndromes characterized by premature aging and increased cancer (de Lange 2005; Muñoz et al. 2006). In particular, TRF2 interacts with Rap1, the WRN and BLM helicases, the DNA–PK complex, the Mre11/Nbs1/Rad50 complex, PARP1 and PARP2 poly(ADP)ribosylases, and the ERCC1/XPF endonuclease, among others (Li et al. 2000; Zhu et al. 2000, 2003; Opresko et al. 2002; Dantzer et al. 2004).
We have recently generated mice that overexpress TRF2 under the 5′-regulatory region of the keratin 5 (K5) gene, which targets TRF2 to various stratified epithelia including the skin (Muñoz et al. 2005). These mice develop skin pathologies reminiscent of the human Xeroderma pigmentosum (XP) syndrome. These pathologies include skin hyperpigmentation and increased UV-induced skin cancer, which are accompanied by severe telomere shortening and increased chromosomal instability (Muñoz et al. 2005). TRF2-induced telomere shortening and chromosomal instability are mediated by XPF, a nuclease involved in UV-damage repair that is mutated in XP patients (de Laat et al. 1999; Petit and Sancar 1999). These findings specifically link TRF2 function in vivo to UV-damage repair and UV-induced carcinogenesis. In agreement with this notion, human skin carcinomas show elevated levels of hTRF2 (Muñoz et al. 2005).
Of notice, TRF2 is overexpressed in several other human tumors, such as breast carcinomas, liver hepatocarcinomas, and lung carcinomas (Matsutani et al. 2001; Oh et al. 2005), suggesting a general role of TRF2 in tumorigenesis. Here, we demonstrate that increased TRF2 expression acts as a potent oncogenic insult in vivo. In particular, we show that K5TRF2 mice develop both spontaneous and carcinogen-induced epithelial tumors.
The fact that K5TRF2 mice show increased cancer in spite of having critically short telomeres is in marked contrast to telomerase-deficient mice with short telomeres, which are resistant to skin carcinogenic treatments (González-Suárez et al. 2000). To elucidate the mechanisms by which TRF2 promotes tumorigenesis, we generated K5TRF2 mice in a telomerase-deficient genetic background, K5TRF2/Terc−/− mice. Here, we show that increased TRF2 expression in the absence of telomerase activity results in a dramatic acceleration of epithelial carcinogenesis coinciding with increased telomere dysfunction and chromosomal instability. Interestingly, telomeric recombination and ALT (alternative telomere maintenance mechanism)-associated PML bodies (APBs) were significantly increased by TRF2 overexpression, suggesting the activation of alternative telomere maintenance pathways in these mice. Together, the results presented here indicate that TRF2 overexpression acts as a potent oncogenic insult in vivo through the induction of severe chromosomal instability and the activation of telomere recombination. These effects of TRF2 are shown to be independent of telomerase activity, suggesting that telomerase inhibition may not be effective in the treatment of tumors with high levels of TRF2 expression.
Results
TRF2 acts as a potent oncogene in vivo in a skin model of multistage carcinogenesis
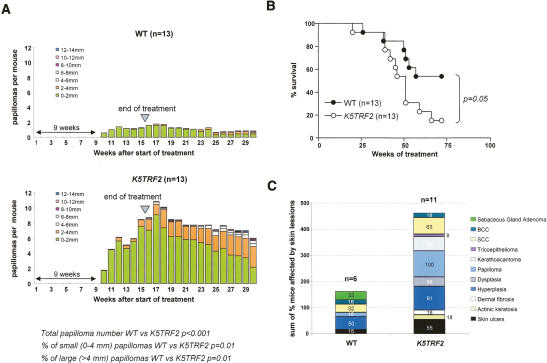
To study the impact of increased TRF2 expression in epithelial tumor formation, we used a classical model of chemically induced skin multistage carcinogenesis based in tumor initiation using a single dose of 7,12-dimethylbenz(a)anthracene (DMBA) and tumor promotion with two weekly doses of 12-O-tetradecanoylphorbol 13-acetate (TPA) during 15 wk (Materials and Methods) (Balmain et al. 1984). K5TRF2 mice developed significantly more papillomas than wild-type mice (p < 0.001) (Fig. 1A), and K5TRF2 papillomas grew to bigger lesions than wild-type papillomas (Fig. 1A). K5TRF2 mice also showed a higher mortality compared with wild-type controls (log rank test, p = 0.05) (Fig. 1B). A detailed histopathological analysis of the sacrificed mice revealed increased preneoplastic and neoplastic skin lesions in treated K5TRF2 mice compared with wild-type littermates (Fig. 1C). Together, these results indicate that TRF2 overexpression is oncogenic in the DMBA/TPA skin carcinogenesis model.
Figure 1.
K5TRF2 mice are more susceptible to multistage carcinogenesis protocols. (A) Average number of papillomas per mouse at the indicated times after the start of DMBA/TPA treatment. The treatment was interrupted at week 15 (arrow). The time at which the first lesions appeared is also indicated. n = number of mice of each genotype. Statistical comparisons using the Wilcoxon-Mann-Whitney rank sum test of papilloma number and size between wild-type and K5TRF2 mice during the 30-wk duration of the experiment are indicated. (B) Survival of DMBA + TPA-treated wild-type and K5TRF2 mice. Statistical significance of the difference in survival between genotypes using the log rank test is indicated. (C) Percentage of wild-type and K5TRF2 mice showing the indicated skin lesions after full histopathological analysis.
Telomerase deficiency accelerates loss of viability and spontaneous epithelial carcinogenesis in K5TRF2/Terc−/− mice
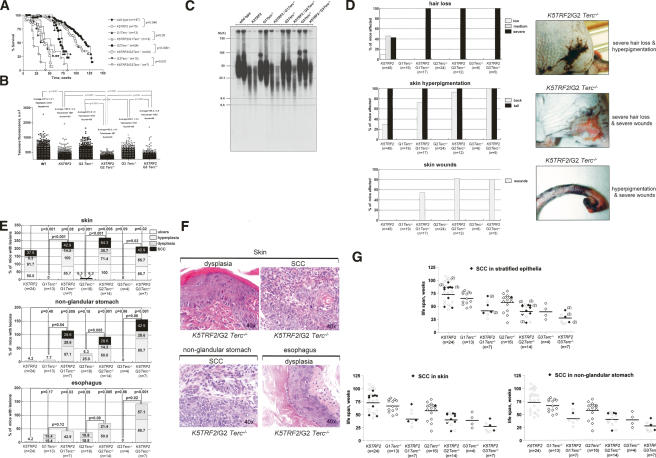
To study the impact of telomerase in TRF2-induced carcinogenesis in vivo, we obtained increasing generations of K5TRF2 mice in a telomerase-deficient (Terc−/−) background, K5TRF2/Terc−/− mice (Fig. 2A). Single Terc−/− mice showed decreased survival due to premature loss of viability with increasing mouse generations (G1 to G3), as previously described (Fig. 2A; Lee et al. 1998; Herrera et al. 1999; González-Suárez et al. 2000; Blasco 2005; Garcia-Cao et al. 2006). K5TRF2/Terc−/− mice showed an even lower survival than the single Terc−/− mice for every mouse generation (G1 to G3; log rank test, p ≤ 0.05 for all comparisons) (Fig. 2A). This lower survival coincided with an increase incidence of tumors, severe skin atrophies, and infections in K5TRF2/Terc−/− mice at the time of death, which were not present to the same extent in the single Terc−/− controls, whose main cause of death was atrophies of the small intestine (Supplementary Fig. 1). To address whether this lower survival of K5TRF2/Terc−/− mice compared with Terc−/− mice was associated with a further telomere shortening (Muñoz et al. 2005), we measured telomere length using quantitative fluorescence in situ hybridization (Q-FISH) on skin sections from the tail (Materials and Methods). K5TRF2 mice showed shorter telomeres than wild-type controls as previously described (Student’s t-test, p < 0.0001) (Fig. 2B; Muñoz et al. 2005). Increasing generations (G2 and G3) of K5TRF2/Terc−/− mice showed shorter telomeres than the corresponding Terc−/− mice (Student’s t-test, p < 0.0001 for all comparisons) (Fig. 2B) and K5TRF2 controls (Student’s t-test, p < 0.0001 for all comparisons) (Fig. 2B), demonstrating a further shortening of telomeres as the result of TRF2 overexpression in the absence of telomerase. Of notice, third-generation K5TRF2/G3Terc−/− mice did not show further shortening of telomeres compared with second-generation K5TRF2/G2Terc−/− mice (Fig. 2B), in spite of telomerase deficiency for one extra generation. Instead, these mice showed very intense telomere fluorescence signals, which were not present in second-generation K5TRF2/G2Terc−/− mice (Fig. 2B). We confirmed these results by using Southern-based “terminal restriction fragment” (TRF) analysis (Materials and Methods). As shown in Figure 2C, K5TRF2/G3Terc−/− mice showed the appearance of higher-molecular-weight TRF fragments compared with earlier generation G1 and G2 K5TRF2/Terc−/− mice, suggesting the presence of longer telomeres in K5TRF2/G3Terc−/− mice.
Figure 2.
