Abstract
Quorum sensing is a cell–cell communication mechanism that bacteria use to collectively regulate gene expression and, at a higher level, to coordinate group behavior. In the bioluminescent marine bacterium Vibrio harveyi, sensory information from three independent quorum-sensing systems converges on the shared response regulator LuxO. When LuxO is phosphorylated, it activates the expression of a putative repressor that destabilizes the mRNA encoding the master quorum-sensing transcriptional regulator LuxR. In the closely related species Vibrio cholerae, this repressor was revealed to be the RNA chaperone Hfq together with four small regulatory RNAs (sRNAs) called Qrr1–4 (quorum regulatory RNA). Here, we identify five Qrr sRNAs that control quorum sensing in V. harveyi. Mutational analysis reveals that only four of the five Qrrs are required for destabilization of the luxR mRNA. Surprisingly, unlike in V. cholerae where the sRNAs act redundantly, in V. harveyi, the Qrr sRNAs function additively to control quorum sensing. This latter mechanism produces a gradient of LuxR that, in turn, enables differential regulation of quorum-sensing target genes. Other regulators appear to be involved in control of V. harveyi qrr expression, allowing the integration of additional sensory information into the regulation of quorum-sensing gene expression.
Keywords: Quorum sensing, autoinducer, sRNA
Quorum sensing is a process of bacterial cell–cell communication that involves the production, secretion, and detection of extracellular signal molecules called autoinducers (AIs) (Waters and Bassler 2005). Bacteria use quorum sensing to coordinate group behaviors by collectively regulating gene expression. At low cell densities, extracellular AIs are below detectable concentrations, but as bacterial populations grow increasingly dense, extracellular AIs accumulate to the critical concentrations required to trigger changes in target gene expression. In effect, quorum sensing allows bacteria to function as multicellular organisms by enabling them to carry out behaviors such as biofilm formation, symbiosis, type III secretion, virulence, and bioluminescence in unison (Fuqua et al. 1996; McFall-Ngai and Ruby 2000; Miller and Bassler 2001; Hammer and Bassler 2003; Henke and Bassler 2004a; Waters and Bassler 2005).
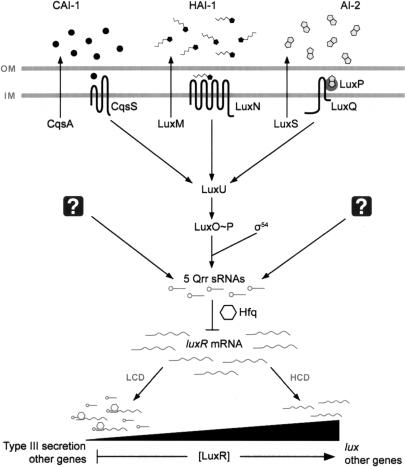
The marine bacterium Vibrio harveyi uses three different AIs—HAI-1, CAI-1, and AI-2—to control the expression of genes responsible for bioluminescence and numerous other traits (Fig. 1). HAI-1, a species-specific signal, is a homoserine lactone synthesized by LuxM (Cao and Meighen 1989; Bassler et al. 1993). CAI-1 is an undefined genus-specific signal produced by CqsA (Miller et al. 2002; Henke and Bassler 2004b). AI-2, a furanosyl borate diester synthesized by the LuxS enzyme, is produced and detected by many bacterial species and is proposed to serve as a universal signal molecule that fosters interspecies cell–cell communication (Bassler et al. 1997; Chen et al. 2002; Miller et al. 2004; Xavier and Bassler 2005). In V. harveyi, each of the three AI signals is detected by a distinct membrane-bound autophosphorylating histidine sensor kinase protein. LuxN detects HAI-1, CqsS detects CAI-1, and LuxQ responds to AI-2 via the periplasmic protein LuxP (Fig. 1; Bassler et al. 1993, 1994a; Freeman et al. 2000; Miller et al. 2002; Henke and Bassler 2004b). All three sensors deliver phosphate to the histidine-containing phosphotransfer protein, LuxU, which in turn transfers the phosphoryl group to the conserved σ54-dependent response regulator protein LuxO (Bassler et al. 1994b; Freeman and Bassler 1999a, b; Lilley and Bassler 2000). Under conditions of low cell density (i.e., when AIs are at negligible concentrations), the three sensors act as kinases, and phosphate is transferred to LuxO. LuxO∼P, in conjunction with σ54, indirectly represses expression of the genes encoding luciferase (lux), so no light is produced under this condition (Freeman and Bassler 1999a; Lilley and Bassler 2000). At high cell densities (i.e., when AIs are at detectable concentrations), the interactions of the sensors with their cognate AIs cause the sensors to switch from kinase mode to phosphatase mode, leading to dephosphorylation of LuxO (Freeman and Bassler 1999a). This sequence of events inactivates LuxO, leading to expression of the lux operon, and light is produced (Miyamoto et al. 1994; Freeman and Bassler 1999a). LuxO functions by indirectly regulating the expression of luxR. LuxR directly activates the lux operon and directly or indirectly controls all other known quorum-sensing target genes (Showalter et al. 1990; Swartzman et al. 1992).
Figure 1.
Model of the V. harveyi quorum-sensing system. Three parallel sensory systems converge to regulate quorum-sensing gene expression by controlling levels of the master transcriptional regulator, LuxR. See text for details. The three AIs are CAI-1 (circles), HAI-1 (pentagons with side chains), and AI-2 (double pentagons). The Qrr sRNAs (lollipops) indirectly regulate LuxR protein levels by destabilizing luxR mRNA (wavy lines), a process that is mediated by the RNA chaperone Hfq (hexagons). The multiple Qrr sRNAs produce an increasing gradient of LuxR protein as the cells transition from low to high cell density. Question marks denote putative regulators that control qrr expression. (OM) Outer membrane; (IM) inner membrane; (LCD) low cell density; (HCD) high cell density.
The molecular events connecting LuxO to downstream target gene expression have been characterized recently in the human pathogen Vibrio cholerae, which is related to V. harveyi. At low cell density, LuxO∼P, together with σ54, activates the expression of the genes encoding four small regulatory RNAs (sRNAs) called Qrr 1–4 (quorum regulatory RNA) (Lenz et al. 2004). The sRNAs destabilize the mRNA of hapR, the equivalent of luxR of V. harveyi. In the absence of HapR, no quorum-sensing-controlled gene expression occurs. At high cell density, dephosphorylation inactivates LuxO, terminating production of the sRNAs, relieving repression of hapR, and enabling the regulation of the downstream quorum-sensing-controlled target genes (Lenz et al. 2004).
sRNAs have recently been recognized as important regulatory elements involved in bacterial and eukaryotic developmental processes (Masse et al. 2003b; Wienholds and Plasterk 2005). In bacteria, sRNAs post-transcriptionally regulate genes involved in responses to conditions such as limited iron, sugar–phosphate stress, cell surface stress, and oxidative stress (Romeo 1998; Zhang et al. 1998; Masse and Gottesman 2002; Vanderpool and Gottesman 2004; Storz et al. 2005). Many bacterial sRNAs require Hfq, an RNA chaperone similar in sequence and structure to eukaryotic Sm proteins involved in RNA splicing (Moller et al. 2002; Zhang et al. 2002). Hfq binds to particular sRNAs and their mRNA targets to promote base-pairing and, in some cases, to facilitate the degradation of the sRNA–mRNA complex (Schumacher et al. 2002; Moll et al. 2003; Geissmann and Touati 2004; Valentin-Hansen et al. 2004; Kawamoto et al. 2006). In the case of V. cholerae quorum sensing, the Qrr sRNAs depend on Hfq to destabilize the hapR mRNA. Importantly, any one of the four Qrr sRNAs in V. cholerae is perfectly sufficient to control hapR, suggesting that the four Qrr sRNAs are functionally redundant (Lenz et al. 2004).
Here we identify and characterize five Qrr sRNAs in V. harveyi. Unlike in V. cholerae, the V. harveyi Qrr sRNAs are not functionally redundant but, rather, act additively to control quorum sensing. Furthermore, mutational analysis reveals that only four of the five sRNAs are required to destabilize the luxR mRNA. In V. cholerae, we have proposed that multiple redundant sRNAs function as an ultrasensitive switch to control the transition from low to high cell density. We propose that, in V. harveyi, the multiple sRNAs function to translate increasing AI concentrations into a precise gradient of LuxR protein. LuxR, in turn, induces a gradient of expression of quorum-sensing target genes. We further propose that the multiple sRNAs serve as the focal point through which additional metabolic cues feed into the quorum-sensing circuit to regulate global gene expression.
