Abstract
The yeast GCN5 (yGCN5) transcriptional coactivator functions as a histone acetyltransferase (HAT) to promote transcriptional activation. Here, we present the high resolution crystal structure of the HAT domain of yGCN5 and probe the functional importance of a conserved glutamate residue. The structure reveals a central protein core associated with AcCoA binding that appears to be structurally conserved among a superfamily of N-acetyltransferases, including yeast histone acetyltransferase 1 and Serratia marcescens aminoglycoside 3-N-acetyltransferase. A pronounced cleft lying above this core, and flanked by N- and C-terminal regions that show no sequence conservation within N-acetyltransferase enzymes, is implicated by cross-species conservation and mutagenesis studies to be a site for histone substrate binding and catalysis. Located at the bottom of this cleft is a conserved glutamate residue (E173) that is in position to play an important catalytic role in histone acetylation. Functional analysis of an E173Q mutant yGCN5 protein implicates glutamate 173 to function as a general base for catalysis. Together, a correlation of the yGCN5 structure with functionally debilitating yGCN5 mutations provides a paradigm for understanding the structure/function relationships of the growing number of transcriptional regulators that function as histone acetyltransferase enzymes.
Gene activation is a tightly regulated process that relies on the coordinated activities of several different proteins. Sequence-specific transcriptional activators play an important role in nucleating gene expression by recruiting the basal transcriptional machinery, which includes TATA box-binding proteins, TAFs (TATA box-binding protein-associated proteins), polymerase, and other protein cofactors, to DNA (1, 2). Under physiological conditions, the transcriptional machinery has to deal with a DNA template that is complexed with histones that form nucleosomes and that are assembled into higher order chromatin (3). Because chromatin assembly strongly represses transcription (4, 5), DNA regulatory proteins must physically destabilize nucleosome/DNA interactions to facilitate transcriptional activation.
The relatively recent findings that histone acetyltransferase (HAT) and histone deacetyltransferase enzymes are proteins that had been previously characterized as transcriptional cofactors has revealed a direct mechanistic link between chromatin modification and transcriptional regulation (6–10). Specifically, a number of transcriptional cofactors with HAT activity have been identified, including GCN5 {from Tetrahymena [11] yeast [yGCN5] [12], human [hGCN5 (13) and hP/CAF (14)], Drosophila [15] and Arabidopsis [GenBank accession no. 2642602]}, yeast ESA1 (16), and human CBP/p300 (17), TAFII250 (18), Tip60 (19), ACTR (20), and SRC-1 (21). In addition, the catalytic subunits of distinct histone deacetylase complexes shown to mediate transcriptional repression have been identified, including yeast and mammalian RPD3/HDAC2 (22, 23), yeast HDA1 and phd1 (24, 25), maize HD1-A and HD2 (26–28), and human HDAC1 and HDAC3 (29–31).
Of the HAT enzymes identified to date, yGCN5 is currently the best characterized. Recombinant GCN5 has been shown to efficiently acetylate free histones with a strong preference for lysine 14 of histone H3 and a somewhat lower preference for lysines 8 and 16 of histone H4 (32). Interestingly, acetylation of nucleosomal histones requires that GCN5 be part of one of two distinct multiprotein complexes called Ada and SAGA (Spt-Ada-GCN5-acetyltransferase) (33, 34). Moreover, in the context of these multiprotein complexes, GCN5 displays an acetylation preference for histones H3 and H2B (33). Extensive deletion and mutagenesis studies of yGCN5 have identified a HAT region, within residues 95–261, that is sufficient for acetylase activity and essential for GCN5-mediated transcriptional activation (35–38). This region of GCN5 has recently been shown to harbor three relatively small submotifs (15–33 residues) with homology to a biologically diverse family of GCN5-related N-acetyltransferases (GNATs) (39). To obtain a detailed view of the mechanism of histone acetylation by yGCN5, we report here the 1.9-Å crystal structure of its HAT domain.
MATERIALS AND METHODS
Overexpression and Purification.
The DNA sequence encoding the HAT domain of yGCN5 (residues 99–262) was amplified by PCR as an AseI/HindIII fragment and was subcloned into an NdeI/HindIII-digested pRSET vector (Invitrogen) for overexpression. The plasmid was transformed into Escherichia coli strain BL21(DE3) and was overexpressed by induction with 0.5 mM isopropyl β-d-thiogalactoside and growth at 15°C. The protein was purified with sequential use of SP-Sepharose (Amersham Pharmacia) cation-exchange, CoA agarose (Sigma), and Superdex 75 (Amersham Pharmacia) gel filtration chromatographies. Purified protein was concentrated to 12 mg/ml by using a Centriprep 10 (Amicon) concentrator and was judged to be >99% pure by SDS/PAGE.
Crystallization and Data Collection.