Decreased survival, severe skin lesions, and accelerated carcinogenesis in K5TRF2/Terc−/− mice. (A) Survival of mice of the indicated genotype as a function of age. Statistical significance of the differences in survival using the log rank test is indicated. (B) Telomere fluorescence as determined by Q-FISH in skin sections of the indicated genotypes. Between 45 and 100 keratinocyte nuclei and 500–2000 telomere dots from one to two mice of each genotype were analyzed by Q-FISH. Average fluorescence in arbitrary units (auf) and standard error are shown. Statistical significance is indicated for each comparison. Telomere fluorescence values should be compared within each experiment due to variations in fluorescence intensity between experiments. (C) Telomere length as determined by TRF in adult skin keratinocytes of the indicated genotypes. Note increased TRF size in third-generation K5TRF2/G3Terc−/− mice compared with earlier K5TRF2/Terc−/− generations. (D) Quantification of skin pathologies, as well as representative examples, in G1 to G3 K5TRF2/Terc−/− mice and the K5TRF2 and G1 to G3 Terc−/− controls. (E) Percentage of mice of the indicated genotype showing preneoplastic (hyperplasia, dysplasia) and neoplastic lesions (SCC) in different stratified epithelia at the time of death. Frequency of skin ulcers in sacrificed mice is higher than that shown in D since we only included moribund mice, while D shows the incidence of lesions in all the mouse cohort including healthy animals. (F) Representative examples (40× magnification) of the indicated preneoplastic and neoplastic lesions in stratified epithelia from K5TRF2/Terc−/− mice. The image corresponding to “skin dysplasia” shows irregular growth pattern with hyperkeratosis, squamous cell hyperplasia, hyperchromatic nuclei, and prominent nucleoli. The image corresponding to a “skin SCC” shows nests of squamous cells with invasive growth of the dermis, as well as numerous mitotic figures and nuclear atypias. The image corresponding to a “nonglandular stomach SCC” shows endophytic evidence of invasive growth of the muscularis mucosae, with some hyperchromatic cells with prominent nucleoli and numerous mitotic figures. The “esophagus” dysplasia shows diffuse thickening of the squamous epithelium and dysplastic basal cell proliferation. Note that in SCC, the tumoral cells interrupt the basal membrane in both the skin and the nonglandular stomach. (G) Onset of SCC with age in increasing generations of K5TRF2/Terc−/− mice and the K5TRF2 and G1 to G3 Terc−/− controls. Each rectangle represents a mouse. White rectangles indicate mice without SCC at the time of death; black rectangles indicate mice with one or more SCC at the time of death. The number in parentheses indicates the number of SCC of independent origin per mouse. The upper graph summarizes SCC in skin and nonglandular stomach; parentheses indicate the number of independent SCC per mouse in different locations.
Along with their decreased survival and shorter telomeres, K5TRF2/Terc−/− mice showed more severe skin pathologies compared with K5TRF2 and Terc−/− controls, including complete hair loss, massive skin hyperpigmentation, and the presence of very large skin wounds, which corresponded to necrotic skin ulcerations (Fig. 2D). These results suggest that degenerative skin pathologies produced by TRF2 overexpression are further aggravated by telomere shortening in the absence of telomerase activity.
Next, we studied spontaneous tumors with increasing K5TRF2/Terc−/− generations compared with K5TRF2 and Terc−/− controls. In particular, we studied preneoplastic and neoplastic lesions in the skin, the esophagus, and the nonglandular stomach, where the K5 promoter is expressed (Murillas et al. 1995; González-Suárez et al. 2001, 2002). As previously described by us, K5TRF2 transgenics showed an increased incidence of preneoplastic (hyperplasia, dysplasia) and neoplastic (squamous cell carcinoma, SCC) lesions in the skin (Fig. 2E,F; Muñoz et al. 2005), and of benign lesions (hyperplasia) in the nonglandular stomach and the esophagus (Fig. 2E,F). Interestingly, G1 to G3 K5TRF2/Terc−/− mice developed more tumoral lesions in the different stratified epithelia than single Terc−/− and K5TRF2 controls (Fig. 2E,F). Furthermore, the onset of SCC in the skin and the nonglandular stomach was accelerated with increasing generations of K5TRF2/Terc−/− mice compared with single K5TRF2 controls, as shown by the presence of SCC lesions in young G2 and G3 K5TRF2/Terc−/− mice at their time of death (Fig. 2G). These lesions were rarely developed by the corresponding Terc−/− controls (Fig. 2G; Supplementary Fig. 1). These results indicate that TRF2 increased expression is a potent oncogenic insult that along with telomerase deficiency and dysfunctional telomeres dramatically accelerates epithelial carcinogenesis. It is important to note that TRF2 overexpressing mice and p53-deficient mice (Chin et al. 1999; Artandi et al. 2000) are the only two known situations where severe telomere dysfunction in the absence of telomerase does not act as a tumor suppressor mechanism but instead accelerates carcinogenesis.
Telomerase deficiency dramatically accelerates UV-induced epithelial carcinogenesis in K5TRF2 mice
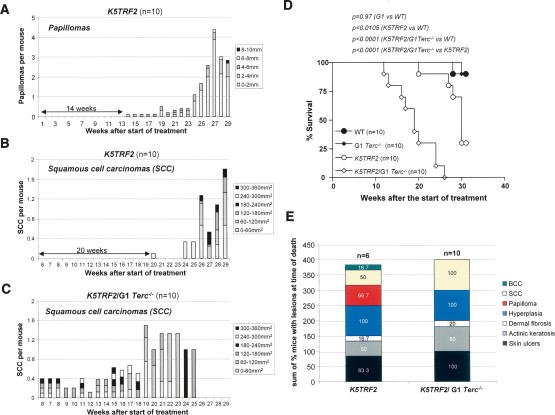
The early onset of epithelial tumors with increasing generations of K5TRF2/Terc−/− mice compared with single Terc−/− and K5TRF2 controls suggests a contribution of both telomerase deficiency and dysfunctional telomeres to TRF2-induced tumorigenesis. Next, we studied whether telomerase deficiency per se also cooperated with TRF2 overexpression in accelerating tumorigenesis in young animals from the first generation (K5TRF2/G1Terc−/−) compared with K5TRF2 controls. To this end, we subjected K5TRF2/G1Terc−/− and K5TRF2 mice to a chronic UV-irradiation protocol previously shown to induce tumors in K5TRF2 mice (Muñoz et al. 2005). Control K5TRF2 mice developed papillomas at week 14 after the start of treatment, which continued to grow in number and size during the following weeks (Fig. 3A). In addition, K5TRF2 mice developed SCC starting at week 20 (Fig. 3B), which eventually resulted in decreased survival of the mouse cohort compared with wild-type controls (log rank test, p < 0.0105) (Fig. 3D; Muñoz et al. 2005). Absence of telomerase in K5TRF2/G1Terc−/− mice resulted in accelerated UV-induced carcinogenesis, with all K5TRF2/G1Terc−/− mice developing SCC as early as 6 wk after the start of treatment in the absence of previous papilloma development (Fig. 3C). Furthermore, K5TRF2/G1Terc−/− mice showed a decrease survival compared with wild-type and K5TRF2 controls, with all K5TRF2/G1Terc−/− mice dead at week 26 after the start of treatment due to large tumors (log rank test, p < 0.0001 for both comparisons) (Fig. 3D). Detailed histopathology of skin lesions in the sacrificed mice revealed different stages of skin carcinogenesis ranging from pretumoral lesions and benign tumors to SCC and basal cell carcinomas (BCC), with 100% of K5TRF2/G1Terc−/− mice showing SCC at the time of death (Fig. 3E). The increased UV-induced carcinogenesis shown by K5TRF2/G1Terc−/− mice, together with the earlier onset of spontaneous tumors in these mice compared with single K5TRF2 mice, strongly supports the notion that telomerase deficiency accelerates TRF2-induced epithelial carcinogenesis.
Figure 3.
Telomerase deficiency accelerates UV-induced carcinogenesis in K5TRF2/Terc−/− mice compared with K5TRF2 controls. (A) Total number of papillomas per mouse in K5TRF2 mice after chronic UVB irradiation. (B) Number of carcinomas per mouse in K5TRF2 mice after chronic UVB irradiation. (C) Number of carcinomas per mouse in K5TRF2/Terc−/− mice after chronic UVB irradiation. (D) Survival curve of chronically irradiated K5TRF2/Terc−/− and K5TRF2 cohorts. All irradiated K5TRF2/Terc−/− mice were dead at week 26 after the start of the treatment due to large skin tumors, while control K5TRF2 mice showed longer survival. The statistical significance of differences in survival using the log rank test is indicated. (E) Percentage of K5TRF2/G1Terc−/− and K5TRF2 mice showing the indicated skin lesions at time of death after full histopathological analysis. Of notice, 100% of K5TRF2/Terc−/− mice presented SCC lesions at the time of death.
