Abstract
The coiled-coil coactivator (CoCoA) is involved in transcriptional activation of target genes by nuclear receptors and the xenobiotic aryl hydrocarbon receptor, as well as target genes of the Wnt signaling pathway, which is mediated by the lymphocyte enhancer factor (LEF)/T cell factor transcription factors and the coactivator β-catenin. The recruitment of CoCoA by nuclear receptors is accomplished by the interaction of the central coiled-coiled domain of CoCoA with p160 coactivators; the C-terminal activation domain (AD) of CoCoA is used for downstream signaling, while the function of the N-terminal region was undefined. Here we report that the N-terminus of CoCoA contains another AD, which is necessary and sufficient for synergistic activation of LEF1-mediated transcription by CoCoA and β-catenin. The N-terminal AD contains a p300 binding motif, which is important for synergistic cooperation of CoCoA and p300 as coactivators for LEF1 and β-catenin. p300 contributes to the function of the CoCoA N-terminal AD primarily through its histone acetyltransferase activity. Moreover, in cultured cells endogenous p300 is recruited to the promoter of an integrated reporter gene by the N-terminus of CoCoA. Thus, the coactivator function of CoCoA for nuclear receptors and LEF1/β-catenin involves differential utilization of two different CoCoA ADs.
Keywords: coactivator, CoCoA, β-catenin, p300, LEF1
INTRODUCTION
The transcriptional activation of eukaryotic genes is tightly regulated by a variety of signaling pathways. Extracellular stimuli such as hormones or growth factors initiate a series of molecular responses that lead to the binding of activator proteins to specific DNA sequences in the promoters of their target genes, or the activation of proteins which are already bound to their target genes. The activators recruit coactivator complexes which constitute a signal transduction pathway to transmit the activation signal to the transcription machinery. Coactivators assist in transcriptional activation by two different mechanisms: 1) by causing local remodeling of chromatin through enzymatic action of (e.g.) ATPase, kinase, acetyltransferase and methyltransferase activities; and 2) by recruitment of other coactivators and the basal transcription machinery through protein-protein interactions. The coactivators receive the activation signal from the DNA-bound activators through their signal input domains and transmit the signal to the transcription machinery with their signal output or activation domains (AD) (1). Many coactivators can use a variety of functional domains to cooperate with different activators, and coactivator function can be influenced dramatically by specific promoter and cell context. Thus, elucidation of the function and regulation of coactivator signal input and output domains is critical for understanding the complex mechanisms of transcriptional regulation.
In this study, we focus on the coiled-coil coactivator (CoCoA), the product of the calcoco1 gene. Although CoCoA was originally discovered as a coactivator for nuclear receptors (2) and the aryl hydrocarbon receptor (3), it is also involved in the transcriptional activation of target genes in the Wnt/β-catenin pathway (4). The Wnt/β-catenin signaling cascade is important in developmental processes such as cell fate determination and axis formation. Deregulation of this pathway has been associated with carcinogenesis in a variety of tissues (5,6). Activation of this pathway by extracellular Wnt ligands results in reduced degradation and thus increased cellular accumulation of β-catenin, followed by its nuclear translocation. In the nucleus β-catenin binds directly to and serves as a coactivator for T cell factor/lymphocyte enhancer binding factor (TCF/LEF) transcriptional activator proteins to turn on transcription of specific target genes (7,8). β-catenin serves as a scaffold protein, which recruits downstream coactivators, including the p160 nuclear receptor coactivator GRIP1 (9,10), the protein acetyltransferases p300 and CBP (11-13), the arginine methyltransferase CARM1 (14), and the Brg1 ATPase subunit of the Swi/Snf chromatin-remodeling complex (15).
CoCoA consists of a large central coiled-coil domain flanked by a strong C-terminal AD and an N-terminal region of undefined function (2). When CoCoA functions as a coactivator for nuclear receptors or the aryl hydrocarbon receptor, the coiled-coil domain of CoCoA serves as a signal input domain by binding to the basic helix-loop-helix-Per-Arnt-Sim (bHLH–PAS) domain found in the N-terminal region of the p160 nuclear receptor coactivator or the aryl hydrocarbon receptor and its heterodimer partner ARNT. In these contexts, the potent C-terminal AD of CoCoA is used as a signal output domain, i.e. it transmits the activating signal to the transcription machinery and is essential for the coactivator function of CoCoA with nuclear receptors and with the aryl hydrocarbon receptor (2,3). In contrast, during activation of the canonical Wnt-signaling pathway, CoCoA directly binds to β-catenin with both its N- and C-terminal regions, suggesting that both terminal regions of CoCoA can function as signal input domains when cooperating with β-catenin (4). However, the C-terminal AD of CoCoA is dispensable when CoCoA cooperates with β-catenin, while the N-terminus, a region with previously undefined function, is essential, suggesting that the N-terminus functions as a signal output domain (or AD) in the context of β-catenin. Given the importance of the N-terminal region of CoCoA in β-catenin-mediated transcription, we investigated the molecular mechanism of downstream signaling by this newly defined signal output domain.
RESULTS
CoCoA N-terminus has autonomous transcriptional activation activity
CoCoA functions as a secondary coactivator in LEF1-mediated transcriptional activation through its interaction with β-catenin, which binds directly to LEF1 (4). Moreover, the N-terminal region of CoCoA, which is dispensable when CoCoA cooperates with GRIP1 and nuclear receptors as a secondary coactivator, is essential when CoCoA cooperates with β-catenin. We therefore tested whether a short N-terminal fragment (amino acids 1-190) of CoCoA is sufficient by itself (i.e. providing both signal input and signal output functions) to serve as a secondary coactivator for β-catenin in transient transfection assays. Full-length CoCoA and β-catenin synergistically enhanced LEF1-mediated expression of transiently transfected luciferase reporter plasmid pGL3OT, which contains TCF/LEF-responsive elements (Fig. 1A, assays 1–4). The CoCoA N-terminal fragment (amino acids 1-190) also cooperated with β-catenin to enhance LEF1-mediated transcriptional activation, although with reduced activity compared with full length CoCoA (Fig. 1A, assays 5–6). To further test the activity of CoCoA on a more physiological gene target, we employed a 293 cell line (STF) with a stably integrated luciferase reporter gene which contains seven TCF/LEF-binding sites (16). Transient transfection of CoCoA alone did not activate the stably integrated luciferase reporter, while overexpression of low levels of β-catenin alone caused a moderate enhancement (Fig. 1B, assays 1–3). Both full-length CoCoA and the N-terminal fragment of CoCoA cooperated synergistically with β-catenin to a similar extent to enhance transcriptional activation of the integrated reporter gene (assays 4–7). Thus, the N-terminal region of CoCoA is necessary and sufficient for synergistic coactivator function with β-catenin, indicating that the N-terminal region of CoCoA contains both signal input and signal output domains.
Fig. 1. CoCoA N-terminus has an activation function and serves as a coactivator for LEF1/β-catenin.
