Abstract
Background
Skin cancer accounts for 1/3 of all newly diagnosed cancer. Although seldom fatal, basal cell carcinoma (BCC) is associated with severe disfigurement and morbidity. BCC has a unique interest for researchers, as although it is often locally invasive, it rarely metastasises. This paper, reporting the first whole genome expression microarray analysis of skin cancer, aimed to investigate the molecular profile of BCC in comparison to non-cancerous skin biopsies. RNA from BCC and normal skin specimens was analysed using Affymetrix whole genome microarrays. A Welch t-test was applied to data normalised using dCHIP to identify significant differentially-expressed genes between BCC and normal specimens. Principal component analysis and support vector machine analysis were performed on resulting genelists, Genmapp was used to identify pathways affected, and GOstat aided identification of areas of gene ontology more highly represented on these lists than would be expected by chance.
Results
Following normalisation, specimens clustered into groups of BCC specimens and of normal skin specimens. Of the 54,675 gene transcripts/variants analysed, 3,921 were differentially expressed between BCC and normal skin specimens. Of these, 2,108 were significantly up-regulated and 1,813 were statistically significantly down-regulated in BCCs.
Conclusion
Functional gene sets differentially expressed include those involved in transcription, proliferation, cell motility, apoptosis and metabolism. As expected, members of the Wnt and hedgehog pathways were found to be significantly different between BCC and normal specimens, as were many previously undescribed changes in gene expression between normal and BCC specimens, including basonuclin2 and mrp9. Quantitative-PCR analysis confirmed our microarray results, identifying novel potential biomarkers for BCC.
Background
It is estimated that the incidence of cutaneous basal cell carcinoma is increasing worldwide by up to 10% per year [1] and it currently accounts for approximately 80% of all non-melanoma skin cancer – with highest rates in elderly men and increasing incidence in young women [2]. Several sub-types of BCC have been identified. These include nodular-ulcerated BCC (the most frequently occurring type; often with ulceration ("rodent ulcer")); superficial BCC (often multiple); sclerosing BCC (cancer cells surrounded by dense fibrosis and so resemble scars; highest recurrence rate of BCCs after treatment); cystic BCC (uncommon; tumour undergoes central degradation to form a cystic lesion); linear BCC (recently recognised clinical entity with increased risk for aggressive histopathology); and micronodular BCC (small tumour nests; often with subclinical growth). Although BCC only occasionally (0.003–0.55% of cases [3-5]), results in metastasis and is seldom fatal, BCC is often locally invasive with destructive growth and may be associated with severe disfigurement and morbidity as a result of local tissue destruction or due to necessary surgery. Furthermore, people with BCC are at higher risks of developing further BCCs and other malignancies, including squamous cell carcinomas, malignant melanomas, and possibly also non-cutaneous malignancies [1].
Current selection of best treatment for BCC is based on accurate diagnosis and sub-classification of these cancers, mainly on histomorphology/pathology of H&E stained sections [6]. Features associated with recurrence and metastasis are also considered – including tumour diameter >2 cm, location in the central part of the face or ear, present for long duration, incomplete excision, aggressive growth pattern (based on histology) and perinuclear or perivascular involvement [2]. Genes reported to be associated with susceptibility to BCC include CYP2D6, GST-T1, vitamin D receptor, and TNF; with UVB irradiaton known to cause mutations in the p53 tumour suppressor gene, leading to the development of this cancer [1].
With the exception of a single study of BCCs using a small cDNA microarray – representing 1,718 genes [7] – investigations aimed at identifying BCC biomarkers and understanding the molecular events involved in this disease have, in general, been limited to one-at-a-time studies, built on chance analyses of proteins or mRNAs. Examples of such protein analysis have identified CD10 [8,9], p63 [10], low expression levels of CD44 [11] to be associated with the presence of BCC, generally with absence of ICAM-1 and LFA-3 adhesion molecule expression [12] and with Ki67 expression levels differing between BCCs that recur, compared to those that do not recur [13]. RT-PCR analysis has indicated 1,25-dihydroxyvitamin D(3) receptor mRNA levels to be increased in BCCs compared to normal skin [14], while qPCR quantification of gli1 transcripts has been found to discriminate BCC (and trichoepithelioma) from other skin cancers [15].
While such studies have indicated the importance and relevance of gene expression analyses in BCC, the number of gene products simultaneously analysed have been very limited. In order to increase our understanding of the molecular events involved in the development/expression of BCC, here we report our findings from whole genome microarray analysis of BCC and normal skin specimens.
Results
Quality Control of Microarray Data
As indicated in Table 1, quality control (Q.C.) analysis of all 25 microarray data sets (from 20 BCC and 5 normal skin specimens) indicated an average percentage present call of 42.68% (+/- 5.79 SD). This would be of the order expected for high quality RNA from cell lines from many origins (Affymetrix Inc. "Genechip® Expression Analysis Data Analysis Fundamentals" [16], indicating that these results are acceptable for further analysis. Again, based on cell line Q.C. parameters, the accepted background levels are <100, while 52.31 +/- 4.08 was found in this study; acceptable noise levels are <3, here we report 1.67 +/- 0.4; and the acceptable scaling factor is <3 fold between data sets being compared. This was generally, but not always, achieved. The acceptable 3'/M ratio of <3 was achieved in 18/25 cases.
Table 1.
Q.C. Analysis of Microarray Results
| Specimen I.D. | Present Call (%) | Background | Noise | Scaling Factor | 3'/M Ratio GAPDH |
| BCC26 | 49.9 | 52.6 | 1.82 | 0.94 | 14.56 |
| JT2 | 50.2 | 63.6 | 2.05 | 0.795 | 1.41 |
| JT3 | 49.9 | 53.85 | 1.74 | 1.037 | 1.27 |
| BCC4 | 48.8 | 55.72 | 1.87 | 0.868 | 1.82 |
| JT6 | 41.8 | 50.33 | 1.56 | 2.431 | 2.46 |
| JT11 | 39 | 48.93 | 1.58 | 3.173 | 2.17 |
| JT8 | 43 | 46.94 | 1.48 | 2.463 | 1.95 |
| JT9 | 46.5 | 53.85 | 1.71 | 1.434 | 1.66 |
| T16 | 43.4 | 51.87 | 1.62 | 1.896 | 3.25 |
| JT12 | 38.1 | 53.09 | 1.7 | 2.709 | 2.86 |
| JT13 | 47.1 | 54.46 | 1.67 | 1.374 | 1.67 |
| JT4 | 46.5 | 55.73 | 1.77 | 1.18 | 1.66 |
| JT5 | 45.8 | 53.86 | 1.75 | 1.453 | 1.86 |
| T24 | 38.5 | 57.3 | 1.83 | 2.905 | 3.0 |
| T25 | 34.1 | 56.57 | 1.77 | 3.988 | 3.1 |
| T28 | 34.3 | 56.32 | 1.79 | 3.771 | 3.6 |
| JT7 | 39.6 | 48.25 | 1.56 | 3.098 | 2.52 |
| T11 | 43.3 | 50.10 | 1.61 | 2.416 | 2.1 |
| T19 | 32.2 | 54.24 | 1.7 | 4.539 | 3.1 |
| T22 | 29.1 | 49.14 | 1.59 | 6.639 | 5.86 |
| N1 | 46.6 | 46.38 | 1.46 | 1.571 | 1.67 |
| N2 | 44.0 | 47.85 | 1.56 | 1.565 | 1.8 |
| N3 | 42.5 | 50.84 | 1.63 | 2.105 | 2.46 |
| N5 | 46.7 | 48.96 | 1.54 | 1.417 | 1.85 |
| N6 | 46.2 | 46.92 | 1.44 | 1.821 | 2.39 |
Note: Acceptable Q.C. cut-offs, based on high quality cell line RNA analysis, for background is <100; noise <3; scaling factor <3 fold between specimens being compared; 3'/M ratio <3 (Affymetrix Inc. "Genechip® Expression Analysis Data Analysis Fundamentals" [16]).
Data Analysis
Approximately 7% (3,921/54,675) of the probe sets representing transcripts on the microarray were significantly differentially expressed between BCC and normal skin specimens (Tables 2 &3; [see Additional Files 1 &2] for further information). The scatter plot of data differentially expressed ≥1.2 fold (Fig. 1) indicates an even, normal distribution of data. As shown in Table 4, of the 2,108 up-regulated by ≥1.2 fold, genes involved in many crucial aspects of cellular biology, including metabolism, transcription, cell cycle regulation, cell adhesion, cell migration, cell proliferation and cell motility were amongst the largest groups of genes affected, while oxidative phosphorylation, lipid metabolism, translation, and apoptosis were among the main categories down-regulated ≥1.2 fold in BCCs compared to normal skin (Table 5). Of the 748 probesets representing transcripts up-regulated by ≥2 fold, approximately 11 were described as cloned cDNAs, 122 were ESTs, 6 were described as hypothetical genes, and 46 encoded hypothetical proteins. Of the 484 transcripts showing ≥2 fold down-regulation, twenty-five represented hypothetical proteins, 11 cloned cDNAs/RIKENS and 49 ESTs.
Table 2.