TRF2 overexpression in the absence of telomerase increases chromosomal instability and DNA damage
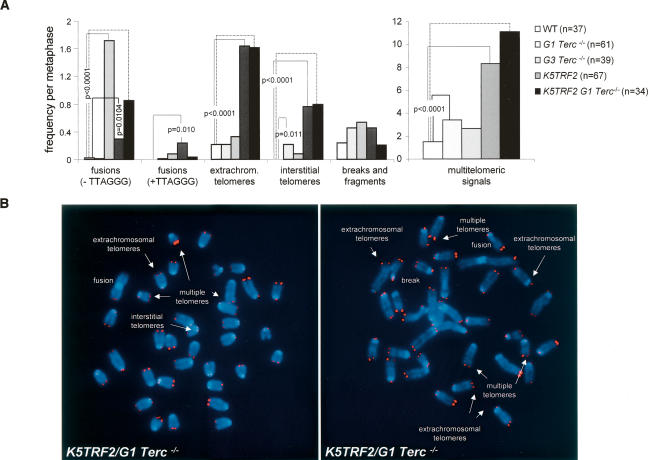
We showed previously that K5TRF2 keratinocytes have increased end-to-end fusions coincidental with critically short telomeres (Muñoz et al. 2005). In addition, K5TRF2 cells show increased chromosomes with multiple telomere signals at the ends, extrachromosomal telomere signals, and chromosomes with interstitial telomeres (Fig. 4A,B; Muñoz et al. 2005). To address the possible involvement of telomerase and short telomeres in these chromosomal aberrations, we performed cytogenetic analysis of keratinocytes derived from K5TRF2, G1 and G3 Terc−/−, K5TRF2/G1Terc−/−, as well as wild-type mice using Q-FISH (Materials and Methods). End-to-end fusions lacking TTAGGG signals at the fusion point were dramatically increased in K5TRF2/G1Terc−/− cells compared with single K5TRF2 and G1 Terc−/− controls (Fig. 4A,B), in agreement with shorter telomeres in these cells (Fig. 2C). As a control for critically short telomeres, G3 Terc−/− cells also showed increased end-to-end fusions lacking telomere signals at the fusion point compared with wild-type cells (Fig. 4A), suggesting that these fusions are produced by telomere shortening. In contrast, the occurrence of multiple telomere signals at chromosome ends (multitelomeric signals), of extrachromosomal telomere signals, as well as the presence of interstitial telomeres, was similarly elevated in K5TRF2/G1Terc−/− and K5TRF2 cells but not in G3 Terc−/− cells with short telomeres (Fig. 4A), suggesting that they were the consequence of increased TRF2 expression. These results indicate that TRF2 is a potent inducer of chromosomal instability. On one hand, TRF2 induces telomere shortening and end-to-end fusions, which are further increased in combination with telomerase deficiency. In addition, TRF2 overexpression induces extrachromosomal telomeres, interstitial telomeres, and multitelomic signals at chromosome ends, which are independent of telomere length and telomerase activity. These findings suggest a model of TRF2-induced tumorigenesis in which TRF2 increased expression induces severe telomere dysfunction and chromosomal instability that are further aggravated in the absence of telomerase.
Figure 4.
Increased chromosomal instability in K5TRF2/Terc−/− mice. (A) Quantification of frequency of chromosomal aberrations per metaphase in the indicated genotypes. Note that end-to-end fusions lacking TTAGGG repeats at the fusion point (−TTAGGG) are increased in G3 Terc−/− and K5TRF2/G1Terc−/− cells compared with K5TRF2 controls, suggesting that they are the consequence of telomere shortening. Extrachromosomal telomere signals, multitelomeric signals, and interstitial telomeres are specifically increased in K5TRF2 and K5TRF2/Terc−/− cells but not in G3 Terc−/− cells, suggesting that they are the consequence of TRF2 overexpression. Statistically significant differences are indicated. Up to six independent keratinocyte cultures were analyzed per genotype. n = number of metaphases analyzed. (B) Representative examples of the indicated chromosomal aberrations (indicated by white arrows).
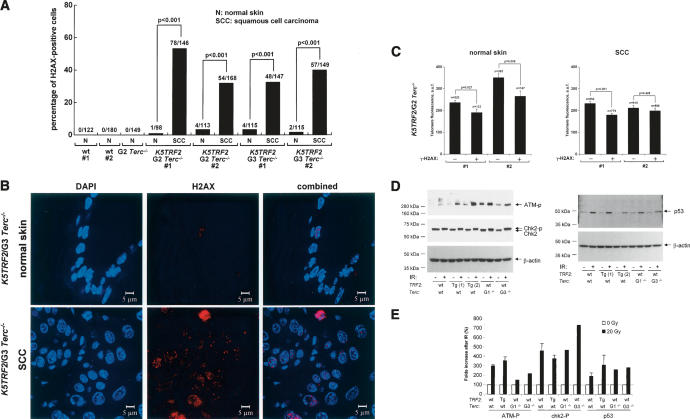
To address whether dysfunctional telomeres and chromosomal instability in K5TRF2/Terc−/− mice contribute to TRF2-induced carcinogenesis, we quantified the presence of γH2AX foci in skin tumors from G2 and G3 K5TRF2/Terc−/− mice compared with nontumoral sections from the same animals. Dysfunctional telomeres have been previously shown to form γH2AX DNA damage foci (d’Adda di Fagagna et al. 2003; Takai et al. 2003), a hallmark of double-strand breaks (DSB) (Modesti and Kanaar 2002). We have previously shown that single K5TRF2 mice show elevated levels of γH2AX DNA foci in the skin compared with wild-type mice (Muñoz et al. 2005). In agreement with this, normal skin from G2 and G3 K5TRF2/Terc−/− mice showed increased γH2AX DNA foci compared with wild-type skin (Fig. 5A), and this was further increased in G2 and G3 K5TRF2/Terc−/− tumors (p < 0.001 for comparisons between normal skin and SCC) (Fig. 5A,B). We previously showed that γH2AX foci in K5TRF2 overexpressing cells colocalize with the TRF1 telomeric protein, suggesting that they correspond to short/dysfunctional telomeres (Muñoz et al. 2005). In accordance with this, skin cells positive for γH2AX DNA foci showed shorter telomeres than γH2AX-negative cells (Fig. 5C), supporting the idea that these foci were the result of telomere shortening and severe telomere dysfunction (Muñoz et al. 2005). Furthermore, in agreement with increased DNA damage in K5TRF2/Terc−/− tumors, we detected an increase in anaphase bridges both in spontaneous and UV-induced K5TRF2/Terc−/− tumors compared with the normal surrounding tissue (Supplementary Fig. 2).
Figure 5.
Increased γH2AX foci in K5TRF2 cells and K5TRF2/Terc−/− tumors. (A) Quantification of percentage of nuclei containing γH2AX foci in normal skin of the indicated genotypes, as well as various SCC in G2 and G3 K5TRF2/Terc−/− mice (n = 2) compared with the normal surrounding tissue. The number of γH2AX-positive cells out of the total number of cells analyzed is indicated. Statistical significance of differences between the SCC and the normal surrounding tissue are indicated. (B) Representative examples of γH2AX staining in a K5TRF2/G3Terc−/− SCC compared with the corresponding normal surrounding tissue. Magnification bars correspond to 5 μm. (C) Telomere fluorescence as determined by Q-FISH directly on skin cells that are positive (+) or negative (−) for γH2AX foci. #1 and #2 correspond to two independent pairs of normal skin and the SCC in K5TRF2/G2Terc−/− mice. Note shorter telomeres in γH2AX-positive than in γH2AX-negative cells. The statistical significance of differences in telomere length is indicated. (D) Western blot analysis of ATM-P, ChK2, and p53 in keratinoctytes of the indicated genotypes before (−) and after (+) ionizing radiation. Phosphorylated Chk2 (Chk2-P) is detected as a change in the Chk2 mobility in the gel. Actin was used as a loading control. (E) Quantification of ATM-P, ChK2-P, and p53 levels in keratinoctytes of the indicated genotypes before (−) and after (+) ionizing radiation after normalization to actin levels. Note a similar ATM phosphorylation, ChK2 phosphorylation, as well as p53 accumulation after ionizing radiation in the different genotypes.
The fact that γH2AX foci were increased in TRF2-induced tumors indicates the activation of a DNA damage response in these tumors in response to telomere dysfunction. In this regard, it has been previously shown that TRF2 can bind and inhibit ATM (Karlseder et al. 2004; Bradshaw et al. 2005). To address this in our skin model, we have studied the ATM DNA damage signaling pathway (ATM phosphorylation, ATM-P; Chk2 phosphorylation, Chk2-P; p53 amounts) in K5TRF2 keratinocytes before and after treatment with ionizing radiation to activate this pathway (Fig. 5D,E). In addition, we have measured p53 levels by confocal immunofluorescence in normal and tumoral tissue from different generation K5TRF2/Terc−/− mice (Supplementary Fig. 3a–d). We found a normal activation of the ATM DNA damage signaling pathway as indicated by ATM-P, Chk2-P, and p53 accumulation in K5TRF2 cells in response to ionizing radiation (Fig. 5D,E), as well as a normal expression of these factors in the absence of irradiation in keratinocytes (Fig. 5D,E). Furthermore, quantification of p53 levels by confocal immunofluorescence in G2 and G3 K5TRF2/Terc−/− spontaneous tumors, as well as in the normal surrounding skin, showed that p53 levels are increased in normal skin from G2 and G3 K5TRF2/Terc−/− mice compared with skin from wild-type and G2 Terc−/− single mutant controls (Supplementary Fig. 3a–d), in agreement with shorter telomeres and increased telomere damage in these mice. Tumoral tissue from the same mice showed variable levels of p53 expression compared with the normal surrounding tissue (Supplementary Fig. 3a–d). In particular, two out of four K5TRF2/Terc−/− tumors showed decreased p53 compared with the normal surrounding tissue, while the other tumors maintained p53 expression, suggesting a possible alteration of p53 function in some K5TRF2/Terc−/− tumors. Together, these results suggest a normal DNA damage response to short telomeres as well as to direct ionizing radiation in K5TRF2 mice and cells. These results are in agreement with previous observations from our group showing that TRF2-overexpressing skin keratinocytes showed a normal sensitivity to γ-irradiation (Muñoz et al. 2005). It is possible that TRF2 may have differential effects on DNA damage signaling in human and mouse cells, or that its effects may vary depending on the cell type (Karlseder et al. 2004; Bradshaw et al. 2005).