Crystals of yGCN5 were obtained with a protein concentration of 5 mg/ml at 4°C by using the hanging drop method containing a reservoir solution of 400 mM (NH4)2SO4 and 20–25% polyethylene glycol 8,000. Crystals appeared in 2–3 days, grew to average dimensions of 100 × 200 × 300 μM, and were harvested in 300 mM (NH4)2SO4, 25% polyethylene glycol 8,000, and 25% glycerol before flash freezing in propane for data collection. Diffraction data were collected from a single crystal at −180°C by using an Rigaku (Tokyo) Raxis IV image plate detector with Cu Kα radiation [focused with Molecular Structure (Houston)/Yale mirrors] produced from a Rigaku (Tokyo) RU-300 generator and were processed and scaled by using denzo and scalepack (40) (Table 1).
Table 1.
Crystallographic data and refinement statistics
| Crystal parameters | Data collection statistics | ||
| Unit cell dimensions | Resolution range | 50.0–1.9 Å | |
| a = 40.04 Å, b = 66.51 Å, c = 80.19 Å | Total reflections | 181,066 | |
| α = 90.00°, β = 97.71°, γ = 90.00° | Unique reflections | 31,832 | |
| Rsym | 3.0% | ||
| Space group | P2 | I/sigma(I) | 23.6 |
| Asymmetric unit | Two molecules | Completeness | 96.4% |
| Refinement statistics | |||
| Resolution range | 50.0–1.9 Å | rms values | |
| Reflections (Fo > 2σ) | 31,686 | Bond length, Å | 0.007 |
| Final model | Bond angles, ° | 1.25 | |
| Protein atoms | 2,702 | NCS molecules, Å | 0.89 |
| Waters atoms | 216 | B-factors, Å | 1.40 |
| Glycerol atoms | 12 | Average B-factors, Å2 | |
| R-factors* | Protein | 16.5 | |
| Rworking | 19.5% | Water | 21.8 |
| Rfree | 23.6% | Glycerol | 23.5 |
R-factor: Rworking = Σ| |Fo| − |Fc| |/Σ|Fo|; Rfree = ΣT| |Fo| − |Fc| |/ΣT|Fo|, where T is a test data set of 10% of the total reflections randomly chosen and set aside before refinement.
Structural Refinement.
The structure of yGCN5 was solved by molecular replacement using the program amore (41), with a partially refined model of the Tetrahymena thermophilia GCN5 HAT domain (in which all nonconserved side chains were alanized) as the search model (J.R.R, R.C.T., J.Z., Y. Mo, X. Li, S.L.B., C.D.A., and R.M., unpublished work). Rotation and translation searches produced two solutions in the asymmetric unit with a correlation coefficient of 45% and an R-factor of 48%. The two molecules were refined with x-plor (42) from 8.0 Å to 3.0 Å with rigid body and positional refinement and strict noncrystallographic symmetry constraints between the two HAT domains in the asymmetric unit. The program o (43) was used to place additional side-chains and to make model adjustments by using sigmaA-weighted 2|Fo| − |Fc| and |Fo| − |Fc| difference Fourier maps. Subsequent rounds of positional, simulated annealing (44), bulk solvent correction (45), and atomic B-factor refinement in x-plor followed by model building in o permitted the resolution range to be extended from 50.0 Å to 1.9 Å while gradually releasing the noncrystallographic symmetry restraints until they were removed entirely. Toward the end of refinement, water and two glycerol molecules were modeled into strong |Fo| − |Fc| peaks, which were refined to make feasible hydrogen bonds and have low atomic B-factors. The quality of the final model was verified with simulated annealing omit maps (46) <10% of the model at a time. The final model has excellent geometry (Table 1), with none of the nonglycine residues lying in disallowed regions of the Ramachandran plot (47).
GCN5 Mutant Study.
A single amino acid substitution, changing Glu to Gln (codon GAA to CAA), was made at residue 173 of full-length yGCN5 in pRSETA-yGCN5 (13) by using the QuikChange mutagenesis kit (Stratagene). Subcloning, overexpression, and purification of the E173Q yGCN5 HAT domain (residues 99–262) then was carried out essentially as described herein for the native yGCN5 HAT domain. The full-length mutant gene (GCN5 E173Q) was amplified by PCR and was subcloned as an XbaI/BglII fragment into yeast low-copy plasmid pPC87 (36) for expression from the constitutive ADH1 promoter. Transformants of GCN5-deleted strain SB303 [a trp1Δ derivative of FY1370 (48); MAT α gcn5Δ∷HIS3 his3Δ200 leu2Δ1 trp1Δ ura3–52] containing pPC87 empty vector, pPC87-yGNC5 (36), or pPC87-yGCN5 E173Q were tested for growth in synthetic dextrose minimal media in liquid cultures and on plates. Liquid cultures were incubated with shaking at 30°C, and log-phase growth was measured spectrophotometrically at 1.5-hr intervals. Streaked plates were incubated at 30°C for 2.75 days.
For in vivo transcription assays, the cells described above were additionally transformed with low copy GAL4-VP16FA activator plasmid (49) and pLGSD5 β-galactosidase reporter plasmid (50) or with low copy LexA-GCN4 activator plasmid (51) and LexA-8x reporter (13). These cells were grown to an optical density of 0.8 at 600 nm, and extracts were prepared and assayed for β-galactosidase activity and protein concentration as described (52).