Results
Identification of five sRNAs in V. harveyi
We used a bioinformatics approach to identify four quorum-sensing-regulated sRNA genes (qrr1–4) in V. cholerae. We could not perform the analogous analysis in V. harveyi because its genome has not been fully sequenced. However, we did identify one quorum-sensing sRNA gene in V. harveyi (qrr1) located upstream of and adjacent to luxO, as this DNA sequence was available (Lenz et al. 2004). We suspected that, as in V. cholerae, additional quorum-sensing regulatory sRNAs would exist in V. harveyi. To identify them, we performed a genetic screen based on differential fluorescence induction and fluorescence-activated cell sorting. Our rationale was that LuxO∼P would induce the expression of the sRNA genes at low cell density (Fig. 1). We exploited this relationship by introducing a V. harveyi random promoter-gfp library into an Escherichia coli strain carrying V. harveyi luxO D47E (Freeman and Bassler 1999a), a LuxO allele that mimics the constitutively active form of LuxO, LuxO∼P. The E. coli transformants were sorted for those producing high levels of GFP, suggesting that they harbored plasmids with promoters directly activated by LuxO∼P. To verify this assumption, and to eliminate constitutively active promoters, the gfp-expressing plasmids were isolated and transformed into an E. coli strain carrying a luxO-null allele (luxO D47A) (Freeman and Bassler 1999a). Again, the cells were sorted, this time for those exhibiting low gfp expression. As a final step, these GFP-containing plasmids were isolated and once again introduced into the E. coli strain carrying luxO D47E. Cells displaying high gfp expression were isolated and the plasmids were sequenced. This screen identified V. harveyi qrr2, qrr3, and qrr4 based on their similarities to other available Vibrio qrr sequences. We anticipated identifying five qrr sRNA genes in V. harveyi because our previous bioinformatics analysis identified five candidate qrr genes in Vibrio vulnificus and Vibrio parahaemolyticus, two Vibrio species that are more closely related to V. harveyi than to V. cholerae. We identified V. harveyi qrr5 by scanning the partially sequenced V. harveyi genomic DNA sequence (available at http://www.tigr.org).
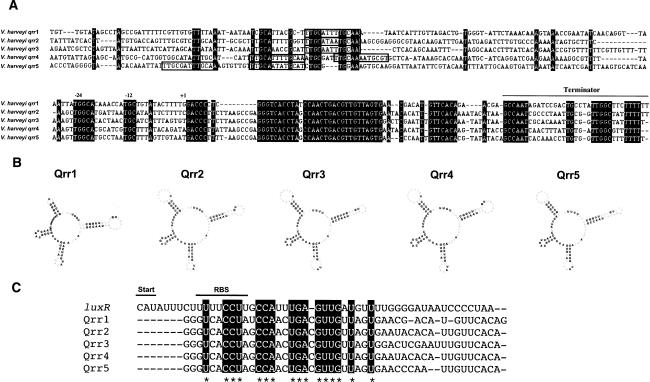
The five V. harveyi qrr sRNA genes have high sequence identity and the sRNAs have similar predicted secondary structures (Fig. 2A,B; Hofacker 2003). Qrr1 is a slightly shorter RNA than the other four and thus has the most distinct predicted secondary structure. The V. harveyi Qrrs belong to a class of trans-acting antisense regulators, meaning that the sRNA genes are encoded in intergenic regions of the genome, not necessarily near the genes encoding their targets (Storz et al. 2005). These sRNAs typically have short regions of complementarity to the 5′ untranslated region (UTR) of their target mRNAs, and pairing within this region can affect the stability of the target messages (Gottesman 2002). We aligned the complement of the luxR 5′UTR with the sequences of all five V. harveyi Qrrs (Fig. 2C). The nucleotide sequences in the regions presumed to be involved in complementary base-pairing between the luxR promoter and the Qrr sRNAs are conserved across all five sRNAs. In all cases, the luxR–Qrr complementary region overlaps the putative ribosome-binding site for luxR, and it presumably takes on a configuration that has been demonstrated to be critical for post-transcriptional regulation in other trans-acting sRNAs (Masse et al. 2003a; Morita et al. 2005, 2006). Although the five qrr gene sequences are highly similar, alignment of their promoter regions shows that very little sequence similarity exists outside of the predicted σ54 consensus binding site and the putative LuxO∼P-binding site (Fig. 2A, the σ54 site is denoted by −12, −24, and the LuxO∼P sites are marked by boxes). The predicted LuxO∼Pbinding site (Lenz et al. 2004), TTGCAW3TGCAA, varies in extent of conservation and in its position relative to the start of transcription of each sRNA gene. Therefore, the expression of each qrr sRNA gene could be differentially regulated by LuxO∼P and possibly by other factors.
Figure 2.
The Qrr sRNAs of V. harveyi. (A) Multiple sequence alignment of the five qrr genes and their upstream regulatory regions. Numbering is based on orthology of flanking genes. Nucleotides in black indicate identity. The putative σ54-binding sites are marked as −12 and −24, the predicted starts of transcription are labeled as +1, and the terminators are indicated by the line above the sequences. Putative LuxO-binding sites are surrounded by boxes. (B) Lowest-energy secondary structural predictions for the V. harveyi Qrr sRNAs as generated by the RNAfold program using default RNA parameters (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). Bold type indicates regions conserved among the V. harveyi Qrr sRNAs. (C) Alignment of the complement of the luxR 5′UTR with a region of the V. harveyi Qrr sRNAs. Regions conserved across all Qrr sRNAs are highlighted and are likely involved in base-pairing to the luxR mRNA. (Start) Translation start site; (RBS) ribosome-binding site.
Differential regulation of qrr genes in V. harveyi
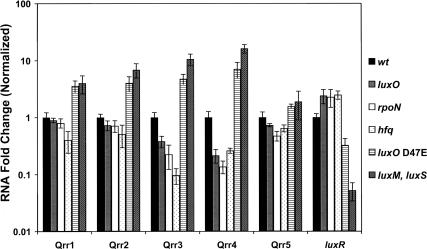
To establish that the V. harveyi qrr genes are regulated by quorum sensing, we used quantitative real-time PCR to measure their transcript levels in wild-type V. harveyi and in various V. harveyi quorum-sensing mutants (Fig. 3). We examined two classes of quorum-sensing mutants: those that are “locked” in high-cell-density mode and those that are “locked” in low-cell-density mode. Both classes are impervious to the presence of AIs and therefore do not exhibit density-dependent gene expression. Qrr RNA levels range from 0.2- to 10-fold lower in the “locked” high-cell-density strains (luxO, rpoN [encoding σ54] and hfq mutants) than in the wild-type strain (Fig. 3). In the absence of LuxO and RpoN, minimal expression of the qrr genes occurs because both proteins are required for activation of their expression (Fig. 1). The low levels of qrr expression in the hfq strain suggest that, in addition to mediating sRNA–mRNA pairing, Hfq is also required to stabilize the Qrr sRNAs. By contrast, high expression of the qrr genes occurs in V. harveyi strains “locked” in low-cell-density mode (Fig. 3, luxO D47E and the luxM, luxS double AI synthase mutant; the triple-synthase mutant was not used because it has a modest growth defect and because CAI-1 has only a slight impact on quorum-sensing-controlled gene expression [Henke and Bassler 2004b; Waters and Bassler 2006]). In a luxO D47E mutant, LuxO is in a constitutively active form; hence, the qrr genes are overexpressed. Likewise, there are high LuxO∼P levels in the double AI synthase mutant; consistent with this, Qrr levels are twofold to 16-fold higher in this strain than in the wild type. As a control, luxR mRNA levels were also quantified in the same strains (Fig. 3). Our model predicts that luxR mRNA varies reciprocally with sRNA levels. Indeed, luxR expression was twofold higher in the “locked” high-cell-density mutants luxO, rpoN, and hfq than in the wild-type strain, and threefold and 20-fold lower in the luxO D47E and luxM luxS mutants, respectively. These luxR levels are in agreement with previously measured LuxR protein and bioluminescence levels for each of the V. harveyi strains shown (Lenz et al. 2004; Waters and Bassler 2006). We note that Qrr5 levels differ marginally in the different quorum-sensing mutants and we return to this point below.