Together, these results suggest increased DNA damage in TRF2-induced tumors.
TRF2 overexpression induces recombination between telomeric sequences
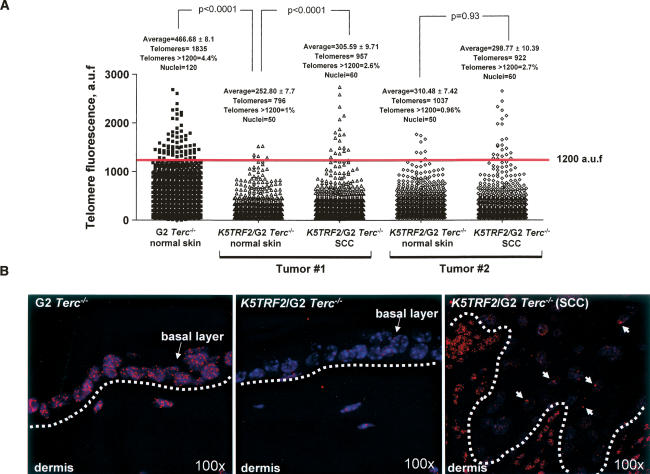
The fact that K5TRF2/Terc−/− mice develop tumors in the absence of telomerase activity and in the presence of very short and dysfunctional telomeres raises the possibility that TRF2 overexpression could be favoring the activation of ALT in these mice (Dunham et al. 2000; Muntoni and Reddel 2005). In order to test this, we first determined telomere length in spontaneous SCC from K5TRF2/G2 Terc−/− mice and in the normal surrounding tissue using Q-FISH (Materials and Methods). As expected, telomeres were shorter in normal skin from K5TRF2/G2Terc−/− mice compared with single G2 Terc−/− controls (Student’s t-test, p < 0.0001) (Fig. 6A,B). Interestingly, K5TRF2/G2Terc−/− tumors (SCC) showed very intense telomere fluorescence signals, which were not present to the same extent in the normal surrounding tissue (Fig. 6A,B). In particular, the percentage of telomere signals above 1200 U of telomere fluorescence was increased to 2.6% and 2.7% in the tumors compared with ∼1% in the normal surrounding tissue (red line in Fig. 6A indicates telomeres with >1200 U of arbitrary fluorescence; see arrows in Fig. 6B). The presence of very intense telomere fluorescence signals in the tumors in the absence of telomerase is consistent with activation of ALT mechanisms in these tumors. Furthermore, these results are in agreement with the elongated telomeres observed both by TRF and Q-FISH in late-generation K5TRF2/Terc−/− mice (Fig. 2B,C).
Figure 6.
Presence of very intense telomere fluorescence signals in K5TRF2/G2Terc−/− tumors compared with normal surrounding tissue. (A) Quantification of telomere fluorescence in tumor sections (SCC) from two independent K5TRF2/G2Terc−/− mice compared with their normal surrounding skin. As control, telomere length of normal skin from G2Terc−/− mice was also determined. More than 50 nuclei and >700 telomere signals from each genotype and condition were analyzed by Q-FISH. Average fluorescence in arbitrary units (auf) and standard error are shown. The percentage of telomere signals with >1200 auf are also indicated with a red line. Statistical significance is indicated for each comparison. The tumoral tissue showed the presence of very intense telomere fluorescence signals, which were not present in the normal surrounding tissue. (B) Representative images of telomere fluorescence in normal skin and tumor sections from the indicated mice. The basal layer of skin keratinocytes is indicated with an arrow and separated from the dermis by a white lane. White arrows indicate very intense telomere signals within the tumoral tissue. Magnification is 100×.
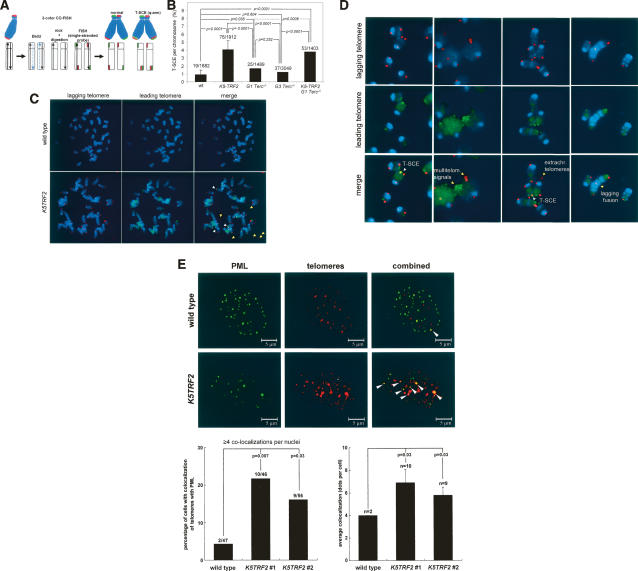
One of the known molecular bases of ALT involves increased recombination between telomeric sequences (Dunham et al. 2000). Therefore, we determined whether increased TRF2 expression unleashed recombination events at telomeres in K5TRF2 mice. To measure recombination events at telomeres, we determined the frequency of telomeric sister-chromatid exchange events (T-SCE) in primary keratinocytes from different genotype mice using two-color chromosome orientation FISH (CO-FISH) (Bailey et al. 2004; Bechter et al. 2004; Gonzalo et al. 2006). The strand-specific nature of the CO-FISH typically yields two telomeric signals of each color (red: lagging; green: leading) per chromosome in the absence of recombination events (Fig. 7A,C). A sister-chromatid exchange within telomeric DNA (T-SCE) leads to the mixture of red and green fluorescence (Fig. 7A). We only counted as positive T-SCE events those that were detected both with the leading and the lagging telomere probes (Fig. 7C,D). Interestingly, K5TRF2 and K5TRF2/G1Terc−/− cells showed increased T-SCE compared with wild type (p < 0.0001 in both cases), and to G1 and G3 Terc−/− controls (p < 0.0001 in both cases) (Fig. 7B), suggesting increased recombination between sister telomeres associated with increased TRF2 expression. Two-color CO-FISH also revealed that the extrachromosomal telomere signals present in K5TRF2 and K5TRF2/Terc−/− cells always involved telomeres produced by lagging-strand synthesis (Fig. 7C,D), suggesting that they may involve a specific mechanism related to lagging-strand processing. In this regard, TRF2 could be inducing the conversion of T-loops into extrachromosomal circles (Wang et al. 2004), which, in turn, could be generated or maintained by mechanisms involving lagging-strand synthesis. Of notice, the presence of extrachromosomal telomeres has also been associated with activation of the ALT pathway (Muntoni and Reddel 2005), in agreement with increased T-SCE in K5TRF2 and K5TRF2/Terc−/− cells. Finally, CO-FISH showed that end-to-end fusions and multitelomeric signals in K5TRF2-overexpressing cells involved both leading- and lagging-strand telomeres (Fig. 7D).
Figure 7.
Increased telomere recombination and APBs in K5TRF2/Terc−/− mice. (A) The scheme depicts the CO-FISH technique to label telomeres produced by either leading-strand (green color) or lagging-strand synthesis (red color). A sister-chromatid exchange within telomeric DNA (T-SCE) at one of the chromosome arms (i.e., q-arm) leads to the splitting of the hybridization signals to both sister chromatid telomeres, resulting in the mixture of red and green fluorescence. (B) Quantification of T-SCE in the indicated genotypes. The number of T-SCE events out of the total number of chromosomes analyzed is indicated on top of each bar. A significant increase in T-SCE is observed in K5TRF2 and K5TRF2/G1Terc−/− cultures compared with wild-type, G1, and G3 Terc−/− controls. Error bars correspond to two to five independent keratinocyte cultures. (C) Representative CO-FISH images of metaphases hybridized with probes against the leading (green fluorescence) and lagging (red fluorescence) telomere. T-SCE events are indicated with white arrows. Note that extrachromosomal telomere signals involving lagging telomeres, multitelomeric signals, and end-to-end fusions are indicated with yellow arrows. (D) Representative CO-FISH images of chromosome aberrations hybridized with probes against the leading (green fluorescence) and lagging (red fluorescence) telomere in a K5TRF2 keratinocyte culture. T-SCE events are indicated with white arrows. Extrachromosomal telomere signals involving lagging telomeres, multitelomeric signals involving both lagging and leading telomeres, as well as a chromatid-type end-to-end fusion involving lagging telomeres are indicated with yellow arrows. The exposition time in K5TRF2 and K5TRF2/Terc−/− keratinocytes was increased compared with wild type in order to be able to detect telomere signals due to the short telomeres present in these genotypes. (E) Confocal microscopy images showing either PML fluorescence (green), telomere fluorescence (red), or combined fluorescence (yellow) in wild-type and K5TRF2 keratinocytes. Arrows indicate colocalization of PML and telomeres. Quantification of percentage of cells showing colocalization of telomeres with PML protein is also shown. A cell was considered positive when it showed four or more colocalization events. An increased frequency of cells showing APBs is observed in two independent K5TRF2 cultures (#1 and #2) compared with wild-type controls. Quantification of the number of APBs per nuclei in the indicated genotypes is also shown. A significant increase in the number of APBs per nuclei is observed in K5TRF2 cultures compared with the wild-type controls.
Another landmark of ALT is the colocalization of PML with telomeres, the so-called APBs (Muntoni and Reddel 2005). As shown in Figure 7E, we detected a significant increase in the percentage of cells showing APBs in two independent K5TRF2 keratinocyte cultures compared with wild-type controls (p < 0.05 in both cases) (Fig. 7E). Furthermore, K5TRF2 cells showed an increased number of colocalization events per nuclei than wild-type controls (p = 0.03 in both cases) (Fig. 7E).