HAT assays on purified yGCN5 and yGCN5 E173Q HAT domain proteins were performed at pH 7.5 essentially as described (33). Assays were performed in duplicate at pH 7.5 with 0.1 μg of GCN5 protein, and reactions were carried out for 30 minutes at 25°C.
RESULTS AND DISCUSSION
Overall Structure of yGCN5.
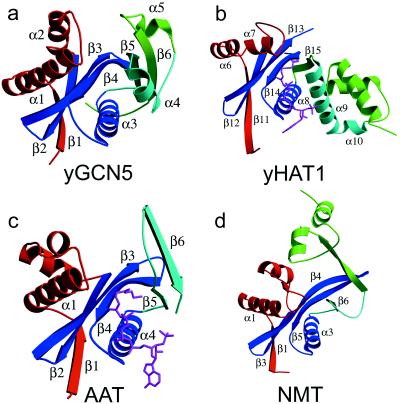
The protein structure has a mixed α/β topology containing five α-helices and six β-strands with a globular fold and approximate dimensions of 43 × 39 × 29 Å (Fig. 1a). The overall topology of the protein is shaped like a vise with the core of the protein lying at the base. The core is formed by two subregions: a three-stranded antiparallel β-sheet (β-strands 2, 3, and 4) that sits on top of an amphipathic helix (α3) with its hydrophobic side facing the sheets and its hydrophilic side facing solvent, and a strand-loop-helix substructure (β5-loop-α4) just adjacent to the β-sheet rich subdomain. The N- and C-terminal ends of the protein constitute the sides of the vise. A β-strand (β1) from the N terminus contributes to the β-sheet network of the core by hydrogen bonding to β2, and a helix-turn helix substructure (α1-turn-α2) sits above one side of the molecule above the core. The C terminus forms the opposing side of the vise and contains a loop-helix-loop-strand substructure (loop-α5-loop-β6). The β6-strand of the C terminus is associated with the core via hydrogen bonds with the β5-strand. A pronounced cleft of approximate dimensions of 10 × 10 × 20 Å is positioned above the core of the protein and flanked by the N- and C-terminal protein segments.
Figure 1.
Structure of yGCN5 and related enzymes. (a) Structure of the yGCN5 HAT domain. The four subdomains of the protein are color-coded; the structurally conserved subdomain that makes up the core (motifs A and D) is colored blue, motif B is colored aqua, and the N-terminal and C-terminal flanking regions are colored red and green, respectively. (b) Structure of residues 130–320 (C terminus) of HAT1 with the same orientation and color coding as in a. The AcCoA cofactor is shown in magenta. (c) Structure of SmAAT with the same orientation and color coding as in a. The CoA cofactor is shown in magenta. (d) Structure of residues 80–260 (N terminus) of NMT with the same orientation and color coding as in a.
Comparison to Other N-Acetyltransferases Reveals a Structurally Conserved Core Domain.
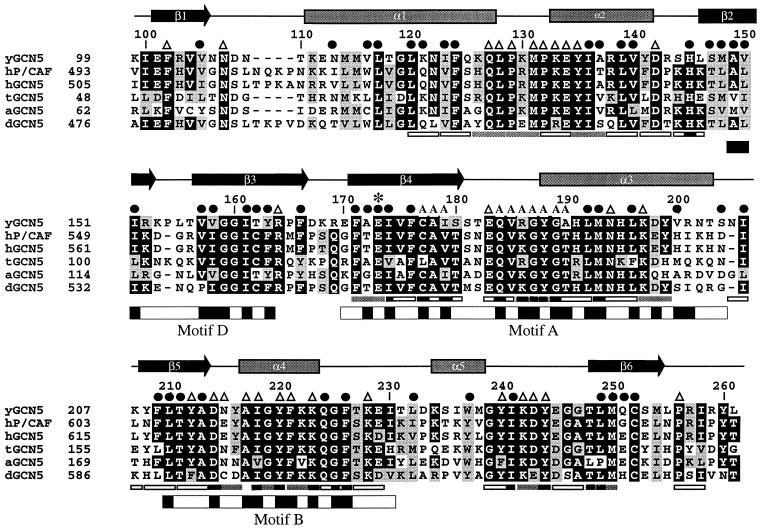
Neuwald and Landsman have recently noted that GCN5 belongs to a functionally diverse subfamily of N-acetyltransferases (called GNATs) containing limited sequence homology within four 15–33 amino acid segments called motifs A, B, C, and D (GCN5, P/CAF, and HAT1 do not harbor statistically significant sequence homology within motif C) (39) (Fig. 2). The recently published crystal structures of the Saccharomyces cerevisiae histone acetyltransferase 1 (HAT1) bound to AcCoA (53) and the Serratia marcescens aminoglycoside 3-N-acetyltransferase (SmAAT) bound to CoA (54) reveal that motifs A, B, and D form a structural core in which the CoA cofactor is bound between the A-D and B submotifs (55) (Fig. 1 b and c). Not surprisingly, motifs A and D show a high degree of structural homology with yGCN5 (rms deviations between Cα atoms for yGCN5 compared with HAT1 and SmAAT of 1.8 Å and 2.0 Å, respectively). Interestingly, the recently published structure of N-myristoyl transferase (NMT) (56, 57), which uses a myristoyl-CoA cofactor to modify the N-terminal glycine of substrate proteins, also shows homology in the region corresponding to motifs A and D of the core domain of HAT1, SmAAT, and yGCN5, even though the NMT protein shows no sequence homology with these N-acetyltransferases (55). The very recently published structure of serotonin N-acetyltransferase (AANAT), another member of the GNAT superfamily, also shows a structurally homologous A and D motif (58). Suprisingly, motif B does not show structural homology between the yGCN5, HAT1, and SmAAT proteins (Fig. 1). In the case of HAT1, this region forms an extended helix (α9) whereas, in the SmAAT structure, this region forms a β-hairpin (β5-turn-β6). Motif B in yGCN5 is formed by a β5–turn-α4 substructure within the core. Interestingly, HAT1 contains a similar strand-turn-helix topology (β15-turn-α9 in HAT1) in a similar geometrical position in the structure, although the HAT1 and yGCN5 structures show relatively poor sequence homology in this region.