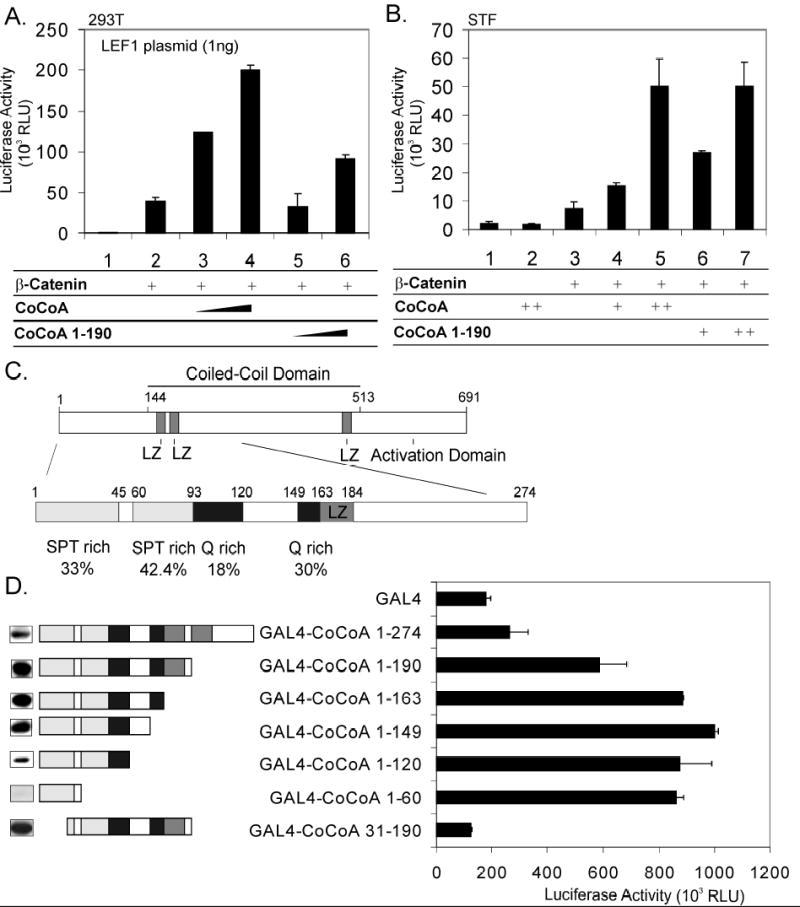
A, 293T cells were transfected in 12-well plates with pGL3OT reporter plasmid (200 ng), pSG5.HA-LEF1 (1 ng), pSG5.HA-β-catenin (5 ng), and pSG5.HA-CoCoA or pSG5.HA-CoCoA (1-190) (200 or 400 ng) as indicated. Cell extracts were assayed for luciferase activity 48 h after transfection. B, STF cells were transfected in 24-well plates with pSG5.HA-β-catenin (2.5 ng), and pSG5.HA-CoCoA or pSG5.HA-CoCoA (1-190) (50 or 100 ng) as indicated. C, Structure of CoCoA N-terminus. LZ, leucine zipper motif. D, 293T cells were transfected in 24-well plates with GK1-Luc reporter plasmid (150 ng) and plasmids encoding Gal4-DBD or Gal4-DBD-CoCoA fusion proteins (100 ng) as indicated. Cell extracts were assayed for luciferase activity (right side) or analyzed by immunoblot with anti-Gal4 antibody (left side) 48 h after transfection.
To begin characterization of downstream signaling by this signal output domain, we tested whether the N-terminal region of CoCoA contains an autonomous activation function. Amino acid sequence analysis of the CoCoA N-terminus revealed Serine/Proline/Threonine-rich and Glutamine-rich regions (Fig. 1C), which are common motifs in ADs (17,18). Gal4 DNA binding domain (DBD) fused to CoCoA (1-190) caused approximately 2.5 fold stimulation of a transiently transfected reporter gene compared to Gal4-DBD alone (Fig. 1D). C-terminal serial deletions of the N-terminal region to amino acid 149 slightly enhanced its activity (up to 5 fold compared with Gal4-DBD). Gal4-CoCoA (1-60) and Gal4-CoCoA (1-149) had similar activities, even though Gal4-CoCoA (1-60) was expressed much more weakly (Fig. 1D, left margin). Interestingly, deletion of the first 30 amino acids, leaving Gal4-CoCoA (31-190), abolished the transactivation activity completely. These results suggest that the minimal activation domain in the CoCoA N-terminus is contained within the first 60 residues, and the first 30 amino acids are necessary for the activity of this AD.
CoCoA N-terminus interacts with p300
To further examine the molecular mechanism which contributes to the activity of the CoCoA N-terminal AD, we looked for candidate proteins which may interact with the first 30 residues of CoCoA. Amino acid sequence analysis revealed a sequence FLNVARTY, which fits the consensus sequence F/YXXΦΦXXΦ (where Φ represents any hydrophobic amino acid and X represents any amino acid). This sequence has been characterized as a binding motif for the KIX domain of p300 and CBP in several transcription factors, including the cAMP-regulated transcription factor CREB (19). The CoCoA sequence (amino acids 17-24) is highly conserved in orthologous CoCoA genes among mammalian and non-mammalian vertebrates and also in homologous mammalian proteins, the nuclear dot protein NDP52 and Tax binding protein TAX1BP1 (Fig. 2A). We next tested the physical interaction of CoCoA N-terminus with the p300 KIX domain and examined the effect of mutations in the putative p300 binding motif. We mutated F17 and V20 in CoCoA, since these are two of the most highly conserved amino acids in the p300 binding motif from CoCoA, NDP52, and TAX1BP1 (Fig. 2A). CoCoA (1-190) strongly interacted with p300 KIX in a GST-pull down assay (Fig. 2B) even in the presence of a high salt concentration (300 mM NaCl). An F17A mutant of CoCoA (1-190) lost this interaction, while V20A and V20P mutants exhibited reduced binding to GST-KIX. Interestingly, CoCoA N-terminus also interacted with the p300 CH3 domain in a GST-pull down assay, although the interaction is only observed under low salt (100 mM NaCl) conditions (data not shown). Thus, even though the N-terminus of CoCoA interacts with multiple regions of p300, KIX may be the primary interaction site.
Fig. 2. CoCoA N-terminus interacts with p300.