Together, these results demonstrate a role for TRF2 in controlling telomere recombination. In particular, TRF2 overexpression results in increased features of ALT (increased T-SCE, increased APBs, increased extrachromosomal telomere signals, and heterogeneous telomere length), which, in turn, may be contributing to telomere maintenance in the absence of telomerase in the K5TRF2/Terc−/− tumors.
Discussion
TRF2 is a key regulator of telomere protection and telomere length (van Steensel et al. 1998; de Lange 2002, 2005; Celli and de Lange 2005; Muñoz et al. 2005), two processes that are essential for chromosomal stability. In addition, TRF2 interacts with several DNA repair factors, as well as DNA damage signaling proteins, most of which are involved in human chromosome instability syndromes characterized by premature aging and increased cancer (Li et al. 2000; Zhu et al. 2000, 2003; Opresko et al. 2002; Dantzer et al. 2004; Blasco 2005). In the past, we have demonstrated that increased TRF2 expression leads to premature skin aging and increased skin tumors in areas exposed to the light, pinpointing a specific role for TRF2 in UV-induced damage repair and UV-induced tumorigenesis. However, the fact that TRF2 is overexpressed in a variety of cancers (skin, lung, liver, breast) (Matsutani et al. 2001; Muñoz et al. 2005; Oh et al. 2005) suggested a more general role of TRF2 in carcinogenesis.
Here, we show that TRF2 overexpression acts as a potent oncogene in vivo by inducing epithelial carcinogenesis, both spontaneously as well as in response to carcinogenic treatments. These results go in line with recent findings suggesting that TRF2 loss of function reduces the tumorigenicity of human malignant cell lines (Biroccio et al. 2006). Furthermore, we show that telomerase deficiency dramatically accelerates epithelial carcinogenesis induced by TRF2 overexpression concomitant with critically short telomeres, severe telomere dysfunction, and increased chromosomal instability. In agreement with this, TRF2-induced tumors showed increased anaphase bridges as well as increased γH2AX foci compared with the normal surrounding tissue. In addition, G2 and G3 K5TRF2/Terc−/− mice show increased levels of p53 expression compared with wild-type controls, suggesting the activation of normal DNA damage signaling in response to dysfunctional telomeres. In support of this, TRF2-overexpressing keratinocytes show a normal activation of the ATM pathway (ATM-P, Chk2-P, p53 accumulation) in response to direct γ-irradiation. These results are in agreement with previous data from our group showing a normal sensitivity of K5TRF2 cells to γ-irradiation (Muñoz et al. 2005), and suggest that increased tumorigenesis in G2 and G3 K5TRF2/Terc−/− mice is not likely to be initiated by cancellation of the ATM pathway as a consequence of TRF2 overexpression (Karlseder et al. 2004; Bradshaw et al. 2005). Finally, G2 and G3 K5TRF2/Terc−/− tumors showed variable p53 levels, which were maintained or decreased compared with the normal surrounding tissue, suggesting a possible alteration of p53 function in these tumors (Chin et al. 1999; Artandi et al. 2000). Together, these results indicate that TRF2 overexpression in the absence of telomerase promotes carcinogenesis by inducing telomere dysfunction and chromosomal instability. Finally, our results indicate that TRF2-induced tumors do not depend on telomerase activity, as epithelial carcinogenesis is accelerated in a telomerase-deficient background.
The fact that K5TRF2/Terc−/− tumors rapidly arise in the context of very short telomeres and telomerase deficiency suggests the activation of alternative telomere maintenance mechanisms in TRF2 mice. In support of this notion, third-generation K5TRF2/G3Terc−/− mice did not show further telomere shortening compared with second-generation K5TRF2/G2Terc−/− cohorts as determined both by Q-FISH and TRF analysis in spite of an additional mouse generation in the absence of telomerase. Furthermore, K5TRF2/G2Terc−/− tumors showed the presence of very intense telomere fluorescence signals, which were absent in the normal surrounding tissue, suggesting the activation of telomere-lengthening mechanisms in these tumors. In this regard, we demonstrate here a role for full-length mouse TRF2 in controlling recombination between telomeric sequences, which may go in line with previous findings showing that a dominant-negative mutant of human TRF2 (Δ-B TRF2) modulates the action of recombination factors at telomeres (Wang et al. 2004), thus supporting a general role for TRF2 in recombination. In particular, TRF2 overexpression in K5TRF2 and K5TRF2/Terc−/− mutant cells significantly increased recombination events between sister telomeres, a known mechanism for telomere maintenance in the absence of telomerase activity. In addition, K5TRF2-overexpressing cells show a higher frequency of APBs, as well as increased extrachromosomal telomere signals, both of which have been also associated with ALT-positive cells. These results demonstrate that TRF2 overexpression unleashes recombination events at telomeres, and therefore may contribute to derepress ALT pathways. Our results do not rule out, however, that TRF2 may be inducing recombination through interaction with other components of the telomere, such as Pot1 or WRN, recently shown to be involved in telomere recombination (Laud et al. 2005; Wu et al. 2006).
Our results support a model in which TRF2 increased expression induces telomere shortening and chromosomal instability, both of which are further aggravated in the absence of telomerase. In addition, TRF2 overexpression leads to derepression of ALT. The combination of both severe chromosomal instability and derepression of telomere recombination may explain, at least in part, the oncogenic effects of TRF2 overexpression. Together, these findings highlight an important role for TRF2 in chromosomal instability-driven carcinogenesis and indicate that TRF2-induced tumors might be refractory to therapeutic strategies based in telomerase inhibition.
Materials and methods
Generation and genotyping of mice
K5TRF2 mice were previously described (Muñoz et al. 2005). To generate K5TRF2/Terc−/− mice, K5TRF2 males from the PM line were first crossed with Terc−/− females (Blasco et al. 1997). K5TRF2/Terc−/− mice were then obtained by crossing wild-type TRF2/Terc+/− males with K5TRF2/Terc+/− females. Genotyping was performed as described (Blasco et al. 1997; Muñoz et al. 2005).
Mouse breeding and housing
Mouse colonies of successive generations (G1 to G3) of K5TRF2/Terc−/− mice, and the corresponding K5TRF2 and Terc−/− controls were generated and maintained at the CNIO under specific-pathogen-free conditions in accordance with the recommendations of the Federation of European Laboratory Animal Science Associations. All mice at the CNIO mouse facility are exposed to day/night cycles (12 h of light and 12 h of dark). The light source used at the CNIO mouse facility is white fluorescent lamps (TLD 36W/840 and TLD 58W/840; Philips), which also emit low doses of radiation between 250 and 400 nm and have a component of UV light.
Histopathology and immunohistochemistry
Skin sections were fixed in 10% buffered formalin embedded in paraffin wax, sectioned at 4 μm and stained with hematoxylin and eosin (H&E). Images were captured with a DP-10 digital camera in an Olympus Vanox microscope at the indicated magnifications.
Immunohistochemistry was performed on deparaffinated skin sections processed with 10 mM sodium citrate (pH 6.0) for 20 min at 95°C. Phosphorylated H2AX foci were detected using a mouse monoclonal anti-phospho-histone H2AX antibody (1:500; from Upstate Biotechnology). After incubation with Cy3-goat anti-mouse antibody (1:400; Jackson ImmunoResearch Laboratories, Inc.) for 30 min at room temperature, slides were mounted in Vectashield with 4′,6-diamino-2 phenylindole (DAPI). Control slides were obtained by replacing the primary antibody with PBS (data not shown). Images were obtained using a confocal ultra-spectral microscope (Leica TCS-SP2-A-OBS-UV).
Tumor induction experiments
Thirteen age-matched (8- to 12-wk-old) mice of each genotype, K5TRF2 and wild type, respectively, were shaved and treated with a single dose of DMBA (0.1 μg/μL in acetone; Sigma). Mice were subsequently treated twice weekly with TPA (12.5 μg in 200 μL of acetone each treatment; Sigma) for 15 wk.
Telomere length analyses on skin sections
For qQ-FISH, paraffin-embedded skin sections were deparaffinated and hybridized with a PNA-telomeric probe, and telomere fluorescence was determined as described (González-Suárez et al. 2000; Muñoz et al. 2005). More than 45 keratinocyte nuclei from each mouse and condition were captured at 100× magnification by using a Leica CTR MIC microscope and a COHU High-Performance CCD camera. Telomere fluorescence was integrated using spot IOD analysis in the TFL-TELO program (kindly provided by Dr. P. Landsdorp, Terry Fox Laboratory, Vancouver, Canada) (Zijlmans et al. 1997).
For quantification of telomere length in cells positive or negative for γH2AX foci, γH2AX immunofluorescence was performed as described above, images were captured at 100× magnification using a Leica CTR MIC microscope and a COHU High-Performance CCD camera, and the image coordinates were saved. Q-FISH analysis was then performed on the same tissue sections and telomere fluorescence was captured in cells with the same coordinates.
Isolation of epidermal keratinocytes from adult mice
Skin from the tail and back was removed surgically by making a longitudinal incision. Adult skin keratinocyes were isolated basically as described (Dlugosz et al. 1995; Muñoz et al. 2005). The skin was cut into 3- to 4-cm2 pieces, placed on autoclaved filter paper (Whatman 3MM), and then floated on 1× trypsin (Sigma) for 2–3 h at 37°C for tail skin and 16 h at 4°C for back skin. The skin was then placed on a dry sterile surface with 4 mL of serum-free Cnt-02 medium (CELLnTEC Advanced Cell Systems AG), and the epidermis was separated from the dermis using forceps. The epidermis was then minced, and the cell suspension was filtered using a nylon gauze (Cell Strainer 0.7 m; Falcon) to remove cornified sheets. Keratinocytes were recovered after centrifugation at 312g for 5 min and frozen for further analysis of telomere length by TRF analysis.