Figure 3.
Regulation of Qrr sRNAs in V. harveyi quorum-sensing mutants. RNA was isolated from the following V. harveyi strains grown to OD600 ∼ 1: BB120 (wild type; black), BB721 (luxO∷Tn5; gray), BNL240 (rpoN∷Cm; white), BNL258 (hfq∷Tn5; dotted), KM83 (luxO D47E; horizontal lined), and KM413 (ΔluxM, ΔluxS, hatched). RNA was measured using quantitative real-time PCR. Measurements were normalized to the wild-type values and fold differences are plotted. Each sample was assayed in quadruplicate.
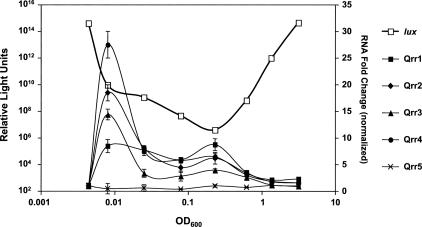
To examine qrr expression in cells transitioning from low to high cell density, wild-type V. harveyi cells were harvested at regular intervals along the growth curve, and bioluminescence and qrr expression were quantified. The quorum-sensing-regulated bioluminescence profile in wild-type V. harveyi exhibits a characteristic pattern: Following overnight growth of the culture, cells are diluted, and subsequently the light output per cell decreases precipitously. This is due to dilution of the AIs below the level required to activate lux expression. As the culture grows, endogenously produced AIs accumulate, and when a threshold AI level is reached, light production commences and increases exponentially (Fig. 4, open squares). Similar to what we observed for luxR and the Qrr sRNAs (Fig. 3), an inverse relationship exists between the expression of the sRNA genes and light production. Specifically, the qrr genes are expressed at low cell density, and their expression decreases as the cells reach high cell density, while light production is minimal at low cell density and increases as the cells achieve high cell density (Fig. 4). Interestingly, at low cell density, each of the sRNA genes is activated to a different degree. Expression of qrr4 shows the most dramatic increase (>25-fold) followed by qrr2, qrr3, and then qrr1 (18-fold, 14-fold, and eightfold, respectively). qrr5 does not exhibit density-dependent gene expression.
Figure 4.
Differential regulation of the Qrr sRNAs. Bioluminescence and Qrr RNA levels were simultaneously measured in wild-type V. harveyi BB120. (Open squares) Light production; (closed squares) Qrr1; (closed diamonds) Qrr2; (closed triangles) Qrr3; (closed circles) Qrr4; (crosses) Qrr5. Relative light units are defined as counts per minute per milliliter per OD600. RNA values are normalized to the first time point (i.e., at the initial dilution). RNA measurements were assayed in quadruplicate.
The V. harveyi Qrr sRNAs function additively to control quorum sensing
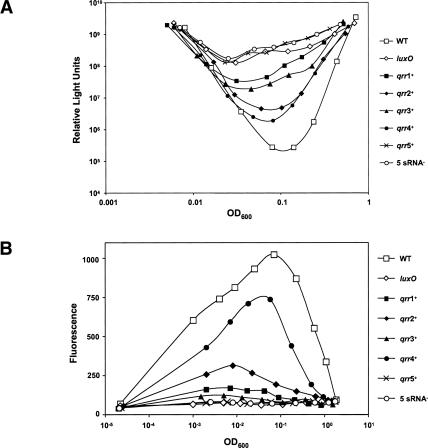
In order to expose the individual roles of the five sRNAs in V. harveyi quorum sensing, we engineered quadruple-deletion mutants that lacked all but one qrr gene and measured some representative quorum-sensing behaviors. We also constructed a quintuple mutant in which all five qrr genes were disrupted. As a reminder, in V. cholerae, the four Qrr sRNAs are completely redundant, and only the simultaneous deletion of all four qrr sRNAs has any effect on repression of hapR expression and, in turn, on quorum-sensing-controlled behaviors (Lenz et al. 2004). To our surprise, the V. harveyi sRNAs are not redundant (Fig. 5A). Rather, we found that the presence of each individual sRNA causes V. harveyi to express a distinct level of density-dependent bioluminescence, suggesting that each sRNA alone is capable of repressing luxR to a particular degree. The extent to which each sRNA represses bioluminescence parallels its strength of promoter expression, as shown in Figure 4. Specifically, in order of descending promoter expression: Qrr4 > Qrr2 > Qrr3 > Qrr1 > Qrr5, an order that exactly matches their individual strengths of lux repression. We interpret this to mean that, because qrr4 is the most highly expressed of the qrr genes, Qrr4 is able to repress luxR to a greater extent than the other Qrrs. Therefore, the density-dependent bioluminescence profile of the qrr4+ mutant most closely resembles that of wild type (Fig. 5A, closed circles). Combinations of single, double, and triple qrr mutants were also assayed for light production, and the single mutants showed the greatest amounts of lux repression, with repression decreasing as additional qrrs were inactivated (data not shown). It is also notable that a strain containing only qrr5 behaves no differently than the quintuple qrr-null mutant, further indicating that qrr5 is not expressed and therefore does not regulate quorum sensing in V. harveyi under the conditions of our experiments. Taken together, these results indicate that the V. harveyi sRNAs act additively to repress luxR expression and that four of the five sRNAs are required for wild-type quorum-sensing behavior.
Figure 5.
The V. harveyi Qrr sRNAs function additively to control quorum sensing. (A) Bioluminescence assays were performed on the following V. harveyi strains: BB120 (wild type; open squares), BB721 (luxO∷Tn5; open diamonds), KT234 (Δqrr2, Δqrr3, Δqrr4, qrr5∷Cm; closed squares), KT212 (Δqrr1, Δqrr3, Δqrr4, qrr5∷Cm; closed diamonds), KT225 (Δqrr1, Δqrr2, Δqrr4, qrr5∷Cm; closed triangles), KT215 (Δqrr1, Δqrr2, Δqrr3, qrr5∷Cm; closed circles), KT133 (Δqrr1, Δqrr2, Δqrr3, Δqrr4; crosses), and KT220 (Δqrr1, Δqrr2, Δqrr3, Δqrr4, qrr5∷Cm; open circles). (B) Fluorescence production from a qrgA-gfp transcriptional fusion carried in trans the same V. harveyi strains as shown in A and in JAF78 (luxO∷Cm; open diamonds).
Many genes are regulated by quorum sensing in V. harveyi, some positively and some negatively. Bioluminescence, which has been our focus up to this point, is positively regulated by quorum sensing. It is of interest, then, to consider Qrr control of a representative negatively regulated gene to examine Qrr function in a related, but distinct, regulatory context. To accomplish this, we monitored gfp expression from a promoter-gfp transcriptional fusion made to a quorum-sensing-repressed gene encoding a GGDEF-containing protein called qrgA (quorum-sensing regulated GGDEF) (Waters and Bassler 2006; in that work, this clone was previously known as #275). The expression pattern of the qrgA-gfp reporter fusion in the different qrr mutants is opposite to that of lux (Fig. 5B). Specifically, in wild type at low cell density, qrgA-gfp expression increases, and subsequently expression decreases as the culture reaches high cell density. Among the qrr mutants, qrgA-gfp expression was highest in the Qrr4+ strain, and decreasing expression was observed in the order Qrr2+, Qrr1+, Qrr3+, Qrr5+. Again, we presume that because qrr4 is the most highly expressed of the five qrr genes, Qrr4 has the most pronounced repressive effect on luxR. Therefore, a LuxR-repressed target is most highly expressed in a Qrr4+ strain. We note that expression of qrgA-gfp is modestly higher in the Qrr1+ strain than in the Qrr3+ strain (Fig. 5B, closed squares vs. closed triangles), and from Figures 4 and 5A we would predict the opposite pattern. However, only extremely low levels of qrgA-gfp expression occur in the Qrr1+ and Qrr3+ strains. This inconsistency is likely due to a lack of sensitivity in our measurements of low output from the reporter. Consistent with results from Figures 4 and 5A, Figure 5B shows that there is no detectable expression of qrgA-gfp in the Qrr5+ strain. Likewise, our controls show that there is also no qrgA-gfp expression in the quintuple sRNA mutant or in a luxO-null mutant. Again, these results suggest that Qrr5 does not function in quorum-sensing-controlled gene regulation. Based on these data, we propose that a balance exists between the combined levels of the Qrr sRNAs and the luxR mRNA and that AIs control V. harveyi quorum-sensing gene expression by altering this balance. At low cell density, the sRNAs outnumber and titrate out the luxR mRNA, while at high cell density this balance is reversed and the luxR mRNA wins out.