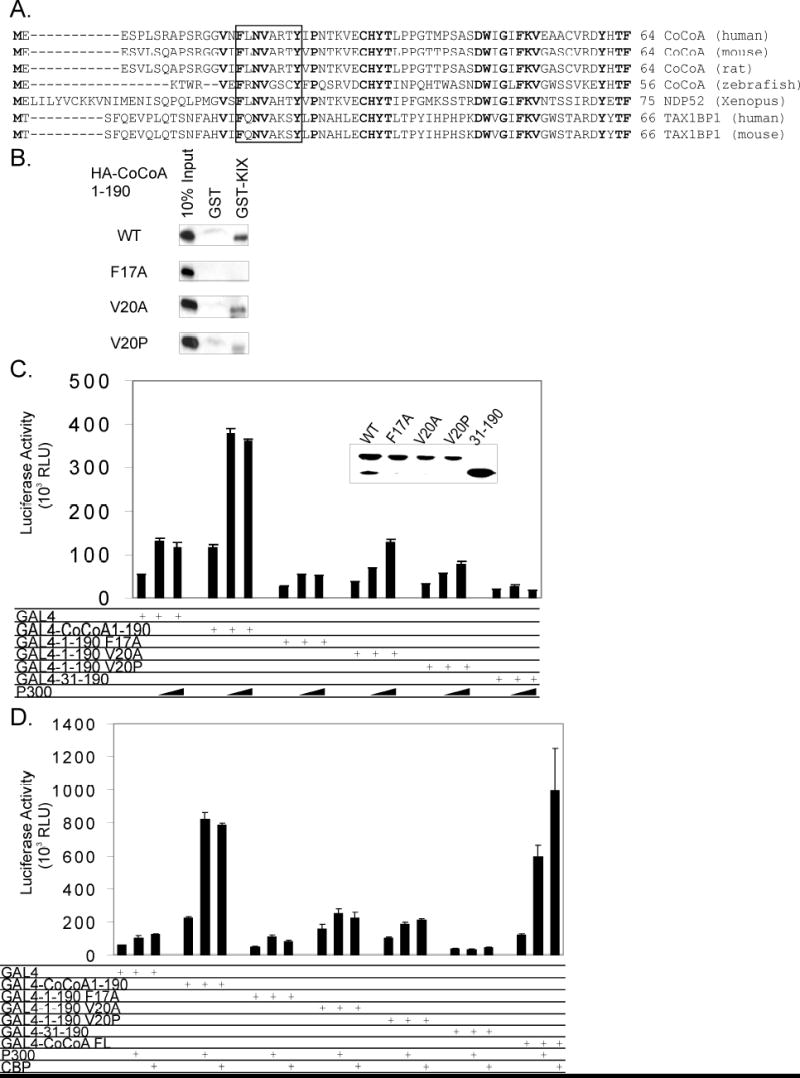
A, Sequence alignment of CoCoA and homologous proteins: human CoCoA (GenBank accession number NM_020898), mouse CoCoA (accession number NM_026192), rat CoCoA (accession number AAH81722), zebrafish CoCoA (accession number BC095162), Xenopus NDP52 (accession number AAG33628), human TAX1BP1 (accession number AAH50358), mouse TAX1BP1 (accession number AAH14798). Box shows the putative KIX-binding motif. Residues that are identical in all listed sequences are shown in bold. B, In vitro translated HA-tagged CoCoA (1-190) wild-type and mutants (F17A, V20A, V20P) were incubated with GST or GST-p300-KIX fusion proteins bound to glutathione-Sepharose beads in the presence of 300 mM NaCl. Bound proteins were eluted and analyzed by immunoblot with anti-HA antibody. C, 293T cells were transfected in 24-well plates with GK1-Luc reporter plasmids (150 ng), Gal4-DBD or Gal4-DBD-CoCoA fusion proteins (200 ng), and pCMV-p300 (50 or 100 ng) as indicated. Cell extracts were analyzed by immunoblot with antibodies against Gal4 (inset) or assayed for luciferase activity 48 h after transfection. D, 293T cells were transfected in 24-well plates with GK1- Luc reporter plasmids (150 ng), Gal4-DBD or Gal4-DBD-CoCoA fusion proteins (100 ng), and pCMV-p300 or CBP (50 ng) as indicated. Cell extracts were assayed for luciferase activity 48 h after transfection.
To investigate the functional implications of the interaction of p300 with the CoCoA N-terminal AD, we tested whether the autonomous activation function is affected by the mutations which reduce p300 binding. Wild-type CoCoA (1-190) fused to Gal4 DBD stimulated expression of a transiently transfected luciferase reporter plasmid containing Gal4 response elements approximately 2 to 4 fold compared to Gal4-DBD alone (Fig. 2C and D). In contrast, the F17A mutation eliminated the autonomous transactivation function of CoCoA N-terminus completely (Fig. 2D), and in some cases even reduced the basal transcription activity of Gal4DBD alone (Fig. 2C). The V20A and V20P mutations also caused severe reductions in the autonomous transactivation function but retained weak activity in some assays (Fig. 2D). Wild type and mutant Gal4 fusion proteins were expressed at similar levels (Fig. 2C, inset). These results corresponded to the GST-pull down assays, in which the V20 mutants retained some autonomous transactivation activity and also retained a weak binding to GST-KIX, while the F17A mutant was inactive in both assays.
Coexpression of p300 enhanced the ability of wild-type CoCoA N-terminus to activate transcription of the luciferase reporter plasmid about 4 fold, compared to the 2 fold stimulation of Gal4-DBD alone (Fig. 2C and D). However, p300 failed to enhance the transactivation function of CoCoA N-terminal mutants above the basal transcription level or the level achieved when Gal4 DBD alone was coexpressed with p300. Interestingly, CREB-binding protein (CBP), a protein highly related to p300 structurally and functionally, shared the ability of p300 to enhance the transactivation activity of Gal4 DBD fused to full-length CoCoA or to CoCoA (1-190) (Fig. 2D). This suggests that p300 and CBP can both contribute to CoCoA N-terminal AD function through interaction with the consensus KIX-binding motif at the extreme N-terminus of CoCoA.
Endogenous p300 is specifically targeted to a chromosomally integrated promoter by CoCoA N-terminus
Chromatin immunoprecipitation assays were used to test whether endogenous p300 is recruited to a specific promoter by the CoCoA N-terminus in cultured cells. Hela cell line (HLR) containing a stably integrated luciferase reporter plasmid (pFL-LUC) was transiently transfected with a plasmid encoding Gal4-DBD or Gal4-DBD-CoCoA (1-149). Sheared chromatin preparations from the transfected cells were immunoprecipitated with antibodies against Gal4 or p300, or with normal IgG; immune complexes were adsorbed with protein A/G-Sepharose beads. The precipitated DNA was analyzed by real-time quantitative-PCR with primers flanking the Gal4 responsive elements which control the integrated luciferase reporter gene. As expected, Gal4-DBD and Gal4-DBD fused to the CoCoA N-terminus were associated with the integrated Gal4 response elements to a similar extent (Fig. 3, left panel). However, promoter occupancy by p300 was enhanced about 8 fold in the presence of Gal4-CoCoA N-terminus compared with Gal4-DBD alone (right panel). IgG and beads with no antibody produced similar backgrounds in the presence of Gal4 or Gal4-CoCoA N-terminus. These results demonstrate the specific recruitment of endogenous p300 to an integrated promoter by interaction with the CoCoA N-terminus.
Fig. 3. p300 is specifically targeted to an integrated promoter by CoCoA N-terminus.
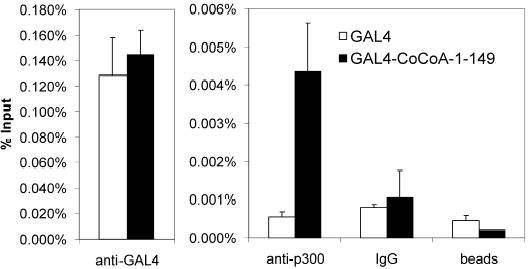
HLR cells in 150-mm dishes were transfected with Gal4-DBD or Gal4-DBD-CoCoA (1-149) (12 μg). 48 h after transfection, Chromatin immunoprecipitation assays were performed with the transfected cell extracts using the indicated antibodies. Quantitative PCR analysis of precipitated DNA was performed using primers that span the Gal4-responsive elements. Results shown are from a single experiment which is representative of three independent experiments.
Contribution of p300 protein acetyltransferase activity to the coactivator function of the CoCoA N-terminus
p300 acetylates histones and other transcription factors and coactivators, and its coactivator function involves both acetyltransferase-dependent and -independent activity when participating in transcriptional activation (20,21). We further tested whether the protein acetyltransferase activity of p300 is important for the enhancement of CoCoA N-terminal activity. Wild-type p300 increased the ability of Gal4-DBD fused to CoCoA (1-190) to activate transcription of a transiently transfected luciferase reporter plasmid containing Gal4 response elements in a dose dependent manner (Fig. 4A). In contrast two p300 deletion mutants with abolished acetyltransferase activity (Δ1471–1522 & Δ1603–1653) caused little or no enhancement of CoCoA N-terminal activity. Immunoblot analyses of parallel transfection experiments indicated that similar expression of wild type and mutant proteins was achieved by using four times as much mutant p300 expression vector as wild type expression vector (Fig. 4A, inset, filled arrowheads). In the reporter gene assays 10 ng of wild type p300 expression plasmid caused a substantial enhancement of luciferase activity, whereas even 300 ng of the mutant p300 expression plasmids had no effect (Fig. 4A, assays 2, 9, & 13). Thus, the mutant p300 proteins lacking acetyltransferase activity failed to enhance the activity of the CoCoA N-terminal AD.