Figure 2.
Sequence alignment of the GCN5 family of HAT domains. The primary sequence of the yeast GCN5 (yGCN5) HAT domain used for the structure determination is shown at the top of the alignment. Sequences from the homologous HAT domains from GCN5 of Arabidopsis, Drosophila, human, and Tetrahymena as well as with human P/CAF are aligned [clustal program (http://www-igbmc.u-strasbg.fr/BioInfo/ClustalW/)] and displayed [boxshade program (http://www.ch.embnet.org/software/BOX_form.html)]. Black and gray backgrounds are used to indicate identical and/or conserved residues found in at least 50% of the proteins at a given position, respectively. Secondary structural elements within the HAT domain of yGCN5 are shown above the sequence alignment. ●, residues that are buried within the core of the protein; A, residues that are implicated from the HAT1 and SmAAT structures to contact the CoA cofactor (53); ▵, solvent exposed residues that are presumably accessible for coenzyme or substrate binding and catalysis; ∗, the position of the putative general base (Glu173) for histone acetylation. Positions of alanine mutations that decrease HAT activity are indicated below the sequence alignment: Triple mutations are indicated with gray bars (38), and single mutations are indicated with black bars (37). Triple alanine mutations that have negligible effect on HAT activity are indicated with open rectangles (all of the single mutations are contained within the triple mutations). The GNAT conserved motifs D, A, and B (39) are indicated below the sequence alignment, with black shading indicating regions of high sequence homology within the GNAT superfamily.
The Core Domain of yGCN5 Binds CoA.
The structural homology within the A and D motifs of HAT1, SmAAT, and yGCN5 and the intimate involvement of the A motif in AcCoA binding by HAT1 and SmAAT implies that yGCN5 binds its AcCoA cofactor in a manner similar to that of HAT1 and SmAAT (Figs. 1 b and c and 2). This is also consistent with the recently derived structure of the human P/CAF HAT domain bound to coenzyme-A (60). The involvement of the B motif in AcCoA binding in HAT1 and SmAAT, coupled to its structural variability between yGCN5, HAT1, and SmAAT, suggests that it may play a role in correctly orienting AcCoA for substrate specific catalysis and/or may play a direct role in substrate specific recognition.
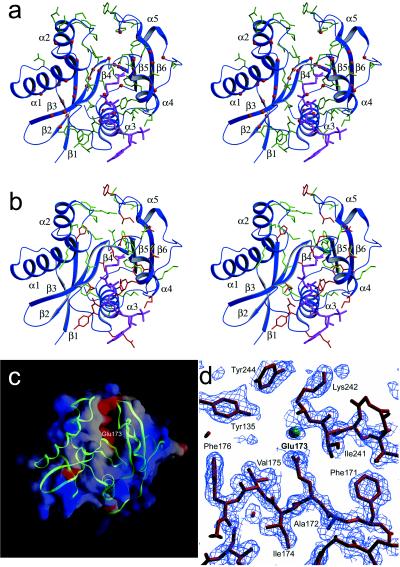
A mapping of residues that are conserved within the GCN5 subfamily of acetyltransferases (Fig. 2) onto the yGCN5 structure reveals that large patches of conservation within the core region (motifs A, D, and B) colocalize to residues that appear either to be involved in stabilizing the hydrophobic interior of the protein or, implicated by the HAT1 and SmAAT structures, to be associated with CoA binding (Fig. 3a). Within motif D, β2 residues Ala149, Val150, Ile151, and β3 residues Val157, Val158, Ile161, Thr162, Tyr163, and Phe166 are highly conserved and are buried in the protein interior, suggesting that these residues play an important role in protein stability (Fig. 2). Within motif A, conserved residues Phe171, Glu173, Ile174, Phe176, Leu192, Met193, and Leu196 also are buried and are therefore also likely to play a role in protein stability. Motif A also bears conserved residues implicated for AcCoA binding, including Ile174, Cys177, Ala178, Ile179, Gln184, Val185, Arg186, Gly187, Tyr188, Gly189, and Ala 190. The majority of conserved residues in motif B (residues 209–214 and 217–226) are solvent-exposed and are thus implicated in playing a role in CoA binding and/or histone substrate binding. Only conserved residues Phe209, Leu210, Ala213, and Phe226 in motif B are buried in the yGCN5 structure and are therefore likely to play a role in protein stability. A mapping of acetylation defective mutations within the core region of yGCN5 underscores the importance of the core region for acetyltransferase function (37, 38). Specifically, the majority of these mutations correlate with residues that the structure suggests are important for protein stability or AcCoA binding (Figs. 2 and 3b). It is striking that, in the context of the Ada and SAGA complexes, the most debilitating mutations in motifs A and B for nucleosomal acetylation lie in the proposed contact points for AcCoA.