Gene Transcripts significantly up-regulated in BCC compared to normal skin specimens (≥1.2 fold; ≥100; p < 0.05).
| probe set | gene | Accession | fold change | Difference | P value |
| 204697_s_at | chromogranin A (parathyroid secretory protein 1) | NM_001275 | 130.34 | 2497.11 | 0.000001 |
| 224590_at | X (inactive)-specific transcript | BE644917 | 69.08 | 427.18 | 0.001884 |
| 214218_s_at | X (inactive)-specific transcript | AV699347 | 62.79 | 621.31 | 0.00072 |
| 242964_at | gb:AI421677/DB_XREF=gi:4267608/DB_XREF=tf54a03.x1/CLONE | AI421677 | 55.54 | 730.97 | 0.000005 |
| 224588_at | X (inactive)-specific transcript | AA167449 | 49.82 | 2086.08 | 0.000552 |
| 204913_s_at | SRY (sex determining region Y)-box 11 | AI360875 | 27.92 | 228.19 | 0.000224 |
| 1560652_at | gb:AL832136.1/DB_XREF=gi:21732679/TID=Hs2.407141.1/CNT=4 | AL832136 | 26.21 | 638.38 | 0.000015 |
| 236029_at | FAT tumor suppressor homolog 3 (Drosophila) | AI283093 | 24.03 | 916.82 | 0.000015 |
| 214913_at | a disintegrin-like and metalloprotease (reprolysin type) | AB002364 | 23.97 | 605.9 | 0.000002 |
| 220345_at | leucine rich repeat transmembrane neuronal 4 | NM_024993 | 22.24 | 270.15 | 0.0061 |
| 233622_x_at | Transcribed locus, weakly similar to XP_375099.1 hypothetical protein | AL162077 | 21.31 | 105.45 | 0.005425 |
| 230863_at | gb:R73030/DB_XREF=gi:847062/DB_XREF=yj94c11.s1/CLONE= | R73030 | 20.05 | 292.94 | 0.029359 |
| 204915_s_at | SRY (sex determining region Y)-box 11 | AB028641 | 19.64 | 524.92 | 0.000059 |
| 204424_s_at | LIM domain only 3 (rhombotin-like 2) | AL050152 | 19.19 | 1287.59 | 0.003562 |
| 236407_at | gb:R73518/DB_XREF=gi:847550/DB_XREF=yj93h12.s1/CLONE= | R73518 | 18.1 | 355.25 | 0.000003 |
| 208025_s_at | high mobility group AT-hook 2///high mobility group AT-hook 2 | NM_003483 | 17.76 | 506.18 | 0.000173 |
| 215311_at | MRNA full length insert cDNA clone EUROIMAGE 21920 | AL109696 | 17.1 | 448.46 | 0.000013 |
| 227671_at | X (inactive)-specific transcript | AV646597 | 16.98 | 383.7 | 0.003059 |
| 218638_s_at | spondin 2, extracellular matrix protein | NM_012445 | 16.91 | 2992.93 | 0 |
| 208212_s_at | anaplastic lymphoma kinase (Ki-1) | NM_004304 | 16.76 | 835.41 | 0.000003 |
| 226346_at | gb:AA527151/DB_XREF=gi:2269220/DB_XREF=ni07b08.s1/CLONE | AA527151 | 15.34 | 534.55 | 0 |
| 204914_s_at | SRY (sex determining region Y)-box 11 | AW157202 | 15.1 | 310.64 | 0.000124 |
| 215443_at | thyroid stimulating hormone receptor | BE740743 | 14.95 | 239.99 | 0.00019 |
| 240460_at | gb:AI190616/DB_XREF=gi:3741825/DB_XREF=qd38e02.x1/CLONE | AI190616 | 14.91 | 168.09 | 0.000564 |
| 1562107_at | hypothetical protein MGC14738 | BC007100 | 14.73 | 284.47 | 0.000263 |
| 213960_at | CDNA FLJ37610 fis, clone BRCOC2011398 | T87225 | 14.18 | 534.88 | 0.000001 |
| 1565936_a_at | LIM domain only 3 (rhombotin-like 2) | T24091 | 13.94 | 162.48 | 0.001811 |
| 229523_at | gb:N66694/DB_XREF=gi:1218819/DB_XREF=yy71g08.s1/CLONE= | N66694 | 13.82 | 614.51 | 0.000001 |
| 210055_at | thyroid stimulating hormone receptor | BE045816 | 13.81 | 181.7 | 0.000595 |
| 224646_x_at | gb:BF569051/DB_XREF=gi:11642431/DB_XREF=602184410T1/ | BF569051 | 13.81 | 1024.01 | 0.000261 |
| 207468_s_at | secreted frizzled-related protein 5 | NM_003015 | 13.54 | 437.41 | 0.001339 |
| 220518_at | target of Nesh-SH3 | NM_024801 | 13.44 | 448.75 | 0.001398 |
| 224997_x_at | H19, imprinted maternally expressed untranslated mRNA | AL575306 | 13.39 | 1131.86 | 0.000494 |
| 222940_at | sulfotransferase family 1E, estrogen-preferring, member 1 | U55764 | 13.29 | 560.15 | 0.000001 |
| 235795_at | gb:AW088232/DB_XREF=gi:6044037/DB_XREF=xc99c09.x1 | AW088232 | 13.26 | 199.49 | 0.000451 |
| 220090_at | chromosome 1 open reading frame 10 | NM_016190 | 12.79 | 736.69 | 0.00414 |
| 238584_at | IQ motif containing with AAA domain | W52934 | 12.68 | 222.06 | 0.001169 |
| 203878_s_at | matrix metalloproteinase 11 (stromelysin 3) | NM_005940 | 12.23 | 685.83 | 0.000417 |
| 1557215_at | Transcribed locus, weakly similar to XP_375935.1 hypothetical protein | AK056212 | 11.92 | 262.54 | 0.003503 |
| 210292_s_at | protocadherin 11 Y-linked///protocadherin 11 X-linked | AF332218 | 11.83 | 211.94 | 0.000324 |
| 1558964_at | FAT tumor suppressor homolog 3 (Drosophila) | AA334950 | 11.22 | 280.34 | 0.000123 |
| 214451_at | transcription factor AP-2 beta (activating enhancer binding protein 2 | NM_003221 | 10.67 | 2196.21 | 0 |
| 209816_at | patched homolog (Drosophila) | AL044175 | 10.59 | 245.97 | 0.001175 |
| 241617_x_at | gb:BE961949/DB_XREF=gi:11764352/DB_XREF=601655369R1 | BE961949 | 10.43 | 1011 | 0.000972 |
| 209815_at | Patched homolog (Drosophila) | BG054916 | 10.34 | 2145.95 | 0.000007 |
| 230496_at | Hypothetical protein FLJ25477 | BE046923 | 10.34 | 131.12 | 0.000467 |
| 205372_at | pleiomorphic adenoma gene 1 | NM_002655 | 10.1 | 1069.14 | 0 |
| 229942_at | gb:AW024890/DB_XREF=gi:5878420/DB_XREF=wu92c11.x1 | AW024890 | 9.98 | 1649.01 | 0 |
| 214297_at | gb:BE857703/DB_XREF=gi:10371993/DB_XREF=7g46a02.x1 | BE857703 | 9.86 | 819.59 | 0.000003 |
| 229669_at | Hypothetical protein LOC339260 | AA166965 | 9.83 | 237.59 | 0.001318 |
Top 50 transcripts – based on fold difference are shown (complete list is supplied as supplementary material).
Table 3.
Gene Transcripts significantly down-regulated in BCC compared to normal skin specimens (≥1.2 fold; ≥100; p < 0.05).
| probe set | gene | Accession | fold change | Difference | P value |
| 239929_at | hypothetical protein FLJ32569 | AA918425 | -28.24 | -3433.29 | 0.014118 |
| 208962_s_at | fatty acid desaturase 1 | BE540552 | -20.2 | -3188.7 | 0.045399 |
| 229476_s_at | thyroid hormone responsive (SPOT14 homolog, rat) | AW272342 | -17.15 | -4660.08 | 0.034919 |
| 207275_s_at | acyl-CoA synthetase long-chain family member 1 | NM_001995 | -15.11 | -1865.7 | 0.039975 |
| 206799_at | secretoglobin, family 1D, member 2 | NM_006551 | -14.97 | -1291.31 | 0.042413 |
| 234513_at | elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like | AF292387 | -13.28 | -670.81 | 0.022544 |
| 221561_at | sterol O-acyltransferase (acyl-Coenzyme A: cholesterol acyltransferase) 1 | L21934 | -12.23 | -998.57 | 0.008422 |
| 206714_at | arachidonate 15-lipoxygenase, second type | NM_001141 | -11.45 | -3891.76 | 0.045644 |
| 214240_at | galanin | AL556409 | -11.12 | -1590.25 | 0.015802 |
| 244661_at | gb:AA946876/DB_XREF=gi:3110271/DB_XREF=oq53c11.s1/CLONE= | AA946876 | -10.56 | -399.74 | 0.017168 |
| 244780_at | sphingosine-1-phosphate phosphotase 2 | AI800110 | -9.79 | -351.6 | 0.024348 |
| 201625_s_at | insulin induced gene 1 | BE300521 | -9.72 | -1591.34 | 0.030044 |
| 231810_at | BRI3 binding protein | BG106919 | -8.9 | -877.2 | 0.021503 |
| 1565162_s_at | microsomal glutathione S-transferase 1 | D16947 | -8.6 | -1071.37 | 0.030259 |
| 238121_at | Transcribed locus, weakly similar to XP_341569.1 similar to ORF4 | AI473796 | -8.29 | -445.99 | 0.042733 |
| 211056_s_at | steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta | BC006373 | -7.88 | -1432.56 | 0.032165 |
| 229957_at | Branched chain keto acid dehydrogenase E1, alpha polypeptide | BF446281 | -7.63 | -1214.3 | 0.025272 |
| 204675_at | steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta | NM_001047 | -7.35 | -2832.7 | 0.027894 |
| 233030_at | adiponutrin | AK025665 | -7.35 | -1220.35 | 0.031698 |
| 231736_x_at | microsomal glutathione S-transferase 1 | NM_020300 | -7.28 | -2497.22 | 0.031804 |
| 205029_s_at | fatty acid binding protein 7, brain | NM_001446 | -7.24 | -524.52 | 0.021739 |
| 201627_s_at | insulin induced gene 1 | NM_005542 | -7.23 | -627.89 | 0.049971 |
| 209522_s_at | carnitine acetyltransferase | BC000723 | -7.23 | -2039.08 | 0.020824 |
| 226064_s_at | diacylglycerol O-acyltransferase homolog 2 (mouse) | AW469523 | -7.1 | -2291.54 | 0.032748 |
| 231156_at | gb:AW242782/DB_XREF=gi:6576459/DB_XREF=xm89g06.x1/CLONE | AW242782 | -7.09 | -339.26 | 0.045195 |
| 223184_s_at | 1-acylglycerol-3-phosphate O-acyltransferase 3 | BC004219 | -7.01 | -1374.94 | 0.045149 |
| 232428_at | monoacylglycerol O-acyltransferase 2 | AK000245 | -6.97 | -182.68 | 0.038707 |
| 1558846_at | Pancreatic lipase-related protein 3 | AL833418 | -6.71 | -2007.72 | 0.044402 |
| 205030_at | fatty acid binding protein 7, brain | NM_001446 | -6.41 | -1864.48 | 0.021392 |
| 205843_x_at | carnitine acetyltransferase | NM_000755 | -6.41 | -1166.98 | 0.020573 |
| 224435_at | chromosome 10 open reading frame 58///chromosome 10 open reading | BC005871 | -6.37 | -1905.14 | 0.007356 |
| 1560507_at | Diacylglycerol O-acyltransferase 2-like 3 | BC039181 | -6.28 | -441.54 | 0.028259 |
| 215726_s_at | cytochrome b-5 | M22976 | -5.95 | -3458.82 | 0.02416 |
| 220431_at | DESC1 protein | NM_014058 | -5.94 | -112.38 | 0.029175 |
| 225716_at | gb:AI357639/DB_XREF=gi:4109260/DB_XREF=qy15b05.x1/CLONE= | AI357639 | -5.94 | -1718.34 | 0.01598 |
| 204388_s_at | monoamine oxidase A | NM_000240 | -5.89 | -362.75 | 0.037779 |
| 45288_at | abhydrolase domain containing 6 | AA209239 | -5.83 | -698.25 | 0.020361 |
| 218804_at | transmembrane protein 16A | NM_018043 | -5.75 | -234.79 | 0.045057 |
| 228479_at | gb:AI094180/DB_XREF=gi:3433156/DB_XREF=qa29b09.s1/CLONE= | AI094180 | -5.7 | -1191.04 | 0.029615 |
| 227804_at | hypothetical protein BC014072 | BE328850 | -5.62 | -363.64 | 0.043913 |
| 234312_s_at | acetyl-Coenzyme A synthetase 2 (ADP forming) | AK000162 | -5.54 | -1297.68 | 0.044265 |
| 1562528_at | RAR-related orphan receptor A | BC040965 | -5.48 | -120.02 | 0.025055 |
| 208964_s_at | fatty acid desaturase 1 | AL512760 | -5.46 | -3305.87 | 0.025757 |
| 213693_s_at | gb:AI610869/DB_XREF=gi:4620036/DB_XREF=tp21e08.x1/CLONE= | AI610869 | -5.43 | -1427.55 | 0.023253 |
| 237507_at | keratin 6 irs3 | AI333069 | -5.43 | -271.6 | 0.008082 |
| 204389_at | monoamine oxidase A | NM_000240 | -5.4 | -180.28 | 0.0359 |
| 214598_at | claudin 8 | AL049977 | -5.27 | -370.2 | 0.01827 |
| 201963_at | acyl-CoA synthetase long-chain family member 1 | NM_021122 | -5.19 | -4459.79 | 0.010049 |
| 218434_s_at | acetoacetyl-CoA synthetase | NM_023928 | -5.17 | -1842.1 | 0.004574 |
| 221552_at | abhydrolase domain containing 6 | BC001698 | -5.06 | -402.38 | 0.024809 |
Top 50 transcripts – based on fold difference are shown (complete list is supplied as supplementary material).