Fig. 4. Contribution of p300 to the activity of the CoCoA N-terminus.
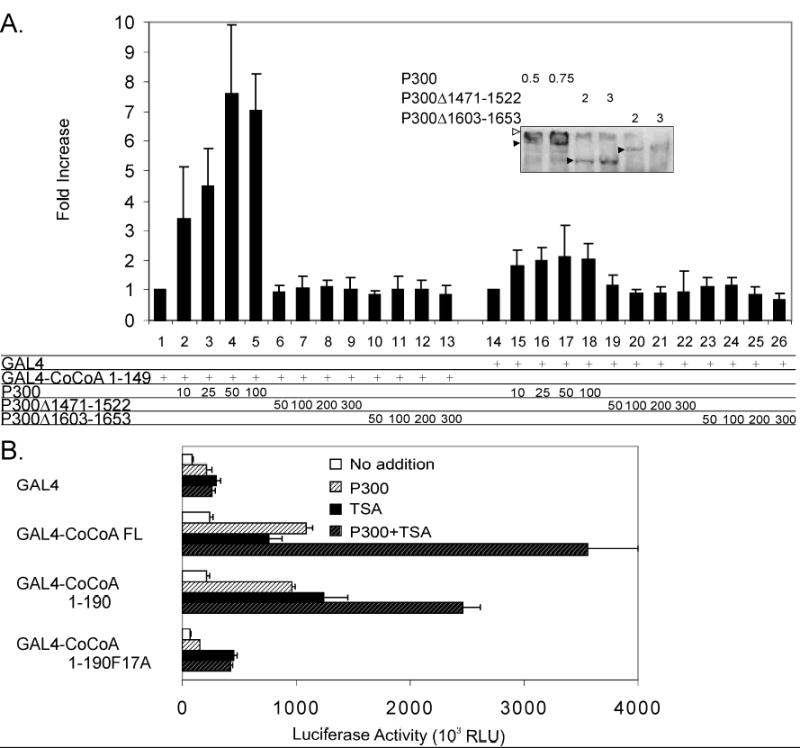
A, 293T cells were transfected in 24-well plates with GK1-Luc reporter plasmid (100 ng), plasmids encoding Gal4-DBD or Gal4-CoCoA (1-190) (75 ng), and pCI-Flag-p300 wild-type or acetyltransferase deletion mutants (Δ1472–1522 and Δ1603–1653) as indicated. Results shown are the mean and standard deviation of five independent experiments. Inset, 293T cells were transfected in 6-well plates with pCI-Flag-p300 wild-type (0.5 or 0.75 μg) or acetyltransferase deletion mutants (2 or 3 μg). Cell extracts were analyzed by immunoblot with anti-Flag antibody. Filled arrowheads show positions of p300 (wild type and mutants). Open arrowhead shows protein trapped at the top of the gel. B, 293T cells were transfected in 24-well plates with GK1-Luc reporter (150 ng), plasmids encoding Gal4-DBD or Gal4-CoCoA wild type or mutants (200 ng), and pCMV-p300 (50 ng) as indicated. 24 h post-transfection, cells were treated with or without TSA (0.2 μM) for 24 h before assaying for luciferase activity.
As a further test of the importance of p300 protein acetyltransferase activity in mediating or enhancing the function of the CoCoA N-terminal AD, we examined the effect of the deacetylase inhibitor Trichostatin A (TSA) on CoCoA or its N-terminal fragment fused to Gal4-DBD. TSA alone enhanced reporter gene activation by CoCoA full-length and by the CoCoA N-terminus (amino acids 1-190) about 4-fold. Addition of p300 in the presence of TSA further enhanced the activity of CoCoA full-length and CoCoA N-terminus, while it had no additional effect on the activity of the N-terminal mutant (F17A) or of GAL4-DBD alone (Fig. 4B). These results suggest that while p300 acetyltransferase-independent activity may modestly enhance the activity of the CoCoA N-terminus, the acetyltransferase activity of p300 plays a key role in the full activation of the CoCoA N-terminus by p300.
Synergistic enhancement of LEF1-mediated transcriptional activation by β-catenin, CoCoA, p300 and GRIP1
When CoCoA functions as a secondary coactivator (i.e. by binding to β-catenin rather than directly to LEF1) to enhance LEF1-mediated transcriptional activation, the N-terminal region of CoCoA is required for cooperation with β-catenin (4). We therefore tested whether mutation in the N-terminal KIX-binding domain of CoCoA, which reduced the N-terminal transactivation function, would affect the secondary coactivator activity of CoCoA. Full-length CoCoA and a C-terminal deletion mutant containing only amino acids 1-500 were effective coactivators when cooperating with β-catenin to enhance LEF1-mediated transcriptional activation of the pGL3OT luciferase reporter gene (Fig. 5A, assays 8–12). In contrast, CoCoA (1-500) containing the F17A mutation in the motif that binds the KIX domain of p300 had no coactivator function (assays 13–14). Immunoblots showed a similar expression level for wild-type and mutant CoCoAs (inset).
Fig. 5. Synergistic enhancement of LEF1-mediated transcriptional activation by β-catenin, CoCoA, p300 and GRIP1.