TRF-based telomere length analysis of adult keratinocytes
Isolated adult epidermal keratinocytes (see above) were included in agarose plugs following instructions provided by the manufacturer (Bio-Rad), and TRF analysis was performed as previously described (Blasco et al. 1997).
Chronic UVB carcinogenesis experiments
The back skin of 6- to 7-wk-old male mice (10 of each genotype) was shaved with electric clippers and irradiated three times per week with UVB at a dose of 1.8 kJ/m2 per exposure during a total of 30 wk. Scoring for tumors was done once a week. Mice were sacrificed when they showed signs of poor health. Different skin sections were fixed in 10% buffered formalin, embedded in paraffin, and subjected to full histopathological analysis.
Isolation and culture of keratinocytes from newborn mouse skin
Skin keratinocyes were isolated as described (Dlugosz et al. 1995; Muñoz et al. 2005). Newborn mice (0–2 d) were sacrificed, soaked in Betadine (5 min), in a PBS antibiotics solution (5 min), in 70% ethanol (5 min), and again in a PBS antibiotics solution (5 min). Using aseptic conditions, limbs and tails were first amputated and then the skin was peeled off using forceps. Skins were then soaked in PBS (2 min), PBS antibiotics solution (2 min), 70% ethanol (1 min), and again in PBS antibiotics solution (2 min). Each skin was carefully floated on the surface of 1× trypsin (Sigma) solution (4 mL on 60-mm cell culture plate) for no more than 16 h at 4°C. Skins were individually transferred to a sterile surface, and the epidermis was separated from the dermis using forceps, minced, and stirred at for 30 min 37°C in serum-free Cnt-02 medium (CELLnTEC; Advanced Cell Systems AG). The cell suspension was then filtered through a sterile Teflon mesh (Cell Strainer 0.7 m; Falcon) to remove cornified sheets. Keratinocytes were collected by centrifugation (160g) for 10 min and seeded on collagen I precoated cell culture plates (BD Biosciences).
Cytogenetics analysis using telomere Q-FISH on metaphases
Exponentially growing primary keratinocytes were incubated with 0.1 μg/mL colcemide (GIBCO) for 5 h at 37°C and then fixed in methanol:acetic acid (3:1). Q-FISH was performed as described (Samper et al. 2000). Images were captured in a linear acquisition mode using a COHU CCD camera on a Leica DMRB microscope. For analysis of chromosomal aberrations, at least 30 metaphases per genotype were analyzed by superimposing the telomere image on the DAPI image using the TFL-telo software.
γ-Irradiation of primary skin keratinocytes and mice
A total of 4 × 105 keratinocytes were seeded in 60-mm collagen I precoated plates and incubated for 24 h at 37°C. For γ-irradiation, we used a 137Cs source (MARK 1-30 irradiator; Shepherd & Associates) at a rate of 2.11 Gy/min. After removing the medium, cells were washed, and fresh medium was added. To analyze the effect of γ-irradiation on protein expression, cells were incubated for 4 h at 37°C and then recovered and frozen for further analysis by Western blot.
Two age-matched (3- to 6-mo-old) mice of each of the indicated genotypes were treated with 20 Gy of γ-irradiation. Two nonirradiated mice of each genotype were used as control. Three hours after irradiation, mice were sacrificed, and after shaving the back, the skin was removed surgically, cut into 1- to 2-cm2 pieces, and immediately frozen in liquid nitrogen and conserved at −80°C.
Western blots
Whole-cell extracts were prepared from primary keratinocytes as described (Muñoz et al. 2005). Back skin was homogenized in lysis buffer (50 mM Tris at pH 8, 5 mM EDTA, 350 mM NaCl, and 0.5% NP-40), containing proteases and a cocktail of phosphatase inhibitors by using an ULTRA-TURRAX homogenizer (IKA-Werker). The homogenates were centrifuged for 15 min at 4°C, and whole-cell extracts were recovered from supernatants. Protein concentration was determined using the Bio-Rad DC Protein Assay (Bio-Rad). Twenty-five micrograms to 70 μg of each extract was separated in 4%–20% gradient SDS–polyacrilamide gels by electrophoresis. After transfer, the membranes were incubated with an anti-ATM pS1981 polyclonal antibody (1:1000; Rockland Inc.), anti-chk2 monoclonal antibody (1:1000; Upstate Biotechnology), anti-p53 polyclonal antibody CM5 (1:500; Novocatra Laboratories Ltd), and anti-β-actin monoclonal (1:10.000; Sigma). Antibody binding was detected after incubation with a secondary antibody coupled to horseradish peroxidase using enhanced chemiluminescence.
Quantification of p53 expression by confocal immunofluorescence
High-throughput quantitative image analysis of p53 expression levels was performed on confocal images using the Metamorph platform (version 6.3r6; Molecular Devices). The DAPI image was used to define the nuclear area and the Cy3 image for quantification of p53 fluorescence. The DAPI images were signal-intensity thresholded, segmented, and converted to 1-bit binary images. The binary DAPI mask was applied to the matching Cy3 to obtain a combined image with p53 fluorescence information for each nucleus. Cy3 fluorescence intensity (p53 fluorescence) was measured as “average gray values” units (arbitrary units of fluorescence). A code of four colors was used to classify the nuclei according to their average p53 fluorescence. p53 fluorescence ranges were determined using the p53 staining of a p53−/− skin as a negative control. Finally, p53 fluorescence values for each histological region were exported to Excel, and the frequency histograms were generated.
Telomere recombination measurements using CO-FISH
Exponentially growing primary keratinocytes were subcultured in the presence of 5′-bromo-2′-deoxyuridine (BrdU; Sigma) at a final concentration of 1 × 10−5 M, and then allowed to replicate their DNA once for 36 h at 37°C. Colcemid was added at a concentration of 1 μg/mL during the last hour. Cells were then recovered and metaphases were prepared as described (Samper et al. 2000). CO-FISH was performed as described (Bailey et al. 2004; Gonzalo et al. 2006) using first a (TTAGGG)7 probe labeled with Cy3 and then a second (CCCTAA)7 probe labeled with Rhodamine Green (Applied Biosystems). Metaphase spreads were captured on a Leitz Leica DMRB fluorescence microscope. The exposure time in K5TRF2 and K5TRF2/Terc−/− keratinocytes was increased compared with wild type in order to be able to detect telomere signals due to the short telomeres present in these genotypes.
Confocal PML immunofluorescence-FISH experiments
Keratinocytes (5 × 104) were fixed for 20 min at 4°C with 4% PFA in PBS and permeabilized with 0.1% Triton X-100 in PBS for 7 min at room temperature. After blocking with 10% BSA (in PBS) for 20 min at 37°C, cells were incubated with rabbit anti-mouse PML polyclonal antibody (gift from Paul Freemont, Cancer Research UK, London, UK) diluted 1/1000 in blocking solution overnight at 4°C. After washing twice with 0.05% Triton X-100 in PBS for 5 min, cells were incubated with a secondary goat anti-rabbit IgG conjugated with Alexa 488 and diluted 1/300 in blocking solution for 1 h at room temperature. After immunostaining, telomeric FISH was performed as described (Samper et al. 2000) with minor modifications. Briefly, slides were fixed for 2 min in 4% formaldehyde, dehydrated in ethanol series (70%, 90%, and 100%), and air-dried. Hybridization with cy3-labeled telomeric PNA probe was performed as described (Samper et al. 2000). After hybridization, slides were washed twice with 50% formamide, 10 mM Tris (pH 7.2), and 0.1% BSA for 15 min, followed by three washes with 0.1 M Tris, 0.15 M NaCl (pH 7.5), and 0.08% Tween 20 for 5 min. Slides were then dehydrated with ethanol series and air-dried. Finally, slides were counterstained with DAPI (0.2 μg/mL) in Vectashield (Vector Laboratories). Images were captured in a confocal microscope (Leica TCS SP2) and analyzed by the Leica confocal software.
Statistical analysis
A t-student test with “two-tails,” “two samples of unequal variance” (or Welch’s correction) was used to calculate the statistical significance of the observed differences in telomere length. Microsoft Excel version 2001 and Graphpad Instat version 3.05 were used for the calculations.
To calculate the statistical significance of differences in frequency of chromosomal aberrations and T-SCE, we used the χ2 test. The “two-sided p values” were obtained from a 2 × 2 contingency table analyzed by χ2 test (including Yates’ continuity correction). GraphPad Instat version 2.03 was used for the calculations.
A Fisher’s exact test was used to calculate statistical differences in the percentage of cells showing H2AX foci, APBs, anaphase bridges, p53 expression, as well as the pathologies shown in Figure 2E.
The Wilcoxon-Mann-Whitney rank sum test was used for statistical comparisons of papilloma number and size between wild-type and K5TRF2 mice, as well as for the histograms of p53 abundance in Supplementary Figure 3.
Finally, a log rank test was used to calculate statistical differences in survival of the different mouse cohorts.