Figure 1.
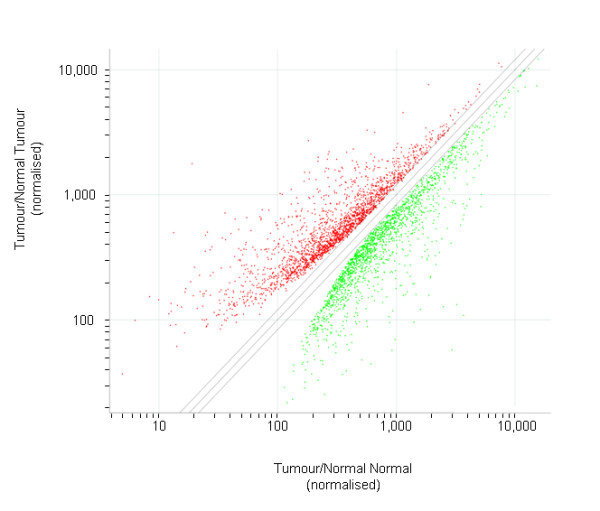
Scatter Plot of the 3,921 gene transcripts identified as significantly differentially expressed (by ≥1.2 fold; ≥100 difference in expression intensity; P < 0.05) between BCC and normal skin specimens. Transcripts significantly up-regulated are shown in red; those down-regulated are shown as green.
Table 4.
GOstat analysis of biological processes.
| GO Number | Observed Transcripts | Total Transcripts | GO Category | P value |
| GO:0019222 | 293 | 3309 | regulation of metabolism | 1.24E-33 |
| GO:0006351 | 264 | 2884 | transcription, DNA-dependent | 5.41E-33 |
| GO:0006355 | 258 | 2809 | regulation of transcription, DNA-dependent | 1.61E-32 |
| GO:0006357 | 55 | 280 | regulation of transcription from RNA polymerase II promoter | 5.40E-31 |
| GO:0008134 | 57 | 346 | transcription factor binding | 6.86E-24 |
| GO:0016563 | 45 | 250 | transcriptional activator activity | 9.36E-22 |
| GO:0003677 | 240 | 2906 | DNA binding | 1.54E-21 |
| GO:0003712 | 44 | 276 | transcription cofactor activity | 3.54E-17 |
| GO:0007049 | 89 | 807 | cell cycle | 9.89E-17 |
| GO:0016043 | 163 | 1986 | cell organization and biogenesis | 8.78E-14 |
| GO:0003713 | 30 | 171 | transcription coactivator activity | 1.08E-13 |
| GO:0008283 | 65 | 560 | cell proliferation | 1.17E-13 |
| GO:0051726 | 61 | 522 | regulation of cell cycle | 5.62E-13 |
| GO:0000074 | 60 | 518 | regulation of progression through cell cycle | 1.51E-12 |
| GO:0005578 | 48 | 377 | extracellular matrix | 3.00E-12 |
| GO:0007155 | 77 | 874 | cell adhesion | 5.65E-08 |
| GO:0008380 | 24 | 173 | RNA splicing | 3.60E-07 |
| GO:0016477 | 18 | 112 | cell migration | 5.56E-07 |
| GO:0016055 | 19 | 128 | Wnt receptor signaling pathway | 2.09E-06 |
| GO:0008219 | 60 | 675 | cell death | 2.29E-06 |
| GO:0007167 | 25 | 207 | enzyme linked receptor protein signaling pathway | 1.13E-05 |
| GO:0030154 | 50 | 550 | cell differentiation | 1.13E-05 |
| GO:0005604 | 13 | 62 | basement membrane | 7.37E-05 |
| GO:0051301 | 22 | 190 | cell division | 0.000156 |
| GO:0042981 | 38 | 418 | regulation of apoptosis | 0.000269 |
| GO:0000904 | 11 | 66 | cellular morphogenesis during differentiation | 0.00262 |
| GO:0043123 | 13 | 91 | positive regulation of I-kappaB kinase/NF-kappaB cascade | 0.00344 |
| GO:0005581 | 11 | 73 | collagen | 0.00597 |
| GO:0030198 | 8 | 43 | extracellular matrix organization and biogenesis | 0.00788 |
| GO:0007154 | 247 | 4475 | cell communication | 0.0122 |
| GO:0005583 | 4 | 11 | fibrillar collagen | 0.0125 |
| GO:0008286 | 5 | 20 | insulin receptor signaling pathway | 0.0182 |
| GO:0016049 | 17 | 174 | cell growth | 0.019 |
| GO:0008361 | 17 | 174 | regulation of cell size | 0.019 |
| GO:0005610 | 2 | 2 | laminin-5 | 0.0205 |
| GO:0008083 | 20 | 221 | growth factor activity | 0.0232 |
| GO:0005588 | 2 | 3 | collagen type V | 0.0477 |
Biological processes that are significantly enriched in our set of 2,108 transcripts found to be significantly up-regulated in BCC compared to normal skin specimens. Shown here is a sub-set of 37 representative significant GO annotations.
Table 5.
GOstat analysis of biological processes.
| GO Number | Observed Transcripts | Total Transcripts | GO Category | P value |
| GO:0006119 | 36 | 138 | oxidative phosphorylation | 2.77E-34 |
| GO:0006091 | 120 | 1003 | generation of precursor metabolites and energy | 3.75E-33 |
| GO:0016491 | 123 | 1170 | oxidoreductase activity | 7.47E-26 |
| GO:0006732 | 44 | 269 | coenzyme metabolism | 5.71E-21 |
| GO:0006629 | 86 | 765 | lipid metabolism | 1.17E-20 |
| GO:0003954 | 24 | 62 | NADH dehydrogenase activity | 8.07E-16 |
| GO:0043037 | 40 | 280 | translation | 4.26E-15 |
| GO:0016651 | 27 | 99 | oxidoreductase activity, acting on NADH or NADPH | 1.63E-13 |
| GO:0016126 | 16 | 31 | sterol biosynthesis | 5.82E-13 |
| GO:0008135 | 29 | 185 | translation factor activity, nucleic acid binding | 8.99E-13 |
| GO:0045045 | 30 | 217 | secretory pathway | 1.31E-10 |
| GO:0006099 | 14 | 45 | tricarboxylic acid cycle | 5.49E-08 |
| GO:0046356 | 14 | 45 | acetyl-CoA catabolism | 5.49E-08 |
| GO:0006511 | 25 | 201 | ubiquitin-dependent protein catabolism | 2.83E-07 |
| GO:0006413 | 17 | 84 | translational initiation | 1.20E-06 |
| GO:0048193 | 19 | 108 | Golgi vesicle transport | 2.00E-06 |
| GO:0006412 | 87 | 1228 | protein biosynthesis | 5.29E-06 |
| GO:0008289 | 35 | 380 | lipid binding | 2.46E-05 |
| GO:0006445 | 15 | 92 | regulation of translation | 7.85E-05 |
| GO:0006888 | 13 | 72 | ER to Golgi vesicle-mediated transport | 9.90E-05 |
| GO:0006915 | 49 | 644 | apoptosis | 0.000214 |
| GO:0008415 | 20 | 190 | acyltransferase activity | 0.000313 |
| GO:0006944 | 7 | 32 | membrane fusion | 0.00289 |
| GO:0016281 | 4 | 9 | eukaryotic translation initiation factor 4F complex | 0.00313 |
| GO:0006984 | 4 | 9 | ER-nuclear signaling pathway | 0.00313 |
| GO:0044242 | 7 | 33 | cellular lipid catabolism | 0.00349 |
| GO:0030503 | 3 | 5 | regulation of cell redox homeostasis | 0.00611 |
| GO:0007050 | 11 | 85 | cell cycle arrest | 0.00717 |
| GO:0016282 | 5 | 27 | eukaryotic 43S preinitiation complex | 0.0303 |
Biological processes that are significantly enriched in our set of 1,813 transcripts found to be significantly down-regulated in BCC compared to normal skin specimens. Shown here is a sub-set of 29 representative significant GO annotations.