A balance between sRNAs and luxR mRNA in V. harveyi
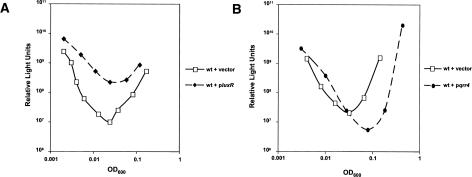
To test the above titration model, we artificially altered the balance between the Qrr sRNAs and luxR mRNA in V. harveyi and monitored the effect on quorum sensing using bioluminescence as the readout (Fig. 6). First, we transformed wild-type V. harveyi with a low-copy plasmid carrying luxR under its native promoter and measured bioluminescence over time (Fig. 6A). Over the growth curve, the strain containing additional copies of luxR produced from threefold to 24-fold more light per cell than did the wild-type strain containing the vector alone. This result suggests that the increased luxR mRNA titrated out the Qrr sRNAs at low cell density, leading to increased LuxR production and, consequently, increased bioluminescence expression. In the reciprocal experiment, we increased the levels of one of the Qrr sRNAs relative to the luxR mRNA. Specifically, we introduced qrr4 into wild-type V. harveyi on the same low-copy plasmid we used in the preceding luxR experiment. Increased expression of qrr4 reduced light production per cell by as much as fourfold throughout growth (Fig. 6B). This result suggests that the balance between the sRNAs and luxR mRNA was shifted in favor of the sRNAs, leading to reduced luxR mRNA and, in turn, to reduced bioluminescence expression. We performed exactly analogous titration experiments with luxR and qrr1–3 and obtained essentially the same results as those shown in Figure 6 for luxR and qrr4. We assume that quorum-sensing targets other than lux would be similarly affected in analogous luxR or qrr multicopy experiments.
Figure 6.
Titration of luxR mRNA and Qrr sRNAs. (A) Bioluminescence assays of wild-type V. harveyi BB120 containing pLAFR2 (solid line, open squares), luxR on pLAFR2 (dotted line, closed diamonds). (B) Wild-type V. harveyi BB120 containing pLAFR2 (solid line, open squares), qrr4 on pLAFR2 (dotted line, closed circles).
Examination of the function of Qrr5
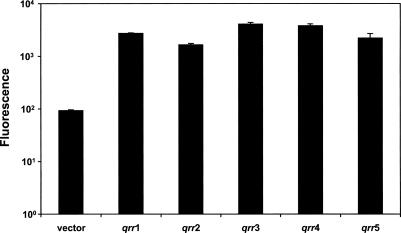
One peculiar finding is that, although similar to the other Qrr sRNAs, Qrr5 appears not to function in V. harveyi (Figs. 4, 5). This finding is especially curious given that V. cholerae does not possess a Qrr5 homolog. It is possible that Qrr5 may be a nonfunctional evolutionary vestige in those Vibrio species that still possess it. To explore this possibility, we constructed transcriptional gfp fusions to all five V. harveyi qrr promoters and transformed them into an E. coli strain carrying luxO D47E. As mentioned, luxO D47E is a constitutively active allele of LuxO. Figure 7 shows that all five of the Qrr promoters are readily activated by luxO D47E in E. coli (15- to 40-fold). No gfp expression was observed from the qrr-gfp fusions in the absence of luxO D47E (data not shown). To account for the finding that qrr5 is expressed in E. coli but not in V. harveyi, we suggest that a repressor function that is not present in E. coli exists in V. harveyi and keeps qrr5 expression off. This possibility is addressed further in the Discussion.
Figure 7.
Expression of qrr genes in E. coli. Fluorescence production from promoter-gfp fusions of each of the five qrr genes in E. coli carrying luxO D47E. E. coli strains: KT1007 (vector control), KT1138 (qrr1-gfp), KT1140 (qrr2-gfp), KT1142 (qrr3-gfp), KT1144 (qrr4-gfp), KT1146 (qrr5-gfp).
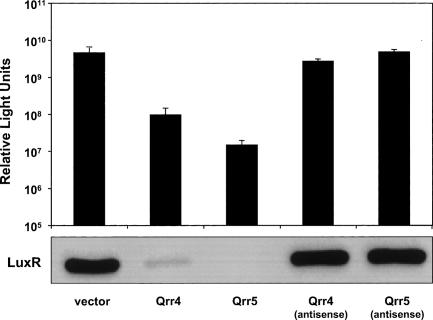
To determine whether Qrr5 can function in V. harveyi, we overexpressed it or its antisense under a constitutive promoter. As controls, we performed the same experiments with Qrr4 and the Qrr4 antisense RNA. When Qrr4 is overproduced, light production per cell decreases ∼100-fold, whereas overproduction of the Qrr4 antisense RNA has no effect on light output (Fig. 8). Similarly, Qrr5, but not the Qrr5 antisense, represses light production following overproduction (>200-fold). Furthermore, Qrr5 functions through the known quorum-sensing pathway because, identically to when qrr4 is overexpressed, LuxR protein levels decrease in cells overexpressing qrr5, as measured by Western blot (Fig. 8, bottom panel). Together, these results show that, if produced, Qrr5 functions analogously to Qrr1–4 in the post-transcriptional regulation of luxR and, in turn, in light production. Thus, it appears that the reason mutation of qrr5 causes no obvious quorum-sensing phenotype in vivo is due to its lack of expression and not to any defect in its function. Consistent with this interpretation, close inspection of the qrr5 promoter region reveals that its predicted LuxO-binding sites each differ from the consensus binding site by one base (TTGCGATTTGCAA, TTGCAATATGCAT). All of the other qrr genes contain at least one perfect LuxO-binding site (TTGCAW3T GCAA). Additionally, the locations of the tandem LuxO-binding sites for qrr5 are 125 and 145 base pairs (bp) upstream of the transcription start site. For the other qrr genes, the LuxO-binding sites lie 95–110 bp upstream of the start sites. Because LuxO likely activates transcription through a DNA looping mechanism (Wyman et al. 1997; Lilley and Bassler 2000), it is possible that the binding site shift, coupled with the alterations in the binding site sequence, severely impair qrr5 expression in vivo. Nonetheless, LuxO∼P is sufficient to activate qrr5 expression in E. coli. This result is discussed further below.
Figure 8.
Overexpression of qrr4 and qrr5 in V. harveyi. Wild-type V. harveyi BB120 was transformed with a vector (KT145) or the vector overexpressing qrr4 (KT153), qrr5 (KT149), qrr4 antisense (KT155), or qrr5 antisense (KT151). (Bottom panel) LuxR protein was measured by Western blotting.