Acknowledgments
Thanks to E. Santos and R. Serrano for mouse care and genotyping, and M. Serrano and members of the Blasco Group for critical reading of the manuscript. R.B. is a predoctoral fellow founded by the Regional Government of Madrid (CAM). P.M. is a Ramon y Cajal senior scientist. M.A.B.’s laboratory is funded by the MCyT (SAF2001-1869, GEN2001-4856-C13-08), by the CAM (08.1/0054/01), European Union (TELOSENS FIGH-CT-2002-00217, INTACT LSHC-CT-2003-506803, ZINCAGE FOOD-CT-2003-506850, RISC-RAD FI6R-CT-2003-508842, Mol. Cancer Med. LSHC-CT-2004-502943), and the Josef Steiner Award 2003.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.406207
References
- Artandi S.E., Chang S., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Chang S., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Gottlieb G.J., Chin L., DePinho R.A., Chin L., DePinho R.A., DePinho R.A. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Bailey S.M., Brenneman M.A., Goodwin E.H., Brenneman M.A., Goodwin E.H., Goodwin E.H. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 2004;32:3743–3751. doi: 10.1093/nar/gkh691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmain A., Ramsden M., Bowden G.T., Smith J., Ramsden M., Bowden G.T., Smith J., Bowden G.T., Smith J., Smith J. Activation of the mouse Harvey-ras gene in chemically induced benign skin papillomas. Nature. 1984;307:658–660. doi: 10.1038/307658a0. [DOI] [PubMed] [Google Scholar]
- Bechter O.E., Zou Y., Walker W., Wright W.E., Shay J.W., Zou Y., Walker W., Wright W.E., Shay J.W., Walker W., Wright W.E., Shay J.W., Wright W.E., Shay J.W., Shay J.W. Telomeric recombination in mismatch repair deficient human colon cancer cells and telomerase inhibition. Cancer Res. 2004;64:3444–3451. doi: 10.1158/0008-5472.CAN-04-0323. [DOI] [PubMed] [Google Scholar]
- Biroccio A., Rizzo A., Elli R., Koering C.E., Belleville A., Benassi B., Leonetti C., Stevens M.F., D’Incalci M., Zupi G., Rizzo A., Elli R., Koering C.E., Belleville A., Benassi B., Leonetti C., Stevens M.F., D’Incalci M., Zupi G., Elli R., Koering C.E., Belleville A., Benassi B., Leonetti C., Stevens M.F., D’Incalci M., Zupi G., Koering C.E., Belleville A., Benassi B., Leonetti C., Stevens M.F., D’Incalci M., Zupi G., Belleville A., Benassi B., Leonetti C., Stevens M.F., D’Incalci M., Zupi G., Benassi B., Leonetti C., Stevens M.F., D’Incalci M., Zupi G., Leonetti C., Stevens M.F., D’Incalci M., Zupi G., Stevens M.F., D’Incalci M., Zupi G., D’Incalci M., Zupi G., Zupi G., et al. TRF2 inhibition triggers apoptosis and reduces tumorigenicity of human melanoma cells. Eur. J. Cancer. 2006;42:1881–1888. doi: 10.1016/j.ejca.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Blasco M.A. Telomeres and human disease: Cancer, ageing and beyond. Nat. Rev. Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- Blasco M.A., Hahn W.C., Hahn W.C. Evolving views of telomerase and cancer. Trends Cell Biol. 2003;13:289–294. doi: 10.1016/s0962-8924(03)00085-0. [DOI] [PubMed] [Google Scholar]
- Blasco M.A., Lee H.-W., Hande P., Samper E., Lansdorp P., DePinho R., Greider C.W., Lee H.-W., Hande P., Samper E., Lansdorp P., DePinho R., Greider C.W., Hande P., Samper E., Lansdorp P., DePinho R., Greider C.W., Samper E., Lansdorp P., DePinho R., Greider C.W., Lansdorp P., DePinho R., Greider C.W., DePinho R., Greider C.W., Greider C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Bradshaw P.S., Stavropoulos D.J., Meyn M.S., Stavropoulos D.J., Meyn M.S., Meyn M.S. Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat. Genet. 2005;37:193–197. doi: 10.1038/ng1506. [DOI] [PubMed] [Google Scholar]
- Celli G.B., de Lange T., de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Chan S.W.-L., Blackburn E.H., Blackburn E.H. New ways not to make ends meet: Telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002;21:553–563. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- Chin L., Artandi S.E., Shen Q., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A., Artandi S.E., Shen Q., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A., Shen Q., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A., Gottlieb G.J., Greider C.W., DePinho R.A., Greider C.W., DePinho R.A., DePinho R.A. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Saretzki G., Carter N.P., Jackson S.P., Carter N.P., Jackson S.P., Jackson S.P. DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- Dantzer X., Giraud-Panis M.J., Jaco I., Ame J.C., Schultz I., Blasco M., Koering C.E., Gilson E., Menissier-de Murcia J., de Murcia G., Giraud-Panis M.J., Jaco I., Ame J.C., Schultz I., Blasco M., Koering C.E., Gilson E., Menissier-de Murcia J., de Murcia G., Jaco I., Ame J.C., Schultz I., Blasco M., Koering C.E., Gilson E., Menissier-de Murcia J., de Murcia G., Ame J.C., Schultz I., Blasco M., Koering C.E., Gilson E., Menissier-de Murcia J., de Murcia G., Schultz I., Blasco M., Koering C.E., Gilson E., Menissier-de Murcia J., de Murcia G., Blasco M., Koering C.E., Gilson E., Menissier-de Murcia J., de Murcia G., Koering C.E., Gilson E., Menissier-de Murcia J., de Murcia G., Gilson E., Menissier-de Murcia J., de Murcia G., Menissier-de Murcia J., de Murcia G., de Murcia G., et al. Functional interaction between poly(ADP-ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol. Cell. Biol. 2004;24:1595–1607. doi: 10.1128/MCB.24.4.1595-1607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W.L., Jaspers N.G., Hoeijmakers J.H., Jaspers N.G., Hoeijmakers J.H., Hoeijmakers J.H. Molecular mechanism of nucleotide excision repair. Genes & Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes & Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Dlugosz A.A., Glick A.B., Tennenbaum T., Weinberg W.C., Yuspa S.H., Glick A.B., Tennenbaum T., Weinberg W.C., Yuspa S.H., Tennenbaum T., Weinberg W.C., Yuspa S.H., Weinberg W.C., Yuspa S.H., Yuspa S.H. Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol. 1995;254:3–20. doi: 10.1016/0076-6879(95)54003-2. [DOI] [PubMed] [Google Scholar]
- Dunham M.A., Neumann A.A., Fasching C.L., Reddel R.R., Neumann A.A., Fasching C.L., Reddel R.R., Fasching C.L., Reddel R.R., Reddel R.R. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao M., O’Sullivan R., Peters A.H., Jenuwein T., Blasco M.A., O’Sullivan R., Peters A.H., Jenuwein T., Blasco M.A., Peters A.H., Jenuwein T., Blasco M.A., Jenuwein T., Blasco M.A., Blasco M.A. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao I., Garcia-Cao M., Tomas-Loba A., Martin-Caballero J., Flores J.M., Klatt P., Blasco M.A., Serrano M., Garcia-Cao M., Tomas-Loba A., Martin-Caballero J., Flores J.M., Klatt P., Blasco M.A., Serrano M., Tomas-Loba A., Martin-Caballero J., Flores J.M., Klatt P., Blasco M.A., Serrano M., Martin-Caballero J., Flores J.M., Klatt P., Blasco M.A., Serrano M., Flores J.M., Klatt P., Blasco M.A., Serrano M., Klatt P., Blasco M.A., Serrano M., Blasco M.A., Serrano M., Serrano M. Increased p53 activity does not accelerate telomere-driven ageing. EMBO Rep. 2006;7:546–552. doi: 10.1038/sj.embor.7400667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Suárez E., Samper E., Flores J.M., Blasco M.A., Samper E., Flores J.M., Blasco M.A., Flores J.M., Blasco M.A., Blasco M.A. Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nat. Genet. 2000;26:114. doi: 10.1038/79089. [DOI] [PubMed] [Google Scholar]
- González-Suárez E., Samper E., Ramírez A., Flores J.M., Martín-Caballero J., Jorcano J.L., Blasco M.A., Samper E., Ramírez A., Flores J.M., Martín-Caballero J., Jorcano J.L., Blasco M.A., Ramírez A., Flores J.M., Martín-Caballero J., Jorcano J.L., Blasco M.A., Flores J.M., Martín-Caballero J., Jorcano J.L., Blasco M.A., Martín-Caballero J., Jorcano J.L., Blasco M.A., Jorcano J.L., Blasco M.A., Blasco M.A. Increased epidermal tumors and increased skin, wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 2001;20:2619–2630. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Suárez E., Flores J.M., Blasco M.A., Flores J.M., Blasco M.A., Blasco M.A. Cooperation between p53 mutation and high telomerase transgenic expression in spontaneous cancer development. Mol. Cell. Biol. 2002;22:7291–7301. doi: 10.1128/MCB.22.20.7291-7301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S., Jaco I., Fraga M.F., Chen T., Li E., Esteller M., Blasco M.A., Jaco I., Fraga M.F., Chen T., Li E., Esteller M., Blasco M.A., Fraga M.F., Chen T., Li E., Esteller M., Blasco M.A., Chen T., Li E., Esteller M., Blasco M.A., Li E., Esteller M., Blasco M.A., Esteller M., Blasco M.A., Blasco M.A. DNA methytransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Goytisolo F.A., Blasco M.A., Blasco M.A. Many ways to telomere dysfunction: In vivo studies using mouse models. Oncogene. 2002;21:584–591. doi: 10.1038/sj.onc.1205085. [DOI] [PubMed] [Google Scholar]