In order to identify specimen similarity/diversity in our group of 25 skin specimens, condition tree clustering (using Pearson's correlation coefficient as similarity metric) and principal component analysis (PCA) were performed using GeneSpring software. As indicated in Fig. 2(A), while Pearson's correlation coefficient does not suggest a significant difference (i.e. no values <0.9) between the 25 specimens analysed, the 5 normal specimens form a discrete cluster in relation to the BCC specimens. (A similar clustering pattern was obtained when Spearman's correlation coefficient was investigated). Two of the BCC specimens, T19 and T22, are apparently different (but not significantly different) to the other 18 BCCs. While the 3'/M ratios were greater than expected (at least in comparison to cell line data) for these specimens, this was also the case for BCC26, T16, T24, T25 and T28, which did not group with T19 and T22. The higher scaling factor resulting from analysis of T19 and T22, compared to all other specimens, may be responsible for/contribute to their apparently somewhat different behaviour as represented on the condition tree. For the purpose of investigating the effects of T19 and T22 on the overall dataset, a re-analysis was performed excluding data relating to those specimens. This resulted in a reduction from 3,921 (i.e. 7.17%) to 3,865 (7.06%) of significantly differentially expressed transcripts between BCC and normal specimens, indicating that the vast majority of these transcripts are unbiased by the slightly different behaviour of T19 and T22 compared to all other BCCs (as described above).
Figure 2.
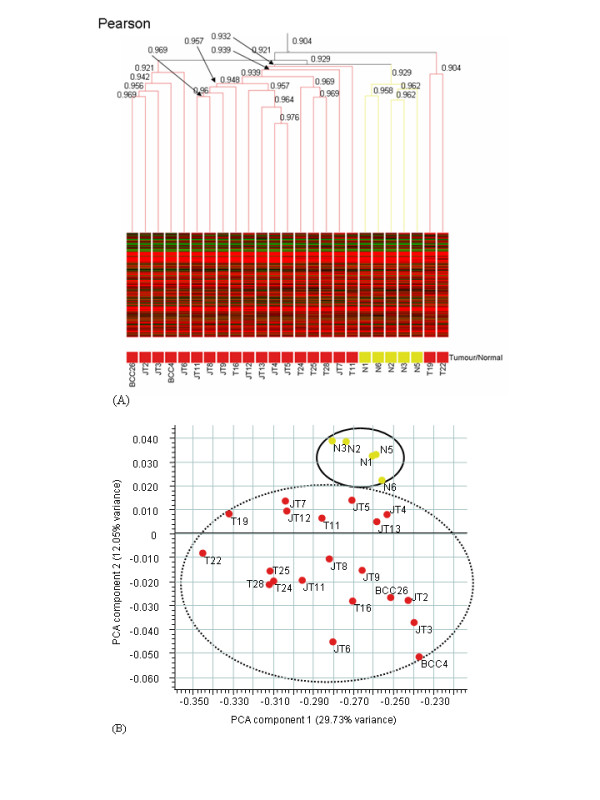
(A) Condition tree distribution of the 20 BCC (red) and 5 normal skin (yellow) specimens (following dCHIP normalisation, similarity measure, Pearson's correlation, clustering algorithm, average linkage). Expression values ≥100 are indicated in red; ≥50 to <100 are indicated in black, and 0 to <50 are indicated in green. (B) Two-dimensional principal component analysis (PCA) plot where red dots represent BCC specimens and yellow dots represent normal skin specimens indicating that while the normal specimens form a loose cluster (solid line oval), the BCC specimens are more "scattered" and varied (broken line oval). The first principal component expression value is 29.73%; the second component expression value is 12.05%.
To further identify the relatedness of the BCC samples to each other, we performed principal components analysis [17] on the entire data set. PCA was carried out on log-transformed data, using mean centering and scaling. As can be seen in Fig. 2(B), while our PCA analysis has divided the specimens into two groups, the results indicate that the BCC sub-group is much more varied than the normal skin group and that the BCC and normal skin specimens do not differ greatly.
Support Vector Machine analysis [18] is a machine learning classification approach which is suitable for application to the dimensionality of microarray data. It operates by examining the expression information of a set of data points whose classification is known (referred to as the "training set"), from which a defined number of classification predictor genes are identified. This predictor genelist can then be applied to a separate set of genes which are known not to be members of the functional class (referred to as the "test set"). The predictor genelist is user-defined, so it is more beneficial to the user for validation if a smaller number of genes comprise the predictor list.
For the purposes of this study, we carried out SVM analysis repeatedly on the tumour vs. normal dataset, in order to identify the lowest number of probesets that discriminated 100% of the time between the two classifications. The gene selection method used was Fisher's Exact Test, the kernel function was Polynomial Dot Product (Order 1) with zero Diagonal Scaling Factor. A minimum number of six identifiers, shown in Table 6, were required for 100% classification of every specimen as either tumour or normal in origin. Attempts to further prioritise these genes resulted in a decrease in the efficiency of the classification, as further analysis aimed at classifying the specimens using less than six transcripts resulted in the mis-classification of normal specimen N5 as a BCC specimen.
Table 6.
Transcripts identified using Support Vector Machine as suitable, as a group, for 100% classification of BCC from normal skin.
| I.D. | Gene Transcript |
| 1553718_at | zinc finger protein 548 |
| 201413_at | hydroxysteroid (17-beta) dehydrogenase 4 |
| 225677_at | B-cell receptor-associated protein 29 |
| 223184_s_at | 1-acylglycerol-3-phosphate O-acyltransferase 3 |
| 203878_s_at | matrix metalloproteinase 11 (stromelysin 3) |
| 225716_at | Full-length cDNA clone CS0DK008YI09 of HeLa cells Cot 25-normalized of Homo sapiens (human) |
qPCR Validation of Microarray Data
Quantitative-PCR (qPCR) analysis for 10 potential endogenous controls was performed on a random selection of four BCC and four normal skin specimens, to select a suitable endogenous control(s) amplifiable in all specimens and where levels of expression did not differ greatly between specimens being analysed. All 10 transcripts studied were found to be expressed in all specimens analysed and, as indicated in Fig. 3 [Additional File 3], expression levels and standard deviation results suggested that the 10 transcripts are likely to be of similar suitability as endogenous controls. Two controls, β-actin and GAPDH, were subsequently selected for amplification in all specimens, in parallel with a selection of 5 target transcripts of interest. The relative expression levels of PTCH1, gli2, Frizzled D2, basonuclin 2, and chromagranin A were analysed by qPCR. As indicated in Table 7, while the fold differences detected by microarray and qPCR methods differed to some extent, the trend (i.e. absent/low expression in normal skin and induced/increased expression in BCCs) was always found.
Table 7.
Validation of microarray data by qPCR
| Transcript | I.D. | Microarray1 Mean Expression Levels in Normal Skin | Microarray1 Mean Expression Levels in BCC | Microarray (fold)2 | qPCR3 Mean Expression Levels in Normal Skin | qPCR3 Mean Expression Levels in BCC | qPCR (fold)2 |
| chromagranin A | NM_001275 | 19.31 | 2516.41 | 130.34 | 0.000 | 1.083 | P |
| gli2 | NM_030379 | 23.51 | 173.73 | 7.39 | 0.000 | 1.662 | P |
| PTCH1 | BG054916 | 229.86 | 2375.81 | 10.34 | 0.025 | 1.455 | 58.2 |
| basonuclin 2 | NM_017637 | 204.42 | 1827.2 | 8.94 | 0.077 | 1.034 | 13.4 |
| frizzled 2 | L37882 | 54.93 | 490.8 | 8.94 | 0.047 | 6.505 | 138.4 |
Note: 1 = Expression levels, Affymetrix arbitrary units; 2 = fold increased expression in BCC compared to normal; 3 = following normalisation of data on mean of β-actin + GAPDH expression and calibrating to a pooled skin specimen control (as described in Materials and Methods); P = induced in BCC, while undetected in normal skin
Discussion
Basal cell carcinoma (BCC), the most common skin cancer in humans, is locally aggressive and relentlessly invasive, but generally does not metastasise [5,19]. Despite this, studies aimed at investigating the molecular mechanisms associated with – possibly responsible for – BCCs, have been very limited. In 2005, Howell et al. [7] reported findings from their analysis of 1,718 transcripts in BCC specimens, using cDNA microarrays. While this study produced very interesting results, as explained by the authors, numbers of transcripts potentially important in BCC – such as PTCH1 and SMO – were not represented on their microarray; limiting their study considerably. Here we have successfully analysed gene expression of BCCs, compared to normal skin, using whole genome microarrays and following extensive analysis of our data, in addition to confirming previous findings, we have identified a number of novel potential biomarkers/therapeutic targets for this disease.