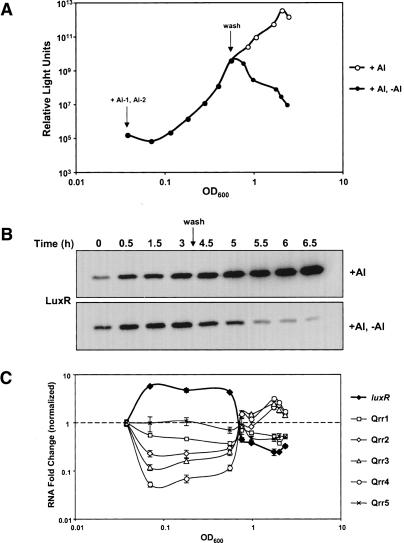
Qrr sRNAs allow a rapid response to changes in cell density
The multiple Qrr sRNAs constitute a post-transcriptional regulatory mechanism that acts at the core of the quorum-sensing circuit in V. harveyi. A sRNA-mediated regulatory mechanism could facilitate extremely rapid transitions between individual and community behaviors. Specifically, unlike proteins, which require transcription and translation, regulatory RNAs, because they are small and only require transcription, can be produced rapidly, and they can likewise be eliminated rapidly through degradation (Masse et al. 2003a). To study how rapidly the Qrrs respond to fluctuations in AI concentrations, we added exogenous AIs to a V. harveyi double AI synthase mutant and subsequently monitored bioluminescence. Additionally, following the initial increase in bioluminescence expression that occurred in response to the AIs, half of the test culture was washed with fresh medium to eliminate AIs and simulate an immediate shift from the high- to the low-cell-density state. The second half of the culture was washed in its own cell-free spent culture fluid so the AI concentration would not change. To follow the molecular events that occur in response to AI changes, we subsequently measured qrr expression, LuxR protein levels, and bioluminescence at different time points. As expected, expression of bioluminescence was triggered by the exogenous addition of AIs. Bioluminescence increased over four orders of magnitude within 3 h following AI addition (Fig. 9A, closed circles). Removal of the AIs resulted in an 80-fold decrease in bioluminescence in 2 h (Fig. 9A, closed circles, after wash). LuxR protein levels showed a pattern similar to that of bioluminescence: Following AI addition, LuxR increased eightfold in 3 h, and upon AI removal, LuxR decreased 20-fold in 2 h (Fig. 9B). Using quantitative real-time PCR, we also established the relationship between qrr and luxR expression over the time course. Within 30 min after AI addition, expression of qrr1–4 decreased maximally, although to varying degrees. Specifically, the different Qrr sRNA levels decreased between twofold and 20-fold within 30 min (Fig. 9C, open symbols), whereas luxR expression increased maximally over fivefold during the same period (Fig. 9C, closed diamonds). By contrast, immediately following removal of AIs, qrr expression increased to above pre-AI-addition levels while luxR expression declined to below initial levels. These results suggest that the sRNAs are instrumental in mediating a rapid response in the form of alterations in luxR levels and, hence, downstream gene expression. We note that, as predicted from the results in Figures 4 and 5, expression levels of qrr5 did not change following addition or removal of AIs (Fig. 9C, crosses).
Figure 9.
Qrr levels fluctuate rapidly in response to changing AI concentrations. (A) Bioluminescence expression in response to AIs. HAI-1 and AI-2 were added at saturating concentrations (5.3 μM and 12.5 μM, respectively) to KM413 (luxM, luxS). Bioluminescence was measured every 30 min for 3 h, after which time the culture was divided in half. One half was washed with fresh medium (closed circles, after wash) and the other half was washed in cell-free spent medium (open circles, after wash), and bioluminescence measurements were taken for 2 h thereafter. (B) LuxR protein levels in response to AIs. Cells were harvested as in A and adjusted for OD600, and LuxR protein levels were determined using Western blotting. (C) Quantitative measurement of Qrr sRNAs and luxR mRNA in response to AIs. RNA was isolated at the time points in A and B and quantified. (Closed diamonds) luxR; (open squares) Qrr1; (open diamonds) Qrr2; (open triangles) Qrr3; (open circles) Qrr4; (crosses) Qrr5. RNA levels were normalized to the first time point, and fold changes are plotted. Samples were assayed in quadruplicate.
Discussion
Bacteria have mechanisms equipping them to respond to fluctuating environmental conditions, such as changes in nutrient levels, acidity, osmolarity, and cell density. In the final case, quorum sensing enables bacteria to perceive changes in their population numbers and to selectively alter gene expression in response to fluctuations in cell density. In Vibrio species, multichannel signal-transduction circuits regulate quorum-sensing-controlled gene expression. In V. cholerae, four sRNAs act redundantly to control quorum-sensing gene expression by destabilizing the mRNA encoding the master quorum-sensing transcriptional regulator, HapR (Lenz et al. 2004). Here, in a closely related but surprisingly differently operating system, we identify five sRNAs in V. harveyi that act additively to control quorum sensing by altering translation of the master transcriptional regulator, LuxR (Fig. 1). Mutational analysis of these sRNAs revealed that only four of the five V. harveyi Qrr sRNAs are required to destabilize the luxR mRNA, and thus to control quorum sensing (Fig. 5).
Bacterial regulatory RNAs participate in a broad range of cellular processes, including carbon storage and utilization, response to oxidative stress, and the transition to stationary phase (Wassarman 2002; Repoila et al. 2003). Many trans-acting sRNAs require the RNA chaperone Hfq, and the pairing of a particular sRNA with its target mRNA can inhibit translation either by occluding ribosome binding or by stimulating degradation of the targeted mRNA (Moller et al. 2002; Moll et al. 2003; Storz et al. 2004). We know that Hfq is required for Qrr-regulated quorum sensing (Lenz et al. 2004), and based on the striking similarity between the V. harveyi Qrr sRNA sequences, and their complementarity to the luxR promoter, we hypothesize that the Qrr sRNAs control luxR expression via a coupled degradation mechanism (Masse et al. 2003a). This stoichiometric mode of action means that regulation by sRNAs is tightly coupled to their synthesis, which in turn is tightly coupled to the activity of particular transcriptional regulators. When sRNA production terminates, the sRNAs are rapidly consumed, so no other mechanism of turnover is required. We propose that, in V. harveyi, high levels of LuxO∼P trigger qrr expression at low cell density. Accumulated Qrr sRNAs pair with luxR mRNA transcripts, and the resulting sRNA–mRNA complexes are subsequently degraded. Thus, the production of multiple sRNAs permits exquisite control over the level of luxR mRNA that can, in turn, be translated into LuxR protein.
The effect of the Qrr sRNAs is of great consequence—initiating, ultimately, a major reprogramming of cell physiology in response to changing cell density. We hypothesize that V. harveyi maintains a tight balance between levels of sRNAs and luxR mRNA, and, in response to changing AI levels, V. harveyi alters target gene expression through the tailored regulation of the genes encoding the multiple Qrr sRNAs (Fig. 1). In turn, the sRNAs collectively enable the cell to maintain a tight rein on levels of LuxR. Other data suggest that all 50 of the quorum-sensing target genes in the V. harveyi regulon are directly or indirectly controlled by LuxR (Waters and Bassler 2006). Thus, Qrr regulation of luxR ultimately dictates the strength and temporal patterns of all the members of the quorum-sensing regulon. We propose that differential gene expression can occur in response to different combinations of AIs because these impinge on LuxR, which is limiting in the cell and has particular affinities for its various target promoters (Waters and Bassler 2006).
Real-time analysis of qrr expression during the transition from low to high cell density shows that each of the five qrr genes is expressed to a different degree, but in a pattern that is reciprocal to that for light production (Fig. 4). This result suggests that each qrr promoter likely has a different affinity for LuxO∼P, which we partially attribute to differences in their LuxO consensus binding sites (Fig. 2). The expression patterns of the five V. harveyi qrr promoter-gfp fusions in E. coli carrying luxO D47E differ from their corresponding expression patterns in V. harveyi. Most notably, qrr5 does not exhibit density-dependent gene expression in V. harveyi (Fig. 4, × marks), and does not affect quorum-sensing behaviors following mutation (Fig. 5), and we know this is not due to limiting levels of LuxO in the cell (K. Tu and B. Bassler, unpubl.). The qrr5 promoter is, however, readily activated in E. coli expressing luxO D47E (Fig. 7, rightmost bar). Moreover, as discussed above, when Qrr5 is artificially overexpressed, it can repress luxR mRNA translation in vivo (Fig. 8). Although we have not yet identified any regulatory role for qrr5 in V. harveyi, we interpret this to mean that, if Qrr5 were expressed in V. harveyi, quorum sensing would be affected. We propose that in V. harveyi, species-specific regulators exist that are not present in E. coli, and they function to control qrr5 expression. Specifically, our data suggest the presence of a repressor that inhibits qrr5 expression in V. harveyi under our experimental conditions.
Other regulators could likewise function at qrr1–4 to uniquely tailor their expression. We note that expression of the five qrrs in E. coli carrying luxO D47E does not follow the same pattern as in V. harveyi (Fig. 7). In V. harveyi, the strength of qrr expression follows in the order qrr4, qrr2, qrr3, qrr1, and qrr5 (Fig. 4), whereas in E. coli the strength of qrr expression follows the order qrr3, qrr4, qrr1, qrr5, and qrr2 (Fig. 7). Consistent with this, initial bioinformatics analysis using hidden Markov models of known transcription factor-binding sites in prokaryotes reveals that there are putative binding motifs in several of the upstream regions adjacent to the Vibrio qrr genes, and these regions differ for the different qrrs. Presumably, these sites function as binding sites for additional regulatory factors. If so, such regulators could furnish additional links from the environment to the system controlling quorum sensing. We are currently performing genetic screens in V. harveyi to identify additional regulators of each of the five sRNA genes and to determine under what conditions qrr5 is expressed in vivo.