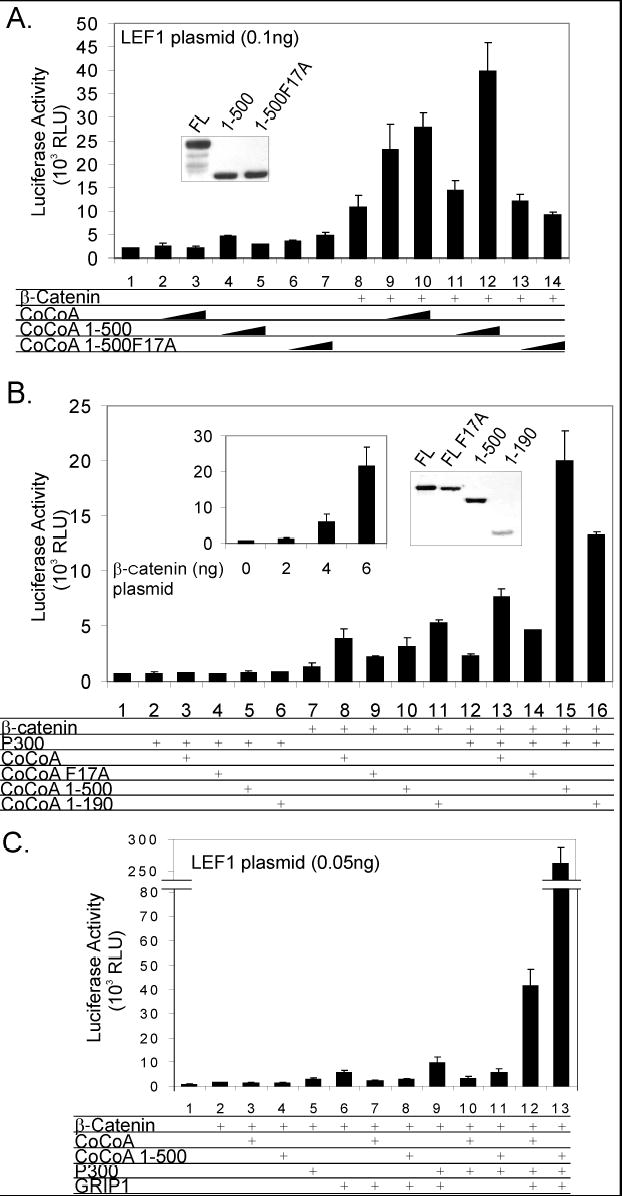
A, 293T cells were transfected in 24-well plates with pGL3OT reporter plasmid (100 ng), pSG5.HA-LEF1 (0.1 ng), pSG5.HA-β-catenin (1 ng), and a pSG5.HA vector encoding CoCoA full-length, CoCoA (1-500), or CoCoA (1-500)F17A (50 or 100 ng) as indicated. Cell extracts were assayed for luciferase activity 48 h after transfection. Inset, COS-7 cells in 12-well plates were transfected with CoCoA full-length, 1-500, or 1-500 F17A (500 ng). Cell extracts were analyzed by immunoblot with anti-HA antibody. B, STF cells were transfected in 24-well plates with pSG5.HA-β-catenin (2 ng), pCMV-p300 (1 ng), and pSG5.HA vectors encoding the indicated CoCoA species (60 ng) as indicated. Cell extracts were assayed for luciferase activity 48 h after transfection. Left inset, 293T cells were transfected with pGL3OT reporter plasmid (100 ng), pSG5.HA-LEF1 (0.1 ng), and the indicated quantities of pSG5.HA-β-catenin. Right inset, COS-7 cells in 12-well plates were transfected with plasmids encoding CoCoA full-length (FL) or the indicated CoCoA fragment or mutant (500 ng). Cell extracts were analyzed by immunoblot with anti-HA antibody. C, 293T cells were transfected in 24-well plates with pGL3OT reporter plasmid (100 ng), pSG5.HA-LEF1 (0.05 ng), pSG5.HA.β-catenin (0.5 ng), pSG5.HA-GRIP1 (50 ng), pCMV-p300 (50 ng), and pSG5.HA-CoCoA or pSG5.HA-CoCoA (1-500) (100 ng) as indicated. Cell extracts were assayed for luciferase activity 48 h after transfection.
To further investigate the cooperation of p300 and CoCoA in transcriptional activation mediated by LEF1 and β-catenin, we used the STF cells which contain a stably integrated luciferase reporter gene controlled by TCF/LEF-binding sites. Overexpression of β-catenin in STF cells enhanced the activation of the integrated luciferase reporter gene in a dose dependent manner (Fig. 5B, left inset). Full-length CoCoA, the CoCoA N-terminal fragment (amino acids 1-190), and a C-terminal deletion mutant of CoCoA retaining amino acids 1-500 all cooperated synergistically with low levels of β-catenin (2 ng of plasmid), while full-length CoCoA with the F17A mutation in the N-terminal KIX-binding domain had only partial secondary coactivator activity (Fig. 5B, assays 7–11). Since p300 and CoCoA each functions as a secondary LEF1 coactivator in cooperation with β-catenin, we tested whether CoCoA and p300 can cooperate synergistically as coactivators in the presence of β-catenin. p300 overexpressed alone or with CoCoA caused little or no activation in the absence of β-catenin (assays 2–6). In the presence of exogenous β-catenin, p300 enhanced the integrated reporter gene expression about 2 fold compared to β-catenin alone (assays 7 and 12). Coexpression of p300 and β-catenin with wild type CoCoA, a CoCoA mutant lacking the C-terminus (1-500) or the N-terminal fragment of CoCoA (amino acids 1-190) synergistically enhanced the expression of the reporter gene (assays 13, 15 and 16), while mutation F17A in the N-terminal KIX binding domain of CoCoA reduced the cooperation between β-catenin, CoCoA and p300 (assays 12–14). Wild type and mutants were expressed at similar levels (Fig, 5B, right inset). Thus, while p300 and CoCoA can each bind to and cooperate with β-catenin, we conclude that the specific interaction between CoCoA and p300 further contributes to transcriptional activation, indicating the importance of their physical and functional cooperation. Furthermore, the N-terminal KIX-binding domain of CoCoA plays a critical role in p300-CoCoA binding and in the synergistic cooperation of β-catenin, CoCoA and p300.
To expand the investigation of the multifaceted participants in the coactivator complex, we included GRIP1, which has also been shown to contribute to LEF1-mediated transcriptional activation by interaction with β-catenin (9,10,22), and has also been shown to cooperate with p300 functionally (20). Since the four coactivators, β-catenin, CoCoA, p300 and GRIP1, can all bind to each other, we therefore tested whether CoCoA could function as an effective coactivator for LEF1 in a four-coactivator system. For this test, we employed transient transfection conditions with low levels of LEF1 expression vectors. It was shown previously with nuclear receptors that these stringent low-activator conditions facilitate observation of synergistic interactions among multiple coactivators (20). Under these conditions, coexpression of β-catenin and LEF1 caused little enhancement in expression of the transiently transfected TCF/LEF1 controlled reporter plasmid (Fig 5C, assay 2). Coexpression of two coactivators (β-catenin and GRIP1) and three coactivators (β-catenin, GRIP1 and p300) caused modest enhancement of about 7 and 13 fold, respectively, in LEF1-mediated transcriptional activation of the reporter gene (assays 6 and 9). Addition of CoCoA as a fourth coactivator further enhanced transcription by an additional 4 fold (compare assays 9 and 12), suggesting the significance of CoCoA in the function of the multiple coactivator complex. Note that overexpression of the C-terminal deletion mutant of CoCoA as a fourth coactivator instead of full-length CoCoA dramatically enhanced transcriptional activation about 7 fold more than wild-type CoCoA (assay 13). This result again indicates the lack of a requirement for the C-terminal region of CoCoA in LEF1-mediated transcriptional activation; it furthermore suggests a possible inhibitory role for the C-terminal region.
Cellular Localization of CoCoA and its fragments
To test whether the strong coactivator activity of the CoCoA C-terminal deletion mutant results from a difference in cellular localization, we used immunofluorescence. The PSORT II server (http://psort.nibb.ac.jp/form2.html) predicted a cellular localization for CoCoA of 43.5% nuclear, 30.4% cytoplasmic and the remaining 27% associated with cytoskeleton or cytoplasmic organelles or vesicles. However, by immunofluorescence with anti-HA antibody in confocal microscopy HA-tagged CoCoA predominantly localized in the cytoplasm of both 293T and SW480 cells (Fig. 6), which agrees with a previously reported expression pattern of human CoCoA (23). A similar expression pattern was also observed in a Hela S cell line with a stably integrated full-length CoCoA expression vector, using CoCoA antisera (data not shown). Deletion of the C-terminal region of CoCoA had little effect on the cellular distribution, which rules out the possibility that the dramatic coactivator function of the C-terminal deletion mutant is a result of increased nuclear localization. Interestingly, an N-terminal fragment of CoCoA (amino acids 1-190) had a substantial increase of nuclear localization in both cell lines. However, mutation in the N-terminal KIX-binding motif (F17A) had no effect on the expression pattern of either full-length CoCoA or CoCoA fragments (data not shown), suggesting that the interaction between p300 and CoCoA has no effect on its cellular localization. In spite of its predominant cytoplasmic localization, CoCoA functions as a transcriptional coactivator and has been shown to localize to specific enhancer elements by chromatin immunoprecipitation assays. This suggests that CoCoA shuttles between nucleus and cytoplasm. In fact, the immunofluorescence data shows that the N-terminal fragment of CoCoA has a weak nuclear localization capability (Fig. 6), and the isolated C-terminal region localizes strongly to the nucleus (data not shown). Thus, CoCoA must contain both nuclear localization and nuclear export signals.