Figure 3.
Implications of the yGCN5 structure for HAT function. (a) Strictly conserved residues within the GCN5 subfamily of acetyltransferases (Fig. 2) that are buried and thus play a role in protein stability are indicated as red balls, and strictly conserved and solvent exposed yGCN5 residues (Fig. 2) (implicated for histone substrate or CoA cofactor binding or catalysis) are shown as green side-chains. AcCoA from the HAT1 structure is placed in the cofactor binding site for reference by superposing the HAT1 core domain. Secondary structural elements are indicated for reference. (b) Mutations in yGCN5 that decrease HAT activity are mapped onto a schematic representation of the yGCN5 HAT domain. Triple mutations are shown as green side chains, and single mutations are shown as red side chains. (c) Electrostatic surface of the yGCN5 HAT domain. Red indicates acidic regions, and blue indicates basic regions. A backbone trace of the protein is superimposed in green. (d) SigmaA-weighted |Fo| − |Fc| omit map around the region of residue Glu173, which is implicated to function as a catalytic base for substrate catalysis. The map was generated by omitting residues within an 8-Å radius of Glu 173 followed by simulated annealing dynamics refinement at a temperature of 1,000 K. The map is contoured at 1.8 sigma. The green sphere indicates a water molecule.
The colocalization of the putative AcCoA binding site to the core region of yGCN5 suggests that AcCoA, in addition to playing a catalytic role in acetylation activity, also may play a structural role in protein stability. This is consistent with the finding that both human GCN5 and P/CAF are stabilized in vitro by the presence of AcCoA or CoA cofactors (59) and by the observation the crystals of HAT1 (53)and SmAAT (54) could only be obtained in the presence AcCoA and CoA cofactors, respectively.
The N- and C-Terminal Segments of yGCN5 Are Implicated for Histone Substrate Binding.
Regions N- and C-terminal to the yGCN5 core region show no sequence homology with other acetyltransferases. Despite this, yGCN5 shows structural homology with the HAT1, SmAAT, and NMT enzymes in the N-terminal β1-turn-α1-loop (Fig. 1). In each protein, the β1-strand forms sheet interactions with the protein core, and, in yGCN5, the α1-loop region is positioned along one side of a pronounced cleft above the protein core and close to the putative active site center (Fig. 1). Interestingly, the β1-turn-α1 component of this region corresponds to the C-motif noted by Neuwald and Landsman to show sequence homology within a subset of N-acetyltransferases (39). Taken together, the structural homology in the α1-loop region of yGCN5 implies that this region may play a general role in histone H3 and H2B substrate recognition and/or catalysis. Consistent with this hypothesis, residues in the α1-loop of yGCN5 are highly conserved within the GCN5 subfamily of acetyltransferases, and mutations in this region significantly decrease acetylation activity (38) (Fig. 2). Specifically, residues Gln127, Leu128, Pro129, and Met131 are both strictly conserved and mutationally sensitive. These residues are also solvent-exposed in the yGCN5 structure and would therefore be available for interaction with histone substrate.
Regions C-terminal to the core of yGCN5 are the most structurally divergent from the HAT1, SmAAT, and NMT enzymes (Fig. 1). In this region of the yGCN5 structure, a highly conserved patch of residues (GYIKDY in residues 239–244) in the loop after the α5 helix and lying along the side of the protein cleft opposite to the α1-loop region are accessible for interaction with histone substrate. The importance of this region of the protein is also consistent with its sensitivity to mutation (37, 38). Specifically, a mutation of KDY to AAA in positions 242–244 (in the loop proceeding the α5 helix) results in a dramatic decrease in the acetylation and transcriptional activation properties of yGCN5. The apparent importance of the loop after the α5 helix in yGCN5 coupled with its lack of structural conservation with the HAT1, SmAAT, and NMT acetyltransferaseas implicates this region playing an important role in substrate-specific binding and/or catalysis. Alternatively, and not mutually exclusive, this region may play a role in protein binding, as many acetyltransferase enzymes are known to function within the context of distinct multiprotein complexes (16–18, 34).