V. harveyi, V. parahaemolyticus, and V. vulnificus all contain qrr5 and are closely related organisms, whereas V. cholerae, which is more distantly related, harbors only qrr1–4. The genes flanking qrr5 are not conserved across the three Vibrio species, but those flanking the other four qrr genes are conserved for the most part. These data suggest that qrr5 emerged most recently by duplication and may be classified as a species-specific qrr.
In order to begin to understand the advantage of partitioning control of quorum sensing over multiple sRNAs versus one sRNA, we sought to determine the role of each sRNA in V. harveyi by deleting each individually and in combinations. Our previous work in V. cholerae established that each of the four Qrrs alone is perfectly sufficient to control quorum sensing. Surprisingly, the V. harveyi sRNAs do not act redundantly. Here, we find that if any one of four (qrr1–4) regulatory sRNAs is present, V. harveyi remains capable of quorum sensing, albeit altered to a particular degree (Fig. 5). Our observations suggest that each sRNA has the ability to repress luxR to a different level, and we suspect that this depends on the particular qrr gene promoter strength. Specifically, qrr4 is more highly expressed than the other qrr genes at low cell density (Fig. 4), and it follows that, when only qrr4 is expressed at low cell density, it has the most pronounced effect on repression of luxR (Fig. 5). qrr2 ranks second in AI-influenced alteration, followed by qrr3, qrr1, and finally, qrr5. The strength of Qrr control over a representative quorum-sensing-repressed target gene follows an identical pattern (Fig. 5B), suggesting that this regulatory hierarchy can be extended to the entire V. harveyi quorum-sensing regulon. Thus, V. harveyi has presumably evolved multiple sRNAs by duplication, and further, qrr1 is likely the original sRNA gene because it is adjacent to luxO. The high sequence similarity between the five qrr genes suggests that they have evolved to maintain complementary base-pairing with the luxR 5′UTR. However, the lack of sequence similarity in the promoter regions of the V. harveyi qrr genes suggests that their specific regulation has been adapted to the differing environmental needs of the organism. Differential regulation of each sRNA gene has the advantage of enabling fine-tuned control of LuxR levels under a variety of environmental circumstances and in the diverse niches V. harveyi occupies. Moreover, the presence of multiple sRNAs functioning additively on a single target could confer increased precision in the control of gene expression by enabling subtle shifts in the control of LuxR. We also suggest that differences in the timing of expression of each qrr (Figs. 5, 6) may influence the accuracy of the control of downstream gene expression in response to fluctuations in AI levels.
The phenomenon of multiple sRNAs regulating a particular developmental biological process is not restricted to prokaryotes. MicroRNAs (miRNAs) confer post-transcriptional gene regulation in plants and animals by binding to complementary sequences in the 3′UTRs of messenger RNAs and directing either degradation of the mRNAs or inhibition of their translation (Ambros 2004). miRNAs constitute one of the most abundant classes of regulators, controlling many developmental processes including cell proliferation, differentiation, and morphogenesis (Ambros 2004). miRNAs are estimated to comprise 1% of predicted genes in higher eukaryotic genomes, and up to 10%–30% of genes might be regulated by miRNAs (Bartel 2004). Analogous to the situation in Vibrios, in eukaryotes, multiple miRNAs can act on a single target. For example, multiple miRNAs of the let-7 family have been shown to function in combination to affect both early and late developmental timing in C. elegans by repressing a shared target (Abbott et al. 2005). Many other genes are also predicted to have multiple miRNAs targeting their expression, some of which may be targeted by as many as six miRNAs (Cui et al. 2006). Taken together, these results suggest that regulatory RNAs have evolved to play important roles in both prokaryotes and eukaryotes to fine-tune gene expression for coordinating critical transitions during development.
Although regulation by multiple sRNAs occurs in both prokaryotes and eukaryotes, it is clear that differences in sRNA-mediated control exist even in closely related species. In the case of the Vibrios, it is difficult to account for the striking difference between how the Qrr sRNAs function in V. harveyi and V. cholerae. Both species couple the information from multiple AIs into a similar shared two-component regulatory circuit to control the transcription of multiple genes encoding sRNAs. The sRNAs, in turn, control translation of the master regulators of quorum sensing, LuxR and HapR, respectively. However, using nearly identical regulatory resources, V. cholerae and V. harveyi have evolved dramatically different mechanistic solutions for luxR/hapR regulation. The V. cholerae sRNAs are functionally redundant, suggesting that, when present, the concentration of each Qrr is at least as high as that of the hapR mRNA. By contrast, the V. harveyi Qrr sRNAs act additively, suggesting that cumulatively their concentrations equal that of luxR mRNA. Some of the downstream quorum-sensing target genes are identical in both species, whereas others are unique to each organism. Although all Vibrios are aquatic, they have evolved distinct lifestyles in the course of colonizing unique environmental niches (Thompson et al. 2004). V. cholerae is a major human pathogen, yet in aquatic environments it is found associated with zooplankton and in the chitinous exoskeletons of crustaceans (Reen et al. 2006). V. harveyi is a serious pathogen of marine fishes and invertebrates but not of humans (Austin and Zhang 2006). Given the different environments and survival needs of these two organisms, it seems naturally advantageous to generate unique solutions for optimizing gene regulation. We propose that related species can accomplish this by tinkering with some master set of basic regulatory components and their interactions.
Finally, we suggest that the V. harveyi Qrr regulators function as a fast-acting, reversible switch in response to AIs. sRNA molecules can be synthesized quickly, and translation is not required, so production is a one-step process. Similarly, Qrr concentrations can be rapidly reduced through degradation (Fig. 9). Thus, we suggest that a competition takes place between the sRNAs and luxR mRNA in the cell. luxR transcription is relatively steady (Showalter et al. 1990; Chatterjee et al. 1996); however, production of sRNAs increases or decreases depending on AI levels (Lenz et al. 2004). Hence, the Qrr sRNAs act like a dial to tune the levels of LuxR to the needs of the community (Fig. 1). Tipping the balance in favor of the Qrr sRNAs causes the cells to exhibit a low-cell-density state and act as individuals, whereas tipping the balance in favor of luxR switches the cells to high-cell-density mode, and the community acts in unison. By virtue of their additivity, their varying affinities for LuxO∼P, and additional regulators that may impinge on their expression, the five Qrr sRNAs in V. harveyi enable numerous subtle regulatory shifts and, consequently, furnish precise, customizable control over quorum-sensing gene expression.
Materials and methods
Bacterial strains and growth conditions
All V. harveyi strains are derived from BB120 (Bassler et al. 1997) and were grown aerobically at 30°C in Luria-marine (LM) broth. E. coli S17-1λpir was used to propagate plasmids at 37°C in LB medium. The following antibiotics were used: ampicillin (Amp), 100 μg/mL; tetracycline (Tet), 10 μg/mL; kanamycin (Kan), 100 μg/mL; chloramphenicol (Cm), 10 μg/mL; gentamicin (Gent), 100 μg/mL; and polymyxcin B (Pb), 50 U/mL. Bacterial growth was monitored by measuring optical density at 600 nm.
DNA manipulations
All DNA manipulations were performed using standard procedures (Sambrook et al. 1989). Herculase Enhanced DNA polymerase (Stratagene) was used for PCR cloning reactions, and Taq polymerase (Roche) was used for all other PCR reactions. dNTPs, restriction endonucleases, and T4 DNA ligase were obtained from New England BioLabs. DNA purification kits were provided by Qiagen. Primer sequences are available on request. V. harveyi deletions were constructed using previously described methods (Datsenko and Wanner 2000) and constructions were introduced onto the V. harveyi chromosome by allelic replacement (Bassler et al. 1993). qrr-gfp promoter fusions were cloned into pSLS3, a derivative of pCMW1 (Waters and Bassler 2006), using the BclI and SalI sites. qrr-gfp plasmids were transformed into JAF1822 carrying luxO D47E on pLAFR2 cosmid. Each promoter contains 275 bp upstream of the predicted +1 sites of the qrr genes. Plasmids were transformed into E. coli in 0.2-cm electroporation cuvettes (USA Scientific) using a Bio-Rad Micro Pulser. Plasmids and cosmids were introduced into V. harveyi by conjugation, and exconjugants were selected using the appropriate antibiotics and polymyxin B. qrr4 and qrr5 overexpression constructs were generated by cloning the sRNA genes into the EcoRI site in pKK177-3R1. Overexpression plasmids were electroporated directly into V. harveyi.