Fig. 6. Cellular localization of CoCoA.
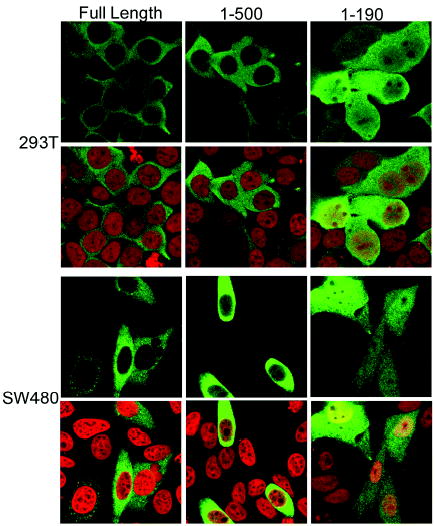
293T and SW480 cells were transfected with plasmids encoding HA-tagged CoCoA full-length or fragments (1 μg) as indicated. 48 h after transfection, cells were stained with anti-HA antibody (green), mounted in the presence of propidium iodide (red) and viewed with confocal microscopy.
DISCUSSION
The N-terminus of CoCoA
The N-terminus of CoCoA consists of regions enriched with amino acids commonly found in ADs, S/P/T-rich (amino acids 1-45 and 60-93) and Q-rich (amino acids 93-120 and 149-163) regions (Fig. 1C) (17,18). Deletion of the coiled-coil domain enhanced the autonomous transactivation activity of the N-terminus, suggesting that the coiled-coil domain may have a negative regulatory effect on the N-terminal AD. Serial C-terminal deletions of the N-terminal region demonstrated that the first 30 amino acids are the core AD.
The N-terminus of CoCoA may possess additional functions other than the transcriptional activation activity. Full-length CoCoA and a C-terminal deletion mutant both showed predominant cytoplasmic localization, while an N-terminal fragment of CoCoA had a significant increase in nuclear localization (Fig. 6). These results suggest that a regulated nuclear localization signal (NLS) is present in the N-terminus and that the central coiled-coil domain may have nuclear export activity which regulates or overrides the N-terminal NLS. Furthermore, the N-terminus (amino acids 1-190) of CoCoA is 70% similar to Xenopus nuclear dot protein 52 (NDP52), which contains the SKIP carboxyl homology (SKICH) domain known for plasma membrane localization (24). This suggests that the N-terminus of CoCoA may contain multiple cellular localization signals.
The multiple functions of the CoCoA N-terminus may be further regulated by post-translational modifications. Human CoCoA was recently shown to be an in vitro substrate of p42 MAPK and of CDK2/Cyclin E with predicted phosphorylation sites in both the N- and C-terminal ADs (23). The large number of serine and threonine residues in the N-terminus may also be potential targets for other kinases. SUMOplot™ (http://www.abgent.com/doc/sumoplot) predicted sumoylation sites in both the N-terminal and the central coiled-coil region, which may be important in regulation of the coactivator function or cellular localization.
The dual function of CoCoA N-terminus as signal input and output domain
Coactivators contribute to transcriptional activation by transmitting the activation signals from the DNA-bound activators to the chromatin and the RNA polymerase II associated transcription machinery. Various regions in each coactivator can serve as either signal input or output domains, or both. The coactivators receive the activating signals from the DNA-binding activators through their interacting domains (signal input domains) and transmit the signals through ADs (signal output domains) to the downstream components of the transcriptional machinery by protein-protein interaction or enzymatic activity which catalyzes post-translational modifications of other proteins (1).
We previously showed that the N-terminus of CoCoA can directly interact with β-catenin and serve as a signal input domain (4). Here we further demonstrated the dual function of CoCoA N-terminus. When β-catenin and CoCoA cooperate as primary and secondary coactivator, respectively (Fig. 1A & B), the CoCoA N-terminus is necessary and sufficient for the coactivator function, i.e. it not only interacts with β-catenin, but also signals downstream to the transcription machinery and enhances transcription, thus fulfilling the requirements for a signal output domain as well as a signal input domain.
To further characterize the molecular mechanism by which this signal output domain functions, we mapped a critical AD in the first 30 amino acids and showed that it contains a p300 KIX-binding motif. p300 is a cofactor which participates in transcriptional activation by many DNA-binding transcriptional activator proteins. p300 may enhance transcription by directly interacting with TFIID, TBP and other components of the basal transcription machinery (25,26); it also acetylates histones and other protein components of the transcription machinery (27–29). In essence, connecting this signal output domain of CoCoA to p300 completes one branch of the complex coactivator signaling pathways that lead from DNA-bound LEF1 to the transcription machinery. This branch of the signal pathway travels through β-catenin, the N-terminal domain of CoCoA, and the acetyltransferase activity of p300 to the transcription machinery (Fig. 7). Obviously, there are other signal branches which travel through β-catenin to GRIP1, CARM1, and directly to p300, since each of these also interacts directly with β-catenin and cooperates synergistically with the others (4,9,10,13–15,22). We found that the N-terminus of CoCoA can interact with p300 and CBP physically and functionally (Fig. 2). CoCoA binds to p300 KIX domain (Fig. 2B) and more weakly to the CH3 domain (data not shown). Moreover, point mutations in the putative KIX-binding motif of the CoCoA N-terminus disrupted this interaction, and strongly reduced the autonomous activation activity of the CoCoA N-terminus (Fig. 2, B, C & D), suggesting the importance of p300 in the output signaling by the CoCoA N-terminus. p300 deletion mutants lacking the acetyltransferase activity caused greatly reduced enhancement of CoCoA N-terminal activity (Fig. 4A), suggesting that while p300 acetyltransferase-independent activity may contribute modestly to the activity of the CoCoA N-terminus, the acetyltransferase activity of p300 is responsible for the major activation effect of p300 on the CoCoA N-terminus. In the presence of CoCoA, p300 may assist in transcriptional activation by either acetylating histones or other known or unknown protein components of the transcription machinery.
Fig. 7. Model for action of β-catenin-CoCoA coactivator complex.