Comparison of our results with those generated by Howell et al. [7] indicated a relatively high level of agreement between these two studies. Many of the transcripts identified by Howell et al. as up-regulated in BCC compared to normal skin were also found to be up-regulated in BCC in our study. Examples of these include collagens (type V, alpha 1 & alpha 2; type IV alpha 1 & 2; type VII alpha 1), topoisomerase IIα, tumour-associated calcium signal transducer 1, profilin 2, calretinin, syndecan 2, and v-myc. Similarly, a high concordance between these two studies was found for transcripts down-regulated in BCCs compared to normal specimens; examples of which include cystatin B, acetyl-Coenzyme acyltransferase 1, 3-hydroxy-3-methylglutaryl-Coenzyme A reductase, glutaredoxin, amyloid β (A4) precursor-like protein, and cytochrome b-5. However, in the case of a limited number of differentially expressed transcripts, the direction of change in expression in BCC compared to normal skin was not in agreement. Examples include ADP-ribosylation factor 3 (down-regulated by 1.67 fold in our study [expression values 941.51 vs. 563.45], but up-regulation reported by Howell et al. [7] and glia maturation factor β (1.42 up-regulated in our analysis [expression values 969.02 vs. 1,377.23], but reported by Howell et al. as down-regulated). While the reasons for these limited number of discrepencies is unknown, it may be due to different splice variants of these transcripts being detected by cDNA compared to oligo microarrays. It is worth noting that the results that differed between our study and that of Howell et al. were generally transcripts that we found to be <2 fold differentially expressed between BCC and normal skin. Unfortunately, as fold changes observed by Howell et al. [7] were not reported in their manuscript and information is not publicly available on transcripts that were present on their microarray, but were not significantly changed, further comparisons between these studies cannot be performed.
The development of BCC is known to be associated with dys-regulation of the hedgehog and Wnt pathways [2,20]. Lack of expression and/or suppressed activation of patched homologue 1 (PTCH1), a tumour suppressor gene that forms part of the hedgehog signaling network [21], has been reported to be fundamental to the development of BCC [22]. Disruption of this tumour suppressor gene results in up-regulated cell proliferation [6]. The accepted mechanism of PTCH1's action is via its binding to another transmembrane molecule, smoothed (SMO), thus suppressing intracellular signaling. Following binding of sonic hedgehog (shh) to PTCH1, this suppressor activity is, however, quenched, resulting in uninterrupted signal transduction by SMO, via GLI transcription factors, and subsequent constitutive activation of target genes, including members of the Wnt pathway [23] and PTCH1, itself [21]. From our analysis of BCC compared to normal skin tissue, while we found no significant changes in shh expression levels, we report an approximately 11 fold increased expression of PTCH1 [expression values 25.65 vs. 271.62], and also increased expression of gli2 (7.39 fold; P = 0.00009) [expression values 23.52 vs. 173.73], respectively. The induced/increased expression of PTCH1 and gli2 found by microarray analysis was confirmed by qPCR analysis of all specimens (see Table 7). It is important to note that the likely reason for the difference in fold expression detected by microarray and qPCR methods is due to their differing baseline sensitivity. Low expression levels detected by microarrays – e.g. gli2 in normal skin – resulting in large fold differences are considered as induction from absent in normal skin compared to present in BCC specimens, when analysed by qPCR. However, although PTCH1 mRNA levels have previously been reported as enhanced in nodular BCC but undetectable in superficial BCC [24], here we report PTCH1 to be detectable in both of these histological types of BCC, with no significant difference in their respective expression values (t-test: p = 0.637).
While the increased expression of gli2 detected in BCCs compared to normal skin may be expected and associated with the development/presence of BCC, the lack of tumour suppressor activity by PTCH1 – despite its increased mRNA levels – may be due to lack of expression of its corresponding protein and/or lack of binding to SMO (levels of which were not significantly different between BCC and normal skin). As PTCH1 may shuttle between the cell membrane and endocytotic vesicles in response to active hedgehog ligand, it is obvious that not only its mRNA expression, but also its protein expression (at the relevant location, binding of SMO, etc.) is necessary to exert its tumour suppressor activity [21]. Furthermore, as loss-of-function mutations of PTCH1 have been identified in 30–40% of sporadic cases of BCC, it may be that the mRNA over-expressed in the BCCs is not coding for a functional protein.
Wnt signaling may be able to regulate a number of the aspects of the biology of tumour cells and thus contribute in several ways to the tumour phenotypes. The strongest link is to the control of proliferation. Knockouts of Wnt signal transduction components, including Wnt5A, can result in proliferative failure [25] while up-regulation of Wnt5A mRNA expression been associated with a range of cancer types, including breast, lung, prostate and malignant melanomas [26,27]. In our study, the involvement of Wnt signaling pathway in BCC is suggested by the significantly increased expression of a number of Wnt family members. These include Wnt5A (3.35 fold; P = 0.00003) [expression values 403.73 vs. 1,353.71], – in agreement with a study by Saldanha et al. [19] where Wnt5A levels were increased in BCCs compared to surrounding skin – and Wnt6 (4.86 fold; P = 0.00002) [expression values 59.7 vs. 290.26]. Increased levels of Wnt ligand binding receptors, Frizzled D2 (8.94 fold; P = 0.000033) [expression values 54.93 vs. 490.8], D7 (2.31 fold; P = 0.000085) [expression values 276.23 vs. 638.34], and D8 (5.89 fold; P = 0.000055) [expression values 44.44 vs. 261.74], and decreased levels of D4 (-2.78 fold; P = 0.02) [expression values 598.41 vs. 215.2], were also found. The increased expression of Frizzled D2 in BCC compared to normal skin was confirmed by qPCR.
In the "canonical" Wnt signaling pathway, secreted ligands bind to Frizzled receptors and regulate the stability of β-catenin. (Given the large number of mammalian Frizzleds and Wnts, considerable ligand-receptor specificity might be expected; however, redundancy of function seems to be the rule [25]). The subsequent accumulation of β-catenin – the central player in the Wnt pathway [28] – in the nucleus, results in its participation in transcriptionally active complexes with members of the LEF/TCF family of transcription factors [29]. While we did not find levels of β-catenin to be significantly different between BCC and normal skin, decreased (-2.1 fold; P = 0.006) [expression values 2,090.09 vs. 992.89], levels of CTNNBIP1, an inhibitor of β-catenin and TCF-4 (ICAT) which would normally prevent β-catenin binding to LEF transcription factors [30-32] and increased levels of LEF1 transcripts (3.42 fold; P = 0.000001) [expression values 513.06 vs. 1,752.32], were found. Levels of Jun (2.34 fold; P = 0.00006) [expression values 875.92 vs. 2,052.07], another transcription factor involved in the Wnt pathway [33], were also found to be increased in BCCs compared to normal skin. Differential expression of other transcription factors associated with cancer has also been found in this study. These include CHES1 (checkpoint suppressor 1) which is apparently involved in repressing expression of genes important for tumorigenesis [34]. CHES1 mRNA has been reported as down-regulated in oral squamous cell carcinoma [35] and in hepatocellular carcinoma [36]. Here we found CHES1 mRNA levels to be significantly (-2.03 fold; P = 0.045) [expression values 920.32 vs. 452.95], down-regulated in BCC compared to normal skin. Not unexpectedly, mRNAs encoding proteins involved in inducing apoptosis were also found to be down-regulated. These include CIDE [37] and CARD15 [38] which are 4.18 fold (P = 0.029) [expression values 415.5 vs. 99.41] and 2.31 fold (P = 0.031) [expression values 313.08 vs. 135.42], down-regulated in BCC compared to normal skin.
Using support vector machine analysis we have identified 6 transcripts that, as a group, enable the accurate classification of all 25 specimens as BCC or normal skin. These include matrix metalloproteinase 11/mmp11 (previously associated with the presence of other cancer types, including oral [39,40], lung [41] and breast [42]); hydroxysteroid (17-beta) dehydrogenase 4/hsd17b4 (changes in expression of which have been associated with breast cancer ([43]); B-cell receptor-associated protein 29/bap29 (a member of the B cell receptor-associated family of proteins [44-46]); 1-acylglycerol-3-phosphate O-acyltransferase 3/agpat3 (which catalyses the acylation of lysophosphatidic acid to form phosphatidic acid, the precursor of all glycerolipids [47]); as well as zinc finger protein 548 and full-length cDNA clone CS0DK008YI09 of HeLa cells Cot 25-normalized of homo sapiens, on which no literature has previously been published. Future studies involving the co-analysis of this group of 6 transcripts in larger cohort of BCC and normal skin specimens should enable validation of their diagnostic relevance.
Molecular events responsible for the quite unique invasive, but non-metastatic, nature of BCCs are not known. However, it is interesting to note that expression of certain genes believed to be involved in malignant invasion and metastasis of another form of skin cancer, i.e. malignant melanoma, apparently differ in their expression patterns in BCCs. AP-2 transcription factor is not expressed in malignant melanoma cells [48], but it is significantly up-regulated (by 10.7 fold) [expression values 227.05 vs. 2,423.26], in BCC specimens compared to normal skin. Conversely, increased expression of EGF-R is associated with melanomas metastasis [48], but its expression is down-regulated (by approximately 1.2 fold) [expression values 76.91 vs. 62.78], in BCCs. Unlike BCCs, breast cancers frequently metastasise. Interestingly, in our microarray study of 104 breast tumours and normal breast tissue (manuscript in preparation) we identified changes in expression patterns of syndecan adhesion receptors (for review: see [49]) i.e. syndecan 1 is up-regulated, and syndecan 2 is down-regulated, in breast tumours compared to normal breast tissue. In this study of non-metastatic BCCs, we found syndecan 1 to be approximately 1.6 fold down-regulated [expression values 3,488.91 vs. 2,124.43], and syndecan 2 to be approximately 3 fold [expression values 160.21 vs. 559.84] up-regulated compared to levels in normal skin tissue. Furthermore, ankyrin (encoding membrane-associated cytoskeletal proteins) binding to membrane molecules has been suggested as necessary for cell adhesion, migration and tumour metastasis [50]. In our breast cancer study we found ankyrin 3 expression levels to be up-regulated compared to normal tissue, while here we report ankyrin 3 levels to be down-regulated (approximately 1.6 fold) [expression values 350.02 vs. 224.48], in BCC compared to normal tissue. While functional studies would be required to determine a causative/direct, rather than an associative, role for transcripts such as AP-2, EGF-R, syndecan 1 &2, and ankyrin 3 in controlling metastasis, the results from this study suggest that expression of the mRNAs may be, in some way, involved in this process.