- Greenberg R.A., Chin L., Femino A., Lee K.H., Gottlieb G.J., Singer R.H., Greider C.W., DePinho R.A., Chin L., Femino A., Lee K.H., Gottlieb G.J., Singer R.H., Greider C.W., DePinho R.A., Femino A., Lee K.H., Gottlieb G.J., Singer R.H., Greider C.W., DePinho R.A., Lee K.H., Gottlieb G.J., Singer R.H., Greider C.W., DePinho R.A., Gottlieb G.J., Singer R.H., Greider C.W., DePinho R.A., Singer R.H., Greider C.W., DePinho R.A., Greider C.W., DePinho R.A., DePinho R.A. Short dysfunctional telomeres impair tumorigenesis in the INK4a(Δ2/3) cancer-prone mouse. Cell. 1999;97:515–525. doi: 10.1016/s0092-8674(00)80761-8. [DOI] [PubMed] [Google Scholar]
- Herrera E., Samper E., Martin-Caballero J., Flores J.M., Lee H.W., Blasco M.A., Samper E., Martin-Caballero J., Flores J.M., Lee H.W., Blasco M.A., Martin-Caballero J., Flores J.M., Lee H.W., Blasco M.A., Flores J.M., Lee H.W., Blasco M.A., Lee H.W., Blasco M.A., Blasco M.A. Disease states associated to telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 1999;18:2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J., Hoke K., Mirzoeva O.K., Bakkenist C., Kastan M.B., Petrini J.H., de Lange T., Hoke K., Mirzoeva O.K., Bakkenist C., Kastan M.B., Petrini J.H., de Lange T., Mirzoeva O.K., Bakkenist C., Kastan M.B., Petrini J.H., de Lange T., Bakkenist C., Kastan M.B., Petrini J.H., de Lange T., Kastan M.B., Petrini J.H., de Lange T., Petrini J.H., de Lange T., de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laud P.R., Multani A.S., Bailey S.M., Wu L., Ma J., Kingsley C., Lebel M., Pathak S., DePinho R.A., Chang S., Multani A.S., Bailey S.M., Wu L., Ma J., Kingsley C., Lebel M., Pathak S., DePinho R.A., Chang S., Bailey S.M., Wu L., Ma J., Kingsley C., Lebel M., Pathak S., DePinho R.A., Chang S., Wu L., Ma J., Kingsley C., Lebel M., Pathak S., DePinho R.A., Chang S., Ma J., Kingsley C., Lebel M., Pathak S., DePinho R.A., Chang S., Kingsley C., Lebel M., Pathak S., DePinho R.A., Chang S., Lebel M., Pathak S., DePinho R.A., Chang S., Pathak S., DePinho R.A., Chang S., DePinho R.A., Chang S., Chang S. Elevated telomere–telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes & Dev. 2005;19:2560–2570. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-W., Blasco M.A., Gottlieb G.J., Greider C.W., DePinho R.A., Blasco M.A., Gottlieb G.J., Greider C.W., DePinho R.A., Gottlieb G.J., Greider C.W., DePinho R.A., Greider C.W., DePinho R.A., DePinho R.A. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Li B., Oestreich S., de Lange T., Oestreich S., de Lange T., de Lange T. Identification of human Rap1: Implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- Matsutani N., Yokozaki H., Tahara E., Tahara H., Kuniyasu H., Haruma K., Chayama K., Yasui W., Tahara E., Yokozaki H., Tahara E., Tahara H., Kuniyasu H., Haruma K., Chayama K., Yasui W., Tahara E., Tahara E., Tahara H., Kuniyasu H., Haruma K., Chayama K., Yasui W., Tahara E., Tahara H., Kuniyasu H., Haruma K., Chayama K., Yasui W., Tahara E., Kuniyasu H., Haruma K., Chayama K., Yasui W., Tahara E., Haruma K., Chayama K., Yasui W., Tahara E., Chayama K., Yasui W., Tahara E., Yasui W., Tahara E., Tahara E. Expression of telomeric repeat binding factor 1 and 2 and TRF1-interacting nuclear protein 2 in human gastric carcinomas. Int. J. Oncol. 2001;19:507–512. [PubMed] [Google Scholar]
- Modesti M., Kanaar R., Kanaar R. DNA repair: Spot(light)s on chromatin. Curr. Biol. 2002;11:229–232. doi: 10.1016/s0960-9822(01)00112-9. [DOI] [PubMed] [Google Scholar]
- Muñoz P., Blanco R., Flores J.M., Blasco M.A., Blanco R., Flores J.M., Blasco M.A., Flores J.M., Blasco M.A., Blasco M.A. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 2005;10:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- Muñoz P., Blanco R., Blasco M.A., Blanco R., Blasco M.A., Blasco M.A. Role of the TRF2 telomeric protein in cancer and ageing. Cell Cycle. 2006;5:718–721. doi: 10.4161/cc.5.7.2636. [DOI] [PubMed] [Google Scholar]
- Muntoni A., Reddel R.R., Reddel R.R. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 2005;14:191–196. doi: 10.1093/hmg/ddi266. [DOI] [PubMed] [Google Scholar]
- Murillas R., Larcher F., Conti C.J., Santos M., Ullrich A., Jorcano J.L., Larcher F., Conti C.J., Santos M., Ullrich A., Jorcano J.L., Conti C.J., Santos M., Ullrich A., Jorcano J.L., Santos M., Ullrich A., Jorcano J.L., Ullrich A., Jorcano J.L., Jorcano J.L. Expression of a dominant negative mutant of epidermal growth factor receptor in the epidermis of transgenic mice elicits striking alterations on hair follicle development and skin structure. EMBO J. 1995;14:5216–5223. doi: 10.1002/j.1460-2075.1995.tb00206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh B.-K., Kim Y.-J., Park C., Park Y.N., Kim Y.-J., Park C., Park Y.N., Park C., Park Y.N., Park Y.N. Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am. J. Pathol. 2005;166:73–80. doi: 10.1016/S0002-9440(10)62233-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A., Harrigan J., Hickson I.D., Bohr V.A., Hickson I.D., Bohr V.A., Bohr V.A. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- Petit C., Sancar A., Sancar A. Nucleotide excision repair: From E. coli to man. Biochimie. 1999;81:15–25. doi: 10.1016/s0300-9084(99)80034-0. [DOI] [PubMed] [Google Scholar]
- Samper E., Goytisolo F.A., Slijepcevic P., van Buul P.P., Blasco M.A., Goytisolo F.A., Slijepcevic P., van Buul P.P., Blasco M.A., Slijepcevic P., van Buul P.P., Blasco M.A., van Buul P.P., Blasco M.A., Blasco M.A. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J.W., Wright W.E., Wright W.E. Telomerase therapeutics for cancer: Challenges and new directions. Nat. Rev. Drug Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A., van Steensel B., Bianchi A., Oelmann S., Schaefer M.R., Schnapp G., de Lange T., van Steensel B., Bianchi A., Oelmann S., Schaefer M.R., Schnapp G., de Lange T., Bianchi A., Oelmann S., Schaefer M.R., Schnapp G., de Lange T., Oelmann S., Schaefer M.R., Schnapp G., de Lange T., Schaefer M.R., Schnapp G., de Lange T., Schnapp G., de Lange T., de Lange T. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H., Smogorzewska A., de Lange T., Smogorzewska A., de Lange T., de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A., de Lange T., Smogorzewska A., de Lange T., de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Wang R.C., Smogorzewska A., de Lange T., Smogorzewska A., de Lange T., de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Wu L., Multani A.S., He H., Cosme-Blanco W., Deng Y., Deng J.M., Bachilo O., Pathak S., Tahara H., Bailey S.M., Multani A.S., He H., Cosme-Blanco W., Deng Y., Deng J.M., Bachilo O., Pathak S., Tahara H., Bailey S.M., He H., Cosme-Blanco W., Deng Y., Deng J.M., Bachilo O., Pathak S., Tahara H., Bailey S.M., Cosme-Blanco W., Deng Y., Deng J.M., Bachilo O., Pathak S., Tahara H., Bailey S.M., Deng Y., Deng J.M., Bachilo O., Pathak S., Tahara H., Bailey S.M., Deng J.M., Bachilo O., Pathak S., Tahara H., Bailey S.M., Bachilo O., Pathak S., Tahara H., Bailey S.M., Pathak S., Tahara H., Bailey S.M., Tahara H., Bailey S.M., Bailey S.M., et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Zhu X.D., Kuster B., Mann M., Petrini J.H., Lange T., Kuster B., Mann M., Petrini J.H., Lange T., Mann M., Petrini J.H., Lange T., Petrini J.H., Lange T., Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- Zhu X.D., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J.H., de Lange T., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J.H., de Lange T., Kuster B., Mann M., Hoeijmakers J.H., de Lange T., Mann M., Hoeijmakers J.H., de Lange T., Hoeijmakers J.H., de Lange T., de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- Zijlmans J.M., Martens U.M., Poon S.S., Raap A.K., Tanke H.J., Ward R.K., Lansdorp P.M., Martens U.M., Poon S.S., Raap A.K., Tanke H.J., Ward R.K., Lansdorp P.M., Poon S.S., Raap A.K., Tanke H.J., Ward R.K., Lansdorp P.M., Raap A.K., Tanke H.J., Ward R.K., Lansdorp P.M., Tanke H.J., Ward R.K., Lansdorp P.M., Ward R.K., Lansdorp P.M., Lansdorp P.M. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]