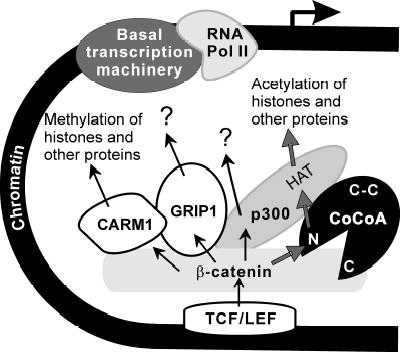
To activate transcription of its target genes DNA-bound TCF/LEF proteins recruit β-catenin which transmits the transactivation signal to the associated promoter by recruiting secondary coactivators, including CoCoA, p300 (or CBP), GRIP1, CARM1 and others not shown here. With the exception of CARM1 each of these coactivators can bind to all of the others, but for simplicity, not all of the binding interactions are represented here. The transcription activating signal from TCF/LEF is represented by arrows arranged in a branching pattern, emanating from TCF/LEF and passing through β-catenin to the secondary coactivators. The synergy among the multiple coactivators demonstrated in this paper indicates that each secondary coactivator makes a distinct contribution to the transcriptional activation process by recruiting or acting upon a distinct downstream target in the chromatin or transcription machinery; not all targets are known at this time. The thicker arrows show at least part of the contribution of CoCoA: the N-terminal region of CoCoA acts as a signal input domain by binding to β-catenin and also acts as a signal output domain by binding to p300. The synergy among β-catenin, CoCoA and p300 requires the protein and histone acetyltransferase activity (HAT) of p300. CoCoA may also contribute by stabilizing the complex through its binding interactions with β-catenin, GRIP1 and p300. GRIP1 may similarly contribute to coactivator complex stability, since it can bind to all of the other coactivator components shown. In addition to HAT activity, other domains of p300 may contribute by recruiting or acting upon additional downstream targets, since p300 is known to bind to TBP and other basal transcription factors. CARM1 contributes through its arginine-specific protein and histone methyltransferase activity.
The importance of the p300-CoCoA binding interaction within the large β-catenin coactivator complex
Transcriptional activation is a dynamic process that involves a series of highly ordered events. In the Wnt- signaling pathway, β-catenin binds to LEF1 and serves as a scaffold protein which recruits a variety of secondary coactivators to assist in transcription of its target genes (4,9,10,13–15,22). CoCoA, p300, and GRIP1 all directly bind to β-catenin and cooperate with β-catenin to enhance transcription (Fig. 5). GRIP1 also directly binds to p300 through its AD1 domain (amino acids 1040-1120) (30), and to CoCoA through the N-terminal basic-helix-loop-helix/Per-Arnt-Sim (bHLH-PAS) domain (2). Here we further demonstrated that the CoCoA N-terminal region can directly interact with p300, linking all of these coactivators directly to one another. Thus, our results indicate that CoCoA, p300, and GRIP1 may form a multisubunit complex to synergistically enhance β-catenin/LEF1-mediated transcription (Fig 5B & C). Conceptually, since each of the three secondary coactivators of this complex can be directly recruited to the promoter region by the primary coactivator β-catenin, loss of binding of one of the secondary coactivators to another might have no significant effect on the transcriptional activity of the complex. On the other hand, the additional molecular interactions might stabilize the complex. As we demonstrated here, CoCoA with a mutation in the N-terminal p300(KIX)-binding motif showed only partial coactivator function in the presence of β-catenin alone or β-catenin and p300 (Fig. 5A & B). This implies that while p300 can still theoretically be recruited to the stably integrated reporter by direct interaction with β-catenin or GRIP1, the ability of CoCoA to interact with p300 still plays an integral role in transcriptional activation.
Differential use of dual p300 binding motifs in the N-terminal and C-terminal regions of CoCoA
Our previous studies demonstrated that the C-terminal AD of CoCoA is absolutely required for synergistic cooperation with GRIP1, CARM1, and p300 in NR-mediated transcription (2), and that the C-terminus of CoCoA can also bind and cooperate with p300 (31). In contrast, a C-terminal deletion mutant of CoCoA showed substantially stronger coactivator activity than full-length CoCoA when cooperating with p300 and GRIP1 in β-catenin/LEF1-mediated transcription (Fig. 5C). This result suggests 1) that the C-terminus of CoCoA is dispensable in the β-catenin coactivator complex, 2) that the C-terminal region contains functions which are repressive in the context of β-catenin-mediated transcriptional activation, and 3) that the coactivator synergy between CoCoA and p300 in the presence of β-catenin is mediated by the N-terminal p300-binding motif of CoCoA, not by the C-terminal p300-binding motif. The fact that CoCoA contains multiple ADs utilized in different coactivator complexes supports the concept that while many transcription factors and cofactors may be widely expressed in cells, the key to regulation of the highly sophisticated process of transcriptional activation may be through differential use of different domains of transcription factors and cofactors in different regulatory settings.
MATERIALS & METHODS
Plasmids
The plasmids, pSG5.HA-LEF1 encoding LEF1 with an N-terminal hemagglutinin (HA) epitope tag, pSG5.HA-β-catenin encoding HA-tagged chicken β-catenin, pSG5.HA-GRIP1, pSG5.HA-CoCoA, pSG5.HA-CoCoA(1-190), pSG5.HA-CoCoA(1-500), and luciferase reporter gene plasmids pGL3OT (for LEF1), GK1-LUC (for Gal4), and pM-CoCoA encoding Gal4 DBD fused to CoCoA were previously described (4). pM-CoCoA(1-274), pM-CoCoA(1-190), pM-CoCoA(1-149) (2), pCMV-p300, pCMV-CBP, pGEX-p300-KIX (32) were also described previously. PCR-amplified CoCoA cDNA fragments were inserted into EcoRI and SalI sites of pM vector (Clonetech). QuikChange site-directed mutagenesis kit (Stratagene) was used to generate point mutations in full-length CoCoA and CoCoA fragments. pCI-FLAG-p300 and its acetyltransferase deletion mutant expression plasmids, pCI-FLAG-p300Δ1472–1522 and pCI-FLAG-p300Δ1603–1653, were kind gifts by Dr. Yoshihiro Nakatani (Harvard University).
Cell culture and Transfection
HEK 293T, COS-7, SW480 and HLR cells (Stratagene) were maintained in Dulbecco’s modified Eagle’s medium (DMEM). STF cells with stably integrated luciferase reporter which contains 7X TCF/LEF-binding sites (kindly provided by Dr. Jeremy Nathans, Johns Hopkins University) (16) were maintained in DMEM F-12. All cells were supplemented with 10% fetal bovine serum and penicillin and streptomycin. Transient transfection of COS-7 cells with TargeFect F-2 reagent (Targeting Systems) were described previously (4). Transfection of HEK 293T, SW480 and STF cells was performed with Lipofectamine 2000 (Invitrogen) as described previously (4,16). The results shown are the means and standard deviations of triplicates from single experiments, which are representative of at least three independent experiments. Luciferase activities were not normalized to internal controls, because expression of so-called constitutive reporter genes is affected by over-expression of many coactivators. Instead, multiple independent experiments with multiple plasmid preparations were used to demonstrate reproducibility.
Protein-Protein Interaction Assays and Immunoblot
GST pull-down assays were performed as described previously (4) using extracts from COS-7 cells transiently transfected with the indicated plasmids. The bound proteins were analyzed by immunoblot with anti-HA antibody (Roche Applied Science).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assays were performed as described previously (4). HLR Cells with stably integrated Gal4-LUC reporter (pFL-LUC) were transfected in 150-mm dishes by Lipofectamine 2000 with expression plasmids as indicated. Soluble chromatin fractions were prepared 48 h post-transfection. Immunoprecipitation was performed using 1 μg of anti-Gal4 antibody 06-262 (Upstate/Chemicon), or 1 μg anti-p300 antibody RW128 (Upstate/Chemicon). The precipitated DNA was quantified using quantitative real-time PCR with primers spanning the Gal4 responsive elements: 5′-GGTACCGAGCTCGAATTCCAGCTT-3′ (forward) and 5′-GCGTATCTCTTCATAGCCTTATGC-3′ (reverse). Quantitative PCR reactions were performed with Brilliant SYBR Green QPCR Master Mix (Stratagene) with the Mx3000P system (Stratagene).