Differential expression, between BCC and normal skin, of many transcripts not previous associated with the presence of BCC was also found during the course of our study. Basonuclin (now termed Basonuclin 1; was first discovered in cultured human epidermal cells [51]) and the more recently discovered basonuclin 2 [52] are zinc finger proteins. Basonuclin 1 is expressed at high levels in proliferating keratinocytes of stratified squamous epithelium. During terminal differentiation of squamous epithelium, basonuclin mRNA and protein disappear from the suprabasal cells [53,54]. In normal cultured human keratinocytes, basonuclin 1 is the predominant transcript – with expression levels approximately 10 fold that of basonuclin 2 [52]. In this study we found that basonuclin 1 levels did not differ significantly between BCC and normal skin, although a previous study of 3 BCC and 2 normal skin specimens indicated increased expression of basonuclin 1 in BCC, dependent on Gli protein expression [55]. In contrast, we report basonuclin 2 levels to be significantly increased in BCCs. This finding was observed with all 4 basonuclin 2 probe sets present on the microarray, indicating a 6.7–9.6 fold increase level (P < 0.00005) of expression [greatest change in expression values being 85.63 vs. 828.01], in BCC. This increased expression of basonuclin 2 in BCC compared to normal skin was confirmed by qPCR analysis. This, we believe, is the first study indicating an association between expression of basonuclin 2 and BCC.
ABCC12/mrp9, identified in 2001 [56] is one of a super-family of 9 ATP-binding cassette (ABC) multiple drug resistant proteins [57]. Mrp9 encodes an approximately 100 kDa protein detectable in breast cancer, normal breast tissue and testis, while an alternative mrp9 transcript – encoding an approximately 25 kDa protein – has been detected in normal brain, skeletal muscle and ovary tissues. Due to the differential levels of expression of mrp9 transcripts in breast tumour and normal tissue, MRP9 has been proposed as an immunotherapy target for breast cancer [58]. The functional relevance of our observation of approximately 8.7 fold greater levels of mrp9 [expression values 21.12 vs. 184.71] in BCC compared to normal skin has yet to be determined; its presence is unlikely to be involvement in drug resistance, as all of the BCCs included in this study were chemotherapy-naive (indeed, except in a limited number of advanced cases of BCC, chemotherapy is not used as a therapy for this disease. In these exceptional cases, excellent response rates have been reported with cisplatin in combination with either 5-fluorouracil or doxorubicin [59]). However, as for breast cancer, mrp9 mRNA may be useful as a member of a panel of BCC biomarkers or as an immunotherapy.
Chromagranin A (ChgA; parathyroid secretory protein 1) is an established tissue marker associated with neuroendocrine differentiation – and indicative of outcome – in non-small cell lung carcinoma [60]. Increased levels of ChgA in serum have been associated with poor prognosis/shortened survival for prostate cancer patients [61]. ChgA protein levels have been proposed to assist in the diagnosis of Merkel cell carcinoma patients who may benefit from oncological therapy [62-64]. Although described as relatively uncommon – using analysis of markers including ChgA – neuroendocrine differentiation in BCC has been reported [65]. In this study we have found ChgA levels to be significantly (130.3 fold; P = 0.000001) [expression values 19.31 vs. 2,516.41], up-regulated in BCCs compared to extremely low levels in normal skin specimens. This is in agreement with the observation of ChgA protein detectable in 55% (11/20) BCC specimens [66]. By qPCR analysis, ChgA was undetected in normal skin but was present in BCC specimens. Interestingly, other neuroendocrine markers, including SNAP-25 (3.24 fold; P = 0.008) [expression values 40.17 vs. 130.25] and neuroendocrine protein 1/7B2 protein (3.48 fold; P = 0.0001) [expression values 46.38 vs. 161.44], were also increased in the BCC specimens. These results indicate ChgA to be a potentially very useful marker for BCC.
In this study we present the first whole genome expression microarray analysis of skin cancer, aimed at investigating the molecular profile of BCC in comparison to non-cancerous skin biopsies. This investigation has not only confirmed previous findings from analyses of limited numbers of transcripts, but it has also identified changes in expression of mRNAs that had never previously been associated with this disease. The results from this work are interesting and exciting, but it is necessary to recognise their preliminary nature. Further analyses, building on our findings, should include independent replication studies so that the true relevance of these findings may be realised.
Conclusion
The success of this study indicates the feasibility and relevance of using whole genome microarrays to study BCC specimens. In addition to confirming previous findings, this work has increased our understanding of molecular differences between BCC and normal skin and has identified a number of novel potential biomarkers for BCC. Future studies including BCC tissue and normal skin tissue from the same individual, thus lowering inter-individual variability and ruling out genetic influences; analyses of age- and gender-matched cases; studies focusing on molecular profiling and comparisons of sub-types of BCC, with due consideration given to disease duration (as early tumours may have a different gene expression profile to prolonged tumours); as well as analysing BCCs that metastasise compared to those that do not, should further increase our understanding of this disease and assist in management of the individual BCC patients. Furthermore, as gene expression may be independent of protein levels, future confirmatory analysis at the protein level would complement these findings.
Materials
Patient Characteristics
This study involved analysis of 20 BCC biopsies from both male and female patients aged between 47 years and 83 years (mean age = 65 +/- 11 years; median = 67 years) at the time of diagnosis. In order to gain an understanding of the most common types of BCC, we elected to include a range of BCC sub-types in this study, rather than to focus on any particular sub-type. For this reason, BCC sub-classifications included were nodular/micronodular, superficial and sclerosing. Tissue specimens from these twenty cases of BCC were procured at Blackrock Clinic and the Bons Secours Hospital, Dublin, examined macroscopically, immediately snap-frozen in liquid nitrogen, and were subsequently stored at -80°C until required for analysis. Five normal skin specimens (from consenting male and female volunteers of a similar age range who do not/never had skin cancer) were also included in these studies.
RNA Extraction
For RNA analyses, dissected tumours that had been snap-frozen in liquid nitrogen and then stored at -80°C until required were homogenised, on ice, in 1 ml TriReagent (Sigma; Poole, England) and total RNA was subsequently isolated according to the manufacturer's instructions. RNA quantity and purity were assessed at 260 nm and 280 nm using a Nanodrop (ND-1000; Labtech. International); an Agilent bioanalyser (Agilent 2100; Agilent Technologies) was used to assess RNA qualitatively after isolation and, subsequently, after biotin-labelling and after fragmentation.
100 ng of each specimen was amplified and labelled using the Affymetrix GeneChip Eukaryotic 2 Cycle Labelling Assays for Expression Analysis, (Affymetrix; 900494) according to the manufacturer's instructions http://www.affymetrix.com/products/reagents/specific/cdna2.affx. Gene expression was examined using whole genome microarrays (Affymetrix; U133 Plus 2.0; 900470).
Microarray hybridization
Hybridisation solution (1 mol/l NaCl, 20 mmol/1 EDTA, 100 mmol/1 2-(N-morpholino) ethanesulfonic acid, and 0.01% Tween 20) was used to pre-hybridise Affymetrix; U133 Plus 2.0 oligonucleotide microarrays for 10 minutes at 45°C and 60 rpm. The pre-hybridisation solution was removed and replaced with 200 μl hybridisation solution containing 0.05 μg/μl fragmented cRNA. The arrays were hybridised for 16 hours at 45°C and 60 rpm. Arrays were subsequently washed (Affymetrix Fluidics Station 400) and stained with streptavidin-phycoerythrin (Stain Buffer, 2 mg/ml acetylated BSA and 10 μg/ml streptavidin R-phycoerythrin; Molecular Probes, Inc., Eugene, OR), and were scanned on an Affymetrix GCS GeneChip GeneArray scanner. Resulting data was analysed using GCOS, dCHIP, and GeneSpring (Agilent Technologies).
Normalisation and Filtering
Cel files obtained from the GCOS server were processed and normalized by dCHIP [67] algorithm. In this normalisation procedure, an array with median overall intensity is chosen as the baseline array against which other arrays are normalized at probe intensity level. Subsequently, a subset of PM probes, with small within-subset rank difference in the two arrays, serves as the basis for fitting a normalization curve. A filter was designed to include a fold change of at least 1.2 fold between normal and BCC specimens, a difference of at least 100 Affymetrix arbitrary units between normal and BCC average values, and a t-test between normal and BCC (with a p-value cut-off <0.05).
Gene Ontology and Pathway Analysis Analysis
In order to establish which gene ontologies (GO) are over-represented in our lists of 2,108 significantly up-regulated and 1,813 significantly down-regulated (in BCCs compared to normal skin) transcripts, we compared these to the list of all human genes from the EBI [68], using Gostat [69]. In brief, for all of the gene transcripts analysed, GOstat determines the associated annotated GO terms and all branches/splits on their connection path. The program then counts the number of appearances of each GO term for the gene transcripts in the list being analysed, as well as in the reference list. For each GO term, a Chi-squared p-value is calculated representing the probability that the observed numbers of counts could have resulted from randomly distributing this GO term between the tested and the reference lists. If the expected value for any analysis is <5, the Chi-squared approximation is considered to be inaccurate. Genmapp [70] was used to identify pathways affected by the differentially-expressed genelist.
Real-time PCR (qPCR)
Following priming with oligo (dT) at 65°C for 5 minutes, followed by 1 minute incubation on ice, cDNA was synthesised from 100 ng total RNA, using Superscript III RNase H- (with increased thermal stability; Invitrogen), RNase OUT Ribonuclease (active against RNase A, B and C; Invitrogen) and a cocktail of dNTPs, by incubating at 50°C for 1 hour, followed by 70°C for 15 minutes, in a 40 μl reaction volume. The cDNA (diluted 1:10), was amplified in 25 μl reactions, by qPCR, using an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA). Following evaluation of 11 potential endogenous controls in a random selection* of 4 BCC and 4 normal specimens, this study involved evaluation, in all 20 BCC and 5 normal skin specimens, of 5 target transcripts normalised to 2 suitable endogenous controls – β-actin and GAPDH – and calibrated against a pooled cDNA of BCC and normal skin specimens, the relative quantity of which was set to 1. The temperature profile of all reactions was 50°C for 2 minutes 95°C for 10 minutes, 40 cycles of 95°C and 60°C for 1 minute. Individual specimens were analysed in triplicate. [* Note: these 4 BCC and normal specimens were analysed by microarrays with all of the other specimens included in this study].