Genetic screen to identify sRNA genes in V. harveyi
A random V. harveyi promoter-gfp library (Waters and Bassler 2006) was transformed into E. coli strain KM1039 carrying luxO D47E on the pLAFR2 cosmid. Transformants were grown 14–16 h and sorted for gfp production on a Becton Dickinson FACS Vantage cell sorter. gfp-positive cells were grown overnight and resorted to enrich the population to near 100% gfp positive. Plasmids were isolated from the gfp-positive cells and transformed into E. coli strain JAF1522 carrying luxO D47A on pLAFR2. Transformants were grown 14–16 h and sorted for lack of gfp expression. Multiple rounds of sorting were performed to enrich for gfp-negative cells. Approximately 10,000 events were sorted and plated. Individual colonies were isolated and analyzed in 96-well plates, and GFP production was measured on a 1420 Victor2 multilabel counter (Wallac). Plasmids were isolated from individual clones expressing low levels of gfp in JAF1522 and transformed into KM1039. gfp expression profiles were screened side by side for candidates that showed differential expression in KM1039 and JAF1822. Gene identities were determined using BLAST-n analysis. DNA fragments containing promoter sequences of the qrr genes were used to probe a V. harveyi cosmid bank for flanking DNA regions to use in mutagenesis experiments.
Bioluminescence assays
V. harveyi bioluminescence expression was measured in an assay that has been described previously (Bassler et al. 1993). Briefly, V. harveyi cultures were grown for 14 h in LM medium at 30°C with aeration and supplemented with antibiotics if necessary to maintain plasmids. The cultures were diluted 1:5000, and light production and OD600 were measured every 45 min thereafter. Relative light units are defined as counts per minute per milliliter per OD600nm. Bioluminescence assays were performed a minimum of three times. The data in the figures are representative of the trials and did not differ significantly. For qrr4 and qrr5 overexpression experiments, cultures were grown in triplicate for 14 h, diluted 1:100, and grown for an additional 3 h, after which bioluminescence measurements were taken.
Western blot analysis
Western blot analysis was performed as previously described (Henke and Bassler 2004a). Membranes were exposed to a LuxR polyclonal antibody (Lenz et al. 2004). LuxR protein was quantified using an Alpha Innotech Fluor Chem SP chemiluminescence image analysis system, ensuring that protein levels were not saturated.
Quantitative real-time PCR analysis
RNA was isolated from V. harveyi using the Ribo-Pure-Bacteria kit (Ambion). RNA was quantified on a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies). Samples were treated with DNase I (Roche) in reactions containing 10× PCR buffer (ABI) and 25 mM MgCl2 (ABI). cDNA was generated in a 100-μL reaction containing 1 μg of RNA, 5× First Strand Buffer (Invitrogen), 100 mM DTT (Invitrogen), 10 mM dNTPs (ABI), random hexamers (Roche), and SuperScript II reverse transcriptase (Invitrogen). Reactions were completed in a thermocycler for 10 min at 25°C, for 50 min at 42°C, and for 10 min at 72°C. To control for genomic DNA contamination, identical reactions were performed in the absence of RT enzyme. Real-time PCR reactions were performed on an ABI Prism 7900HT Sequence Detection System. Reactions were carried out in 384-well optical reaction plates (ABI) in quadruplicate with 2× Sybr Green mix (ABI). Ten microliters were loaded into each well using a Beckman Coulter Biomek FX machine. Real-time PCR primers were designed using Primer Express 2.0 (ABI Software) and are available on request. hfq or rpsL genes were used as endogenous loading controls for the reactions, and RNA levels were quantified using absolute quantification (standard curve analysis).
gfp expression analysis
All gfp expression analyses were performed on a Becton Dickinson FACSAria cell sorter and data were analyzed using FACS Diva software. The gfp allele used as a transcriptional reporter was an unstable variant described previously (Andersen et al. 1998). For monitoring qrgA-gfp expression in V. harveyi, cultures were grown 14 h and diluted 1:250,000, and time points were taken every hour over the growth curve. For qrr-gfp expression in E. coli, cultures were grown in triplicate for 14 h, diluted 1:100, and allowed to grow for an additional 3 h, after which measurements were taken.
AI assay
V. harveyi strain KM413 (luxM, luxS) was grown for 14 h, diluted 1:100, and allowed to continue to grow to OD ∼ 0.1. At that time, HAI-1 (5.3 μM) and AI-2 (12.5 μM) were added. Bioluminescence measurements were taken every 30 min. Cells were collected and used to measure LuxR protein and RNA levels. After 3 h, the culture was divided in half; one half was washed twice in fresh prewarmed medium, and the other half was washed twice with warmed cell-free spent culture fluid. Bioluminescence, LuxR, and RNA were monitored in both cultures for an additional 2 h at 30-min intervals.
Acknowledgments
We thank members of the Bassler laboratory for insightful discussions and suggestions; J. Freeman, J. Henke, B. Lilley, and K. Mok for strain constructions; S. Gottesman, S. Svenningsen, and C. Waters for plasmids; C. DeCoste for flow cytometry assistance; D. Storton and J. Buckles for real-time PCR assistance; and J. Irgon for bioinformatics assistance. We also thank N. Wingreen, P. Koepp, B. Tu, and L. Tu for critical reading of the manuscript. This work was funded by HHMI, NIH grant R01 GM 065859, and NSF grant MCB-0343821.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1502407
References
- Abbott A.L., Alvarez-Saavedra E., Miska E.A., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V., Alvarez-Saavedra E., Miska E.A., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V., Miska E.A., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V., Bartel D.P., Horvitz H.R., Ambros V., Horvitz H.R., Ambros V., Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Andersen J.B., Sternberg C., Poulsen L.K., Bjorn S.P., Givskov M., Molin S., Sternberg C., Poulsen L.K., Bjorn S.P., Givskov M., Molin S., Poulsen L.K., Bjorn S.P., Givskov M., Molin S., Bjorn S.P., Givskov M., Molin S., Givskov M., Molin S., Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin B., Zhang X.H., Zhang X.H. Vibrio harveyi: A significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 2006;43:119–124. doi: 10.1111/j.1472-765X.2006.01989.x. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bassler B.L., Wright M., Showalter R.E., Silverman M.R., Wright M., Showalter R.E., Silverman M.R., Showalter R.E., Silverman M.R., Silverman M.R. Intercellular signalling in Vibrio harveyi: Sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Bassler B.L., Wright M., Silverman M.R., Wright M., Silverman M.R., Silverman M.R. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: Sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 1994a;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- Bassler B.L., Wright M., Silverman M.R., Wright M., Silverman M.R., Silverman M.R. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol. Microbiol. 1994b;12:403–412. doi: 10.1111/j.1365-2958.1994.tb01029.x. [DOI] [PubMed] [Google Scholar]
- Bassler B.L., Greenberg E.P., Stevens A.M., Greenberg E.P., Stevens A.M., Stevens A.M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J.G., Meighen E.A., Meighen E.A. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- Chatterjee J., Miyamoto C.M., Meighen E.A., Miyamoto C.M., Meighen E.A., Meighen E.A. Autoregulation of luxR: The Vibrio harveyi lux-operon activator functions as a repressor. Mol. Microbiol. 1996;20:415–425. doi: 10.1111/j.1365-2958.1996.tb02628.x. [DOI] [PubMed] [Google Scholar]
- Chen X., Schauder S., Potier N., Van Dorsselaer A., Pelczer I., Bassler B.L., Hughson F.M., Schauder S., Potier N., Van Dorsselaer A., Pelczer I., Bassler B.L., Hughson F.M., Potier N., Van Dorsselaer A., Pelczer I., Bassler B.L., Hughson F.M., Van Dorsselaer A., Pelczer I., Bassler B.L., Hughson F.M., Pelczer I., Bassler B.L., Hughson F.M., Bassler B.L., Hughson F.M., Hughson F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Cui Q., Yu Z., Purisima E.O., Wang E., Yu Z., Purisima E.O., Wang E., Purisima E.O., Wang E., Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol. Syst. Biol. 2006;2:46. doi: 10.1038/msb4100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J.A., Bassler B.L., Bassler B.L. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 1999a;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- Freeman J.A., Bassler B.L., Bassler B.L. Sequence and function of LuxU: A two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 1999b;181:899–906. doi: 10.1128/jb.181.3.899-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J.A., Lilley B.N., Bassler B.L., Lilley B.N., Bassler B.L., Bassler B.L. A genetic analysis of the functions of LuxN: A two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol. Microbiol. 2000;35:139–149. doi: 10.1046/j.1365-2958.2000.01684.x. [DOI] [PubMed] [Google Scholar]