The proximity of the N-terminal α1-loop and the C-terminal loop after the α5 helix to opposite sides of the pronounced cleft above the core of the yGCN5 HAT domain and the accessibility of highly conserved (within the GCN5 family) and mutationally sensitive residues that face into the cleft of the structure strongly implicates this region for histone substrate binding (Fig. 2). Significantly, these regions harbor several of the most mutationally sensitive residues (Fig. 2) (37, 38) and have no apparent sequence homology to other acetyltransferases. Consistent with the importance of this cleft region is the putative positioning of the acetate group of the AcCoA cofactor at the base of the cleft in close juxtaposition to where substrate would bind (Fig. 3b) and the highly acidic patch at the base of the cleft (Fig. 3c), which would provide an attractive docking site for the lysine substrate. Comparable acidic patches also have been noted for the putative substrate binding sites for SmAAT (54) and HAT1 (53), and, as in the yGCN5 structure, both patches lie above the protein core domain.
Conservation (AIGYFKKQGF in residues 217–226) (Fig. 2) and mutational sensitivity (Tyr212, Tyr220, Lys223) (37, 38) of solvent-accessible residues around the α4 helix suggests that regions proximal to helix α4 also may be associated with histone substrate contact (Fig. 3 a and b). Moreover, the close proximity of this region to the putative AcCoA binding site suggests that the cofactor itself also may play a structural role in histone substrate binding. Taken together, the data described above are consistent with a model whereby the peptide substrate is bound across the α4-CoA surface and into the cleft above the yGCN5 core and flanked by the N- and C-terminal protein segments.
Glutamate-173 Within the yGCN5 Core Domain Plays an Important Catalytic Role.
Inspection of the putative histone substrate binding cleft of yGCN5 reveals two candidate residues that are in proximity to function as a general base for catalysis: Glu173 within β4 and Asp214 in the loop following β5. Although both residues are strictly conserved within the GCN5 subfamily of HAT enzymes, mutational analysis favors the involvement of Glu173. Specifically, mutation of FAE to AAA in positions 171–173 of yGCN5 was found to be one of the most debilitating mutations within the HAT domain (38) (Fig. 2). In contrast, mutation of Asp214 is only marginally compromised in both transcriptional activation in vivo and histone acetylation in vitro (37). Modeling of the AcCoA cofactor derived from the HAT1 structure into its putative binding site in yGCN5 would place a side chain oxygen of Glu173 (moved to an alternate side chain rotomer) ≈7 Å from the reactive carbonyl carbon of AcCoA, well within range to facilitate proton abstraction from the ɛ amino group of the lysine substrate. Strikingly, a superposition of the yGCN5 core domain with the corresponding core domains of HAT1 and SmAAT shows that Glu255 of HAT1 and Asp110 of SmAAT also may play analogous catalytic roles in these enzymes, respectively. Correlatively, Glu255 of HAT1 is strictly conserved across different species (comparable SmAAT homologues have not been characterized). Interestingly, the recently published structure of AANAT shows that that a histidine (His 122) residue lying in the same approximate position of Glu173 in the core domain of yGCN5 is likely to serve as a general base for catalysis by AANAT (58).
To directly test the importance of residue 173 for catalysis by yGCN5, we mutated it from a glutamate to a glutamine (yGCN5-E173Q) and analyzed its functional properties. Glutamine substitution was made because its sidechain is structurally similar to glutamate but lacks the acidic character required for it to function in a catalytic capacity (Fig. 3d). The overexpression levels, purification properties (including CoA affinity binding), and CD spectra of the yGCN5-E173Q protein were virtually identical to those of yGCN5, suggesting that the protein did not undergo significant structural changes attributable to the glutamate-to-glutamine substitution (data not shown).