Abbreviations
BCC – basal cell carcinoma; qPCR – quantitative-polymerase chain reaction
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
LOD participated in the design and co-ordination of the study, secured financial support for this research, was involved in RNA isolation & study by microarrays and qPCR, data analysis and interpretation, and she drafted the manuscript; JMM participated in RNA isolations, quality assessment, and in preparation of specimens for microarray analysis; PD participated in analysing labeled specimens on microarrays chips and in bioinformatics analysis; EMK performed qPCR analysis; JPM and ER were involved in the bioinformatics analysis; PG and HJ were involved in analysing labeled specimens on microarrays chips; NOD was involved in RNA isolation; NW provided the clinical specimens/anonymised clinical data and was involved in raising financial support for this research; MC participated in the design of the study, and contributed to data interpretation, drafting of the manuscript and he was involved in raising financial support for this research. All authors approved the final manuscript.
Supplementary Material
Table 2 [Additional File 1]. Gene transcripts significantly up-regulated in BCC compared to normal skin specimens
Table 3 [Additional File 2]. Gene transcripts significantly down-regulated in BCC compared to normal skin specimens
Figure 3 [Additional File 3]. qPCR analysis of 10 transcripts in four BCC and four normal specimens as potential endogenous controls for analysis involving validation of microarray data in all specimens.
Acknowledgments
Acknowledgements
This work was supported by funding from Ireland's Higher Educational Authority Program for Research in Third Level Institutes (PRTLI) Cycle 3; Science Foundation Ireland; Dublin City University's Albert College Fellowship; and Dublin City University's Faculty of Science & Health Targeted Research Initiative.
Contributor Information
Lorraine O'Driscoll, Email: Lorraine.odriscoll@dcu.ie.
Jason McMorrow, Email: Jason.mcmorrow@dcu.ie.
Padraig Doolan, Email: Padraig.doolan@dcu.ie.
Eadaoin McKiernan, Email: eadaoin0@yahoo.com.
Jai Prakash Mehta, Email: mehtaj2@mail.dcu.ie.
Eoin Ryan, Email: Eoin.ryan@dcu.ie.
Patrick Gammell, Email: Patrick.gammell@dcu.ie.
Helena Joyce, Email: Helena.joyce@dcu.ie.
Norma O'Donovan, Email: Norma.odonovan@dcu.ie.
Nicholas Walsh, Email: Nicb@dcu.ie.
Martin Clynes, Email: Martin.clynes@dcu.ie.
References
- Wong CS, Strange RC, Lear JT. Basal cell carcinoma. BMJ. 2003;327:794–798. doi: 10.1136/bmj.327.7418.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin AI, Chen EH, Ratner D. Basal-cell carcinoma. N Engl J Med. 2005;353:2262–2269. doi: 10.1056/NEJMra044151. [DOI] [PubMed] [Google Scholar]
- Robinson JK, Dahiya M. Basal cell carcinoma with pulmonary and lymph node metastasis causing death. Arch Dermatol. 2003;139:643–648. doi: 10.1001/archderm.139.5.643. [DOI] [PubMed] [Google Scholar]
- Hafner C, Hartmann A, Knuechel R, Dietmaier W, Landthaler M, Vogt T. Molecular genetic analysis excluded implantation metastasis of basal cell carcinoma. Arch Pathol Lab Med. 2003;127:1221–1224. doi: 10.5858/2003-127-1221-MGAEIM. [DOI] [PubMed] [Google Scholar]
- Ionescu DN, Arida M, Jukic DM. Metastatic basal cell carcinoma: four case reports, review of literature, and immunohistochemical evaluation. Arch Pathol Lab Med. 2006;130:45–51. doi: 10.5858/2006-130-45-MBCCFC. [DOI] [PubMed] [Google Scholar]
- Crowson AN. Basal cell carcinoma: biology, morphology and clinical implications. Mod Pathol. 2006:S127–147. doi: 10.1038/modpathol.3800512. [DOI] [PubMed] [Google Scholar]
- Howell BG, Solish N, Lu C, Watanabe H, Mamelak AJ, Freed I, Wang B, Sauder DN. Microarray profiles of human basal cell carcinoma: insights into tumor growth and behavior. J Dermatol Sci. 2005;39:39–51. doi: 10.1016/j.jdermsci.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Yada K, Kashima K, Daa T, Kitano S, Fujiwara S, Yokoyama S. Expression of CD10 in basal cell carcinoma. Am J Dermatopathol. 2004;26:463–471. doi: 10.1097/00000372-200412000-00004. [DOI] [PubMed] [Google Scholar]
- Pham TT, Selim MA, Burchette JL, Jr, Madden J, Turner J, Herman C. CD10 expression in trichoepithelioma and basal cell carcinoma. J Cutan Pathol. 2006;33:123–128. doi: 10.1111/j.0303-6987.2006.00283.x. [DOI] [PubMed] [Google Scholar]
- Park HR, Min SK, Cho HD, Kim KH, Shin HS, Park YE. Expression profiles of p63, p53, survivin, and hTERT in skin tumors. J Cutan Pathol. 2004;31:544–549. doi: 10.1111/j.0303-6987.2004.00228.x. [DOI] [PubMed] [Google Scholar]
- Baum HP, Schmid T, Schock G, Reichrath J. Expression of CD44 isoforms in basal cell carcinomas. Br J Dermatol. 1996;134:465–468. doi: 10.1046/j.1365-2133.1996.32793.x. [DOI] [PubMed] [Google Scholar]
- Taylor RS, Griffiths CE, Brown MD, Swanson NA, Nickoloff BJ. Constitutive absence and interferon-gamma-induced expression of adhesion molecules in basal cell carcinoma. J Am Acad Dermatol. 1990;22:721–726. doi: 10.1016/0190-9622(90)70097-2. [DOI] [PubMed] [Google Scholar]
- Healy E, Angus B, Lawrence CM, Rees JL. Prognostic value of Ki67 antigen expression in basal cell carcinomas. Br J Dermatol. 1995;133:737–741. doi: 10.1111/j.1365-2133.1995.tb02748.x. [DOI] [PubMed] [Google Scholar]
- Reichrath J, Kamradt J, Zhu XH, Kong XF, Tilgen W, Holick MF. Analysis of 1,25-dihydroxyvitamin D(3) receptors (VDR) in basal cell carcinomas. Am J Pathol. 1999;155:583–589. doi: 10.1016/s0002-9440(10)65153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta N, Hirano T, Kimura T, Hashimoto K, Mehregan DR, Ansai S, Takehara K, Takata M. Molecular diagnosis of basal cell carcinoma and other basaloid cell neoplasms of the skin by the quantification of Gli1 transcript levels. J Cutan Pathol. 2005;32:131–136. doi: 10.1111/j.0303-6987.2005.00264.x. [DOI] [PubMed] [Google Scholar]
- http://www.affymetrix.com/support/downloads/manuals/data_analysis_fundamentals_manual
- Raychaudhuri S, Stuart JM, Altman RB. Principal components analysis to summarize microarray experiments: application to sporulation time series. Pac Symp Biocomput. 2000:455–466. doi: 10.1142/9789814447331_0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MP, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, Ares M, Jr, Haussler D. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci USA. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha G, Ghura V, Potter L, Fletcher A. Nuclear beta-catenin in basal cell carcinoma correlates with increased proliferation. Br J Dermatol. 2004;151:157–164. doi: 10.1111/j.1365-2133.2004.06048.x. [DOI] [PubMed] [Google Scholar]
- Daya-Grosjean L, Couve-Privat S. Sonic hedgehog signaling in basal cell carcinomas. Cancer Lett. 2005;225:181–192. doi: 10.1016/j.canlet.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Cohen MM., Jr The hedgehog signaling network. Am J Med Genet A. 2003;123:5–28. doi: 10.1002/ajmg.a.20495. [DOI] [PubMed] [Google Scholar]
- Boonchai W, Walsh M, Cummings M, Chenevix-Trench G. Expression of beta-catenin, a key mediator of the WNT signaling pathway, in basal cell carcinoma. Arch Dermatol. 2000;136:937–938. doi: 10.1001/archderm.136.7.937. [DOI] [PubMed] [Google Scholar]
- Yamazaki F, Aragane Y, Kawada A, Tezuka T. Immunohistochemical detection for nuclear beta-catenin in sporadic basal cell carcinoma. Br J Dermatol. 2001;145:771–777. doi: 10.1046/j.1365-2133.2001.04468.x. [DOI] [PubMed] [Google Scholar]
- Tojo M, Mori T, Kiyosawa H, Honma Y, Tanno Y, Kanazawa KY, Yokoya S, Kaneko F, Wanaka A. Expression of sonic hedgehog signal transducers, patched and smoothened, in human basal cell carcinoma. Pathol Int. 1999;49:687–694. doi: 10.1046/j.1440-1827.1999.00938.x. [DOI] [PubMed] [Google Scholar]
- Smalley MJ, Dale TC. Wnt signalling in mammalian development and cancer. Cancer Metastasis Rev. 1999;18:215–230. doi: 10.1023/A:1006369223282. [DOI] [PubMed] [Google Scholar]
- Lejeune S, Huguet EL, Hamby A, Poulsom R, Harris AL. Wnt5a cloning, expression, and up-regulation in human primary breast cancers. Clin Cancer Res. 1995;1:215–222. [PubMed] [Google Scholar]
- Iozzo RV, Eichstetter I, Danielson KG. Aberrant expression of the growth factor Wnt-5A in human malignancy. Cancer Res. 1995;55:3495–499. [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1147. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verras M, Sun Z. Roles and regulation of Wnt signaling and beta-catenin in prostate cancer. Cancer Lett. 2006;237:22–32. doi: 10.1016/j.canlet.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Tago K, Nakamura T, Nishita M, Hyodo J, Nagai S, Murata Y, Adachi S, Ohwada S, Morishita Y, Shibuya H, Akiyama T. Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev. 2000;14:1741–1749. [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Weis WI. ICAT inhibits beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol Cell. 2002;10:573–584. doi: 10.1016/S1097-2765(02)00631-7. [DOI] [PubMed] [Google Scholar]
- Stow JL. ICAT is a multipotent inhibitor of beta-catenin. Focus on "role for ICAT in beta-catenin-dependent nuclear signaling and cadherin functions". Am J Physiol Cell Physiol. 2004;286:C745–746. doi: 10.1152/ajpcell.00563.2003. [DOI] [PubMed] [Google Scholar]