- Fuqua C., Winans S.C., Greenberg E.P., Winans S.C., Greenberg E.P., Greenberg E.P. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- Geissmann T.A., Touati D., Touati D. Hfq, a new chaperoning role: Binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Stealth regulation: Biological circuits with small RNA switches. Genes & Dev. 2002;16:2829–2842. doi: 10.1101/gad.1030302. [DOI] [PubMed] [Google Scholar]
- Hammer B.K., Bassler B.L., Bassler B.L. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Henke J.M., Bassler B.L., Bassler B.L. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 2004a;186:3794–3805. doi: 10.1128/JB.186.12.3794-3805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke J.M., Bassler B.L., Bassler B.L. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 2004b;186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto H., Koide Y., Morita T., Aiba H., Koide Y., Morita T., Aiba H., Morita T., Aiba H., Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- Lenz D.H., Mok K.C., Lilley B.N., Kulkarni R.V., Wingreen N.S., Bassler B.L., Mok K.C., Lilley B.N., Kulkarni R.V., Wingreen N.S., Bassler B.L., Lilley B.N., Kulkarni R.V., Wingreen N.S., Bassler B.L., Kulkarni R.V., Wingreen N.S., Bassler B.L., Wingreen N.S., Bassler B.L., Bassler B.L. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Lilley B.N., Bassler B.L., Bassler B.L. Regulation of quorum sensing in Vibrio harveyi by LuxO and σ-54. Mol. Microbiol. 2000;36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- Masse E., Gottesman S., Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E., Escorcia F.E., Gottesman S., Escorcia F.E., Gottesman S., Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes & Dev. 2003a;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E., Majdalani N., Gottesman S., Majdalani N., Gottesman S., Gottesman S. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 2003b;6:120–124. doi: 10.1016/s1369-5274(03)00027-4. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M.J., Ruby E.G., Ruby E.G. Developmental biology in marine invertebrate symbioses. Curr. Opin. Microbiol. 2000;3:603–607. doi: 10.1016/s1369-5274(00)00147-8. [DOI] [PubMed] [Google Scholar]
- Miller M.B., Bassler B.L., Bassler B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Miller M.B., Skorupski K., Lenz D.H., Taylor R.K., Bassler B.L., Skorupski K., Lenz D.H., Taylor R.K., Bassler B.L., Lenz D.H., Taylor R.K., Bassler B.L., Taylor R.K., Bassler B.L., Bassler B.L. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- Miller S.T., Xavier K.B., Campagna S.R., Taga M.E., Semmelhack M.F., Bassler B.L., Hughson F.M., Xavier K.B., Campagna S.R., Taga M.E., Semmelhack M.F., Bassler B.L., Hughson F.M., Campagna S.R., Taga M.E., Semmelhack M.F., Bassler B.L., Hughson F.M., Taga M.E., Semmelhack M.F., Bassler B.L., Hughson F.M., Semmelhack M.F., Bassler B.L., Hughson F.M., Bassler B.L., Hughson F.M., Hughson F.M. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell. 2004;15:677–687. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Miyamoto C.M., Smith E.E., Swartzman E., Cao J.G., Graham A.F., Meighen E.A., Smith E.E., Swartzman E., Cao J.G., Graham A.F., Meighen E.A., Swartzman E., Cao J.G., Graham A.F., Meighen E.A., Cao J.G., Graham A.F., Meighen E.A., Graham A.F., Meighen E.A., Meighen E.A. Proximal and distal sites bind LuxR independently and activate expression of the Vibrio harveyi lux operon. Mol. Microbiol. 1994;14:255–262. doi: 10.1111/j.1365-2958.1994.tb01286.x. [DOI] [PubMed] [Google Scholar]
- Moll I., Afonyushkin T., Vytvytska O., Kaberdin V.R., Blasi U., Afonyushkin T., Vytvytska O., Kaberdin V.R., Blasi U., Vytvytska O., Kaberdin V.R., Blasi U., Kaberdin V.R., Blasi U., Blasi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller T., Franch T., Hojrup P., Keene D.R., Bachinger H.P., Brennan R.G., Valentin-Hansen P., Franch T., Hojrup P., Keene D.R., Bachinger H.P., Brennan R.G., Valentin-Hansen P., Hojrup P., Keene D.R., Bachinger H.P., Brennan R.G., Valentin-Hansen P., Keene D.R., Bachinger H.P., Brennan R.G., Valentin-Hansen P., Bachinger H.P., Brennan R.G., Valentin-Hansen P., Brennan R.G., Valentin-Hansen P., Valentin-Hansen P. Hfq: A bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Morita T., Maki K., Aiba H., Maki K., Aiba H., Aiba H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes & Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T., Mochizuki Y., Aiba H., Mochizuki Y., Aiba H., Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc. Natl. Acad. Sci. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reen F.J., Almagro-Moreno S., Ussery D., Boyd E.F., Almagro-Moreno S., Ussery D., Boyd E.F., Ussery D., Boyd E.F., Boyd E.F. The genomic code: Inferring Vibrionaceae niche specialization. Nat. Rev. Microbiol. 2006;4:697–704. doi: 10.1038/nrmicro1476. [DOI] [PubMed] [Google Scholar]
- Repoila F., Majdalani N., Gottesman S., Majdalani N., Gottesman S., Gottesman S. Small noncoding RNAs, co-ordinators of adaptation processes in Escherichia coli: The RpoS paradigm. Mol. Microbiol. 2003;48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T., Fritsch E.F., Maniatis T., Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Schumacher M.A., Pearson R.F., Moller T., Valentin-Hansen P., Brennan R.G., Pearson R.F., Moller T., Valentin-Hansen P., Brennan R.G., Moller T., Valentin-Hansen P., Brennan R.G., Valentin-Hansen P., Brennan R.G., Brennan R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq–RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter R.E., Martin M.O., Silverman M.R., Martin M.O., Silverman M.R., Silverman M.R. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J. Bacteriol. 1990;172:2946–2954. doi: 10.1128/jb.172.6.2946-2954.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G., Opdyke J.A., Zhang A., Opdyke J.A., Zhang A., Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Storz G., Altuvia S., Wassarman K.M., Altuvia S., Wassarman K.M., Wassarman K.M. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- Swartzman E., Silverman M., Meighen E.A., Silverman M., Meighen E.A., Meighen E.A. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J. Bacteriol. 1992;174:7490–7493. doi: 10.1128/jb.174.22.7490-7493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson F.L., Iida T., Swings J., Iida T., Swings J., Swings J. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 2004;68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P., Eriksen M., Udesen C., Eriksen M., Udesen C., Udesen C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Vanderpool C.K., Gottesman S., Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- Wassarman K.M. Small RNAs in bacteria: Diverse regulators of gene expression in response to environmental changes. Cell. 2002;109:141–144. doi: 10.1016/s0092-8674(02)00717-1. [DOI] [PubMed] [Google Scholar]
- Waters C.M., Bassler B.L., Bassler B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Waters C.M., Bassler B.L., Bassler B.L. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes & Dev. 2006;20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Plasterk R.H., Plasterk R.H. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Wyman C., Rombel I., North A.K., Bustamante C., Kustu S., Rombel I., North A.K., Bustamante C., Kustu S., North A.K., Bustamante C., Kustu S., Bustamante C., Kustu S., Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- Xavier K.B., Bassler B.L., Bassler B.L. Interference with AI-2-mediated bacterial cell-cell communication. Nature. 2005;437:750–753. doi: 10.1038/nature03960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Altuvia S., Tiwari A., Argaman L., Hengge-Aronis R., Storz G., Altuvia S., Tiwari A., Argaman L., Hengge-Aronis R., Storz G., Tiwari A., Argaman L., Hengge-Aronis R., Storz G., Argaman L., Hengge-Aronis R., Storz G., Hengge-Aronis R., Storz G., Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Wassarman K.M., Ortega J., Steven A.C., Storz G., Wassarman K.M., Ortega J., Steven A.C., Storz G., Ortega J., Steven A.C., Storz G., Steven A.C., Storz G., Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]