Immunofluorescence Staining
HEK 293T or SW480 cells were plated on glass cover slides and transfected with 2 μg of HA-tagged CoCoA expression plasmids with Lipofectamine 2000. 48 h post-transfection, cells were fixed with 4% paraformaldehyde, and stained with anti-HA antibody (1:500 dilution; Roche Applied Science) and FITC-conjugated anti-rat IgG (1:250 dilution; Santa Cruz Biotechonology). Cells were treated with RNase A for 30 min and mounted in Vectashield with propidium iodide mounting medium (Vector Labs) and visualized with confocal microscopy in the Microscopy Core at the USC Research Center for Liver Diseases.
Acknowledgments
We thank Mr. Dan Gerke for expert technical assistance, Dr. Jeremy Nathans (Johns Hopkins University) for STF cells, and Dr. Yoshihiro Nakatani (Harvard University) for plasmids expressing FLAG-tagged p300s. This work was supported by grant DK43093 to M.R.S. from National Institutes of Health, and the Microscopy Core Lab of the USC Research Center for Liver Diseases was supported by grant P03 DK48522 from the National Institutes of Health.
Footnotes
This is an un-copyedited author manuscript copyrighted by The Endocrine Society. This may not be duplicated or reproduced, other than for personal use or within the rule of “Fair Use of Copyrighted Materials” (section 107, Title 17, U.S. Code) without permission of the copyright owner, The Endocrine Society. From the time of acceptance following peer review, the full text of this manuscript is made freely available by The Endocrine Society at http://www.endojournals.org/. The final copy edited article can be found at http://www.endojournals.org/. The Endocrine Society disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties. The citation of this article must include the following information: author(s), article title, journal title, year of publication and DOI.
This work was supported by grant DK43093 to M.R.S. from National Institutes of Health, and the Microscopy Core Lab of the USC Research Center for Liver Diseases was supported by grant P03 DK48522 from the National Institutes of Health.
References
- 1.Stallcup MR, Kim JH, Teyssier C, Lee YH, Ma H, Chen D. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J Steroid Biochem Mol Biol. 2003;85:139–145. doi: 10.1016/s0960-0760(03)00222-x. [DOI] [PubMed] [Google Scholar]
- 2.Kim JH, Li H, Stallcup MR. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol Cell. 2003;12:1537–1549. doi: 10.1016/s1097-2765(03)00450-7. [DOI] [PubMed] [Google Scholar]
- 3.Kim JH, Stallcup MR. Role of the Coiled-coil Coactivator (CoCoA) in Aryl Hydrocarbon Receptor-mediated Transcription. J Biol Chem. 2004;279:49842–49848. doi: 10.1074/jbc.M408535200. [DOI] [PubMed] [Google Scholar]
- 4.Yang CK, Kim JH, Li H, Stallcup MR. Differential use of functional domains by coiled-coil coactivator in its synergistic coactivator function with beta-catenin or GRIP1. J Biol Chem. 2006;281:3389–3397. doi: 10.1074/jbc.M510403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morin PJ. beta-catenin signaling and cancer. Bioessays. 1999;21:1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 6.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 7.Billin AN, Thirlwell H, Ayer DE. Beta-catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol Cell Biol. 2000;20:6882–6890. doi: 10.1128/mcb.20.18.6882-6890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu T, DeCostanzo AJ, Liu X, Wang H, Hallagan S, Moon RT, Malbon CC. G protein signaling from activated rat frizzled-1 to the beta-catenin- Lef-Tcf pathway. Science. 2001;292:1718–1722. doi: 10.1126/science.1060100. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Kim JH, Koh SS, Stallcup MR. Synergistic effects of coactivators GRIP1 and beta-catenin on gene activation: cross-talk between androgen receptor and Wnt signaling pathways. J Biol Chem. 2004;279:4212–4220. doi: 10.1074/jbc.M311374200. [DOI] [PubMed] [Google Scholar]
- 10.Song LN, Herrell R, Byers S, Shah S, Wilson EM, Gelmann EP. Beta-catenin binds to the activation function 2 region of the androgen receptor and modulates the effects of the N-terminal domain and TIF2 on ligand-dependent transcription. Mol Cell Biol. 2003;23:1674–1687. doi: 10.1128/MCB.23.5.1674-1687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takemaru KI, Moon RT. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol. 2000;149:249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, Kolligs FT, Hottiger MO, Mosavin R, Fearon ER, Nabel GJ. Regulation of beta -catenin transformation by the p300 transcriptional coactivator. Proc Natl Acad Sci U S A. 2000;97:12613–12618. doi: 10.1073/pnas.220158597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh SS, Li H, Lee YH, Widelitz RB, Chuong CM, Stallcup MR. Synergistic coactivator function by coactivator-associated arginine methyltransferase (CARM) 1 and beta-catenin with two different classes of DNA-binding transcriptional activators. J Biol Chem. 2002;277:26031–26035. doi: 10.1074/jbc.M110865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Q, Wang Y, Dabdoub A, Smallwood PM, Williams J, Woods C, Kelley MW, Jiang L, Tasman W, Zhang K, Nathans J. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116:883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- 17.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 18.Triezenberg SJ. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 19.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y-H, Koh SS, Zhang X, Cheng X, Stallcup MR. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol. 2002;22:3621–3632. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 22.Song LN, Gelmann EP. Interaction of beta-catenin and TIF2/GRIP1 in transcriptional activation by the androgen receptor. J Biol Chem. 2005;280:37853–37867. doi: 10.1074/jbc.M503850200. [DOI] [PubMed] [Google Scholar]
- 23.Wiemann S, Arlt D, Huber W, Wellenreuther R, Schleeger S, Mehrle A, Bechtel S, Sauermann M, Korf U, Pepperkok R, Sultmann H, Poustka A. From ORFeome to biology: a functional genomics pipeline. Genome Res. 2004;14:2136–2144. doi: 10.1101/gr.2576704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gurung R, Tan A, Ooms LM, McGrath MJ, Huysmans RD, Munday AD, Prescott M, Whisstock JC, Mitchell CA. Identification of a novel domain in two mammalian inositol-polyphosphate 5-phosphatases that mediates membrane ruffle localization. The inositol 5-phosphatase skip localizes to the endoplasmic reticulum and translocates to membrane ruffles following epidermal growth factor stimulation. J Biol Chem. 2003;278:11376–11385. doi: 10.1074/jbc.M209991200. [DOI] [PubMed] [Google Scholar]
- 25.Abraham SE, Lobo S, Yaciuk P, Wang HG, Moran E. p300, and p300-associated proteins, are components of TATA-binding protein (TBP) complexes. Oncogene. 1993;8:1639–1647. [PubMed] [Google Scholar]
- 26.Felzien LK, Farrell S, Betts JC, Mosavin R, Nabel GJ. Specificity of cyclin E-Cdk2, TFIIB, and E1A interactions with a common domain of the p300 coactivator. Mol Cell Biol. 1999;19:4241–4246. doi: 10.1128/mcb.19.6.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imhof A, Yang XJ, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 28.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 29.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiology and Molecular Biology Reviews. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen D, Huang S-M, Stallcup MR. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J Biol Chem. 2000;275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- 31.Kim JH, Yang CK, Stallcup MR. Downstream signaling mechanism of the C-terminal activation domain of transcrpitional coactivator CoCoA. Nucleic Acids Res. 2006;34:2736–2750. doi: 10.1093/nar/gkl361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee Y-H, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci USA. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]