- Weeraratna AT. A Wnt-er wonderland – the complexity of Wnt signaling in melanoma. Cancer Metastasis Rev. 2005;24:237–250. doi: 10.1007/s10555-005-1574-z. [DOI] [PubMed] [Google Scholar]
- Scott KL, Plon SE. CHES1/FOXN3 interacts with Ski-interacting protein and acts as a transcriptional repressor. Gene. 2005;359:119–126. doi: 10.1016/j.gene.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Chang JT, Wang HM, Chang KW, Chen WH, Wen MC, Hsu YM, Yung BY, Chen IH, Liao CT, Hsieh LL, Cheng AJ. Identification of differentially expressed genes in oral squamous cell carcinoma (OSCC): overexpression of NPM, CDK1 and NDRG1 and underexpression of CHES1. Int J Cancer. 2005;114:942–949. doi: 10.1002/ijc.20663. [DOI] [PubMed] [Google Scholar]
- Hong Y, Muller UR, Lai F. Discriminating two classes of toxicants through expression analysis of HepG2 cells with DNA arrays. Toxicol In Vitro. 2003;17:85–92. doi: 10.1016/S0887-2333(02)00122-4. [DOI] [PubMed] [Google Scholar]
- Inohara N, Koseki T, Chen S, Wu X, Nunez G. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 1998;17:2526–2533. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine ML, Murillo LS, Morre SA, Winkel EG, Pena AS, van Winkelhoff AJ. CARD15 gene mutations in periodontitis. J Clin Periodontol. 2004;31:890–893. doi: 10.1111/j.1600-051X.2004.00577.x. [DOI] [PubMed] [Google Scholar]
- Soni S, Mathur M, Shukla NK, Deo SV, Ralhan R. Stromelysin-3 expression is an early event in human oral tumorigenesis. Int J Cancer. 2003;107:309–316. doi: 10.1002/ijc.11366. [DOI] [PubMed] [Google Scholar]
- Leivo I, Jee KJ, Heikinheimo K, Laine M, Ollila J, Nagy B, Knuutila S. Characterization of gene expression in major types of salivary gland carcinomas with epithelial differentiation. Cancer Genet Cytogenet. 2005;156:104–113. doi: 10.1016/j.cancergencyto.2004.04.016. [DOI] [PubMed] [Google Scholar]
- Kettunen E, Anttila S, Seppanen JK, Karjalainen A, Edgren H, Lindstrom I, Salovaara R, Nissen AM, Salo J, Mattson K, Hollmen J, Knuutila S, Wikman H. Differentially expressed genes in nonsmall cell lung cancer: expression profiling of cancer-related genes in squamous cell lung cancer. Cancer Genet Cytogenet. 2004;149:98–106. doi: 10.1016/S0165-4608(03)00300-5. [DOI] [PubMed] [Google Scholar]
- Schuetz CS, Bonin M, Clare SE, Nieselt K, Sotlar K, Walter M, Fehm T, Solomayer E, Riess O, Wallwiener D, Kurek R, Neubauer HJ. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006;66:5278–5286. doi: 10.1158/0008-5472.CAN-05-4610. [DOI] [PubMed] [Google Scholar]
- Fiegl H, Millinger S, Goebel G, Muller-Holzner E, Marth C, Laird PW, Widschwendter M. Breast cancer DNA methylation profiles in cancer cells and tumor stroma: association with HER-2/neu status in primary breast cancer. Cancer Res. 2006;66:29–33. doi: 10.1158/0008-5472.CAN-05-2508. [DOI] [PubMed] [Google Scholar]
- Adachi T, Schamel WW, Kim KM, Watanabe T, Becker B, Nielsen PJ, Reth M. The specificity of association of the IgD molecule with the accessory proteins BAP31/BAP29 lies in the IgD transmembrane sequence. EMBO J. 1996;15:1534–1541. [PMC free article] [PubMed] [Google Scholar]
- Schamel WW, Kuppig S, Becker B, Gimborn K, Hauri HP, Reth M. A high-molecular-weight complex of membrane proteins BAP29/BAP31 is involved in the retention of membrane-bound IgD in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2003;100:9861–9866. doi: 10.1073/pnas.1633363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet ME, Cohen-Doyle M, Shore GC, Williams DB. Bap29/31 influences the intracellular traffic of MHC class I molecules. J Immunol. 2004;172:7548–7555. doi: 10.4049/jimmunol.172.12.7548. [DOI] [PubMed] [Google Scholar]
- Lu B, Jiang YJ, Zhou Y, Xu FY, Hatch GM, Choy PC. Cloning and characterization of murine 1-acyl-sn-glycerol 3-phosphate acyltransferases and their regulation by PPARalpha in murine heart. Biochem J. 2005;385:469–477. doi: 10.1042/BJ20041348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary EC, Lev DC, Bar-Eli M. Cellular adhesion pathways and metastatic potential of human melanoma. Cancer Biol Ther. 2002;1:459–465. doi: 10.4161/cbt.1.5.158. [DOI] [PubMed] [Google Scholar]
- Beauvais DM, Rapraeger AC. Syndecans in tumor cell adhesion and signaling. Reprod Biol Endocrinol. 2004;7:3. doi: 10.1186/1477-7827-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon LY, Zhu H, Shao L, Chen YW. Ankyrin-Tiam1 interaction promotes Rac1 signaling and metastatic breast tumor cell invasion and migration. J Cell Biol. 2000;150:177–191. doi: 10.1083/jcb.150.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H, Green H. Basonuclin: a keratinocyte protein with multiple paired zinc fingers. Proc Natl Acad Sci USA. 1992;89:10311–10315. doi: 10.1073/pnas.89.21.10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoutteghem A, Djian P. Basonuclin 2: an extremely conserved homolog of the zinc finger protein basonuclin. Proc Natl Acad Sci USA. 2004;101:3468–3473. doi: 10.1073/pnas.0400268101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H, Green H. Association of basonuclin with ability of keratinocytes to multiply and with absence of terminal differentiation. J Cell Biol. 1994;126:495–506. doi: 10.1083/jcb.126.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Green H. Basonuclin, a zinc finger protein of keratinocytes and reproductive germ cells, binds to the rRNA gene promoter. Proc Natl Acad Sci USA. 1999;96:9628–9632. doi: 10.1073/pnas.96.17.9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Elsam T, Tian Q, Seykora JT, Grachtchouk M, Dlugosz A, Tseng H. Gli proteins up-regulate the expression of basonuclin in basal cell carcinoma. Cancer Res. 2004;64:5651–5658. doi: 10.1158/0008-5472.CAN-04-0801. [DOI] [PubMed] [Google Scholar]
- Tammur J, Prades C, Arnould I, Rzhetsky A, Hutchinson A, Adachi M, Schuetz JD, Swoboda KJ, Ptacek LJ, Rosier M, Dean M, Allikmets R. Two new genes from the human ATP-binding cassette transporter superfamily, ABCC11 and ABCC12, tandemly duplicated on chromosome 16q12. Gene. 2001;273:89–96. doi: 10.1016/S0378-1119(01)00572-8. [DOI] [PubMed] [Google Scholar]
- Lai L, Tan TM. Role of glutathione in the multidrug resistance protein 4 (MRP4/ABCC4)-mediated efflux of cAMP and resistance to purine analogues. Biochem J. 2002;361:497–503. doi: 10.1042/0264-6021:3610497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera TK, Iavarone C, Kumar V, Lee S, Lee B, Pastan I. MRP9, an unusual truncated member of the ABC transporter superfamily, is highly expressed in breast cancer. Proc Natl Acad Sci USA. 2002;99:6997–7002. doi: 10.1073/pnas.102187299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner RF, Casciato DA. Skin Cancer. In: Casciato DA, Lowitz BB, editor. Manual of Clinical Oncology. Fourth. Lippincott, Williams & Wilkins; 2000. pp. 336–348. [Google Scholar]
- Nisman B, Heching N, Biran H, Barak V, Peretz T. The prognostic significance of circulating neuroendocrine markers chromogranin a, pro-gastrin-releasing peptide and neuron-specific enolase in patients with advanced non-small-cell lung cancer. Tumour Biol. 2006;27:8–16. doi: 10.1159/000090151. [DOI] [PubMed] [Google Scholar]
- Ranno S, Motta M, Rampello E, Risino C, Bennati E, Malaguarnera M. The chromogranin-A (CgA) in prostate cancer. Arch Gerontol Geriatr. 2006;43:117–126. doi: 10.1016/j.archger.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Carlei F, Lomanto D, Chimenti S, Pranteda G, Castagna G, Lezoche E, Mariani P, D'Alessandro MD, Gendel SE, Speranza V. Immunocytochemical study of a trabecular carcinoma of the skin (Merkel cell tumor). Case report. Ital J Surg Sci. 1986;16:55–59. [PubMed] [Google Scholar]
- Mount SL, Taatjes DJ. Neuroendocrine carcinoma of the skin (Merkel cell carcinoma). An immunoelectron-microscopic case study. Am J Dermatopathol. 1994;16:60–65. doi: 10.1097/00000372-199402000-00012. [DOI] [PubMed] [Google Scholar]
- Koljonen V, Haglund C, Tukiainen E, Bohling T. Neuroendocrine differentiation in primary Merkel cell carcinoma – possible prognostic significance. Anticancer Res. 2005;25:853–858. [PubMed] [Google Scholar]
- George E, Swanson PE, Wick MR. Neuroendocrine differentiation in basal cell carcinoma. An immunohistochemical study. Am J Dermatopathol. 1989;11:131–135. doi: 10.1097/00000372-198911020-00004. [DOI] [PubMed] [Google Scholar]
- Collina G, Macri L, Eusebi V. Endocrine differentiation in basocellular carcinoma. Pathologica. 2001;93:208–212. [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.ebi.ac.uk/GOA/
- http://gostat.wehi.edu.au/
- http://www.genmapp.org
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 2 [Additional File 1]. Gene transcripts significantly up-regulated in BCC compared to normal skin specimens
Table 3 [Additional File 2]. Gene transcripts significantly down-regulated in BCC compared to normal skin specimens
Figure 3 [Additional File 3]. qPCR analysis of 10 transcripts in four BCC and four normal specimens as potential endogenous controls for analysis involving validation of microarray data in all specimens.