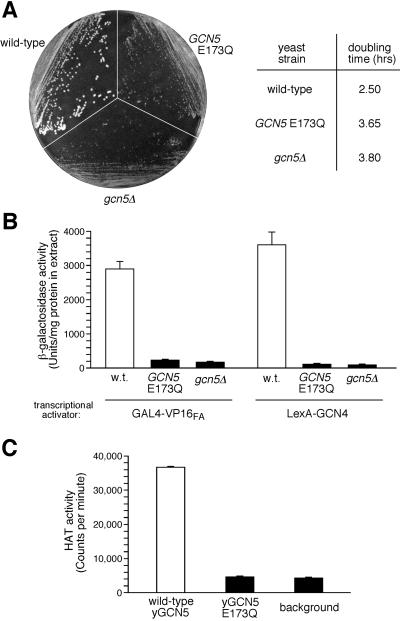
Functional analysis of yGCN5-E173Q demonstrates that glutamate 173 is critical for HAT activity in vitro and yGCN5 function in vivo (Fig. 4). Specifically, in vivo, the E173Q mutation results in a debilitated growth phenotype closely resembling that of gcn5Δ cells (Fig. 4A): Colony size is small on minimal media, and doubling time is ≈1.5× as long as for wild-type cells in liquid culture. Furthermore, yGCN5-E173Q and gcn5Δ showed similar low levels of in vivo transcription with two acidic activators, GAL4-VP16FA and LexA-GCN4 (Fig. 4B); activation of this type is known to directly involve yGCN5 and other transcriptional adaptors (12). In vitro analysis also showed that, although yGCN5 exhibited a significant level of HAT activity, the HAT activity of the E173Q yGCN5 mutant was near background levels (Fig. 4C). The involvement of Glu173 in catalysis by yGCN5 is also consistent with the recent enzymatic analysis of Tanner and coworkers (61). Taken together, the results presented here specifically implicate the carboxyl moiety of glutamate 173 in HAT catalysis by yGCN5, both in heterologously expressed protein in vitro and in its transcription-related function in yeast cells, such as the acetylation of chromatin in the context of the SAGA complex.
Figure 4.
Functional defects of the yGCN5 E173Q mutant and the putative role of Glu173 in catalysis. (A) Growth of yeast cells harboring empty vector (gcn5Δ), wild-type GCN5 plasmid, or GCN5 plasmid with a Glu-to-Gln mutation at residue 173 (E173Q). Strains were streaked on a minimal media plate (Left) or were grown in liquid minimal media at 30°C for determination of log-phase generation (doubling) times of the cells (Right). (B) Transcriptional activation in vivo. Shown are the activities of extracts from cells containing activator plasmid GAL4-VP16FA or LexA-GCN4 and the appropriate β-galactosidase reporter plasmid. (C) HAT activity assays in vitro. Purified yGCN5 and yGCN5-E173Q HAT domain proteins (0.1 μg) were used for liquid HAT assays with 3H-AcCoA and free histone substrates at pH 7.5 and 25°C.
CONCLUSION
The degree of structural conservation between yGCN5, HAT1 (53), SmAAT (54), and NMT (56, 57) suggests that other HAT enzymes, such as CBP and TAFII250, have a structurally homologous core domain associated with AcCoA binding. More significantly, a correlation of the yGCN5 structure with the extensive mutational information (37, 38) reveals the mode of catalysis and implies that nonconserved residues within the core and directly N- and C-terminal to the core are responsible for substrate specific binding. Taken together, the structure presented here provides a mechanistic framework from which to understand the structure/function relationships of the growing number of transcriptional regulators that function as histone acetyltransferase enzymes.
Acknowledgments
The authors thank Robyn Howard for preparing the yGCN5-E173Q HAT domain mutant, Adrienne Clements, Dan King, Xinmin Li, Yi Mo, and Ben Vaessen for useful discussions, and the Christianson and Van Duyne laboratories for technical assistance with data collection. This work was supported by National Institutes of Health grants (GM-52880 to R.M., GM-55360 to S.L.B., and GM-53512 to C.D.A.), a grant from the Fannie E. Rippel Foundation to R.M., a Howard Hughes predoctoral fellowship (70109-522202) to R.C.T., and a National Institutes of Health postdoctoral fellowship (GM-19211) to D.E.S.
ABBREVIATIONS
- yGCN5
yeast GCN5
- HAT
histone acetyltransferase
- HAT1
yeast histone acetyltransferase 1
- SmAAT
Serratia marcescens aminoglycoside 3-N-acetyltransferase
- GNAT
GCN5-related N-acetyltransferases
- NMT
N-myristoyl transferase.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1ygh).
A Commentary on this article begins on page 8807.
References
- 1.Struhl K. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- 2.Zawel L, Reinberg D. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 3.Wolffe A P. Chromatin: Structure and Function. London: Academic; 1992. [Google Scholar]
- 4.Owen-Hughes T, Workman J L. Crit Rev Eukaryotic Gene Expression. 1994;4:401–441. [PubMed] [Google Scholar]
- 5.Paranjape S M, Kamakaka R T, Kadonaga J T. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 6.Workman J L, Kingston R E. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 7.Kadonaga J T. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 8.Mizzen C A, Allis C D. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davie J R. Mol Biol Rep. 1997;24:197–207. doi: 10.1023/a:1006811817247. [DOI] [PubMed] [Google Scholar]
- 10.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe A P. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 12.Marcus G, Silverman N, Berger S, Horiuchi J, Guarente L. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candau R, Moore P A, Wang L, Barlev N, Ying C Y, Rosen C A, Berger S L. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 15.Smith E R, Belote J M, Schiltz R L, Yang X J, Moore P A, Berger S L, Nakatani Y, Allis C D. Nucleic Acids Res. 1998;26:2948–2954. doi: 10.1093/nar/26.12.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith E R, Eisen A, Gu W G, Sattah M, Pannuti A, Zhou J X, Cook R G, Lucchesi J C, Allis C D. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 18.Mizzen C A, Yang X-J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, et al. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto T, Horikoshi M. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 20.Chen H W, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 21.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J X, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, et al. Nature (London) 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 22.Betz R, Gray S G, Ekstrom C, Larsson C, Ekstrom T J. Genomics. 1998;52:245–246. doi: 10.1006/geno.1998.5435. [DOI] [PubMed] [Google Scholar]
- 23.Kadosh D, Struhl K. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y B, Honda A, Yoshida M, Horinouchi S. FEBS Lett. 1998;436:193–196. doi: 10.1016/s0014-5793(98)01124-7. [DOI] [PubMed] [Google Scholar]
- 25.Carmen A A, Rundlett S E, Grunstein M. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 26.Brosch G, Goralik-Schramel M, Loidl P. FEBS Lett. 1996;393:287–291. doi: 10.1016/0014-5793(96)00909-x. [DOI] [PubMed] [Google Scholar]
- 27.Brosch G, Lusser A, Goralik-Schramel M, Loidl P. FASEB J. 1997;11:A936–A936. [Google Scholar]
- 28.Lusser A, Brosch G, Loidl A, Haas H, Loidl P. Science. 1997;277:88–91. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]
- 29.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 30.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Proc Natl Acad Sci USA. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dangond F, Hafler D A, Tong J K, Randall J, Kojima R, Utku N, Gullans S R. Biochem Biophys Res Commun. 1998;242:648–652. doi: 10.1006/bbrc.1997.8033. [DOI] [PubMed] [Google Scholar]
- 32.Kuo M H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Nature (London) 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 33.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, et al. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 34.Grant P A, Sterner D E, Duggan L J, Workman J L, Berger S L. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis C D, Berger S. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Candau R, Zhou J, Allis C D, Berger S L. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo M H, Zhou J X, Jambeck P, Churchill M E A, Allis C D. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Liu L, Berger S L. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuwald A F, Landsman D. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S, editors. Warrington, U.K.: SERC Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 41.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 42.Brunger A T. X-PLOR 3.1: A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992a. [Google Scholar]
- 43.Jones T A, Zou J Y, Cowen S W. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 44.Brunger A T, Krukowski A. Acta Crystallogr A. 1990;46:585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- 45.Jiang J S, Brunger A T. J Mol Biol. 1994;243:100–115. doi: 10.1006/jmbi.1994.1633. [DOI] [PubMed] [Google Scholar]
- 46.Brunger A T, Kuriyan J, Karplus M. Science. 1987;235:458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 47.Kleywegt G J, Jones T A. Curr Biol. 1996b;4:1395–1400. doi: 10.1016/s0969-2126(96)00147-5. [DOI] [PubMed] [Google Scholar]
- 48.Roberts S M, Winston F. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 50.Guarente L, Yocum R R, Gifford P. Proc Natl Acad Sci USA. 1982;79:7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hope I A, Struhl K. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 52.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 53.Dutnall R N, Tafrov S T, Sternglanz R, Ramakrishnan V. Cell. 1998;94:427–438. doi: 10.1016/s0092-8674(00)81584-6. [DOI] [PubMed] [Google Scholar]
- 54.Wolf E, Vassilev A, Makino Y, Sali A, Nakatani Y, Burley S K. Cell. 1998;94:439–449. doi: 10.1016/s0092-8674(00)81585-8. [DOI] [PubMed] [Google Scholar]
- 55.Modis Y, Wierenga R. Structure (London) 1998;6:1345–1350. doi: 10.1016/s0969-2126(98)00134-8. [DOI] [PubMed] [Google Scholar]
- 56.Bhatnagar R S, Futterer K, Farazi T A, Korolev S, Murray C L, Jackson-Machelski E, Gokel G W, Gordon J I, Waksman G. Nat Struct Biol. 1998;5:1091–1097. doi: 10.1038/4202. [DOI] [PubMed] [Google Scholar]
- 57.Weston S A, Camble R, Colls J, Rosenbrock G, Taylor I, Egerton M, Tucker A D, Tunnicliffe A, Mistry A, Mancia F, et al. Nat Struct Biol. 1998;5:213–221. doi: 10.1038/nsb0398-213. [DOI] [PubMed] [Google Scholar]
- 58.Hickman A B, Klein D C, Dyda F. Mol Cell. 1999;3:23–32. doi: 10.1016/s1097-2765(00)80171-9. [DOI] [PubMed] [Google Scholar]
- 59.Herrera J E, Bergel M, Yang X J, Nakatani Y, Bustin M. J Biol Chem. 1997;272:27253–27258. doi: 10.1074/jbc.272.43.27253. [DOI] [PubMed] [Google Scholar]
- 60.Clements, A., Rojas, J. R., Trievel, R. C., Wang, L., Berger, S. L. & Marmorstein, R. (1999) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 61.Tanner, K. G., Trievel, R. C., Kuo, M. H., Howard, R., Berger, S. L., Allis, C. D., Marmorstein, R. & Denu, J. (1999) J. Biol. Chem., in press. [DOI] [PubMed]