Abstract
Most vertebrate mRNAs exit the nucleus with a 200+-residue poly(A) tail and are deadenylated to yield heterogeneous polymers of 50–200 adenosine residues on any given mRNA. We previously reported that Xenopus albumin mRNA and pre-mRNA have an unusually short, discrete 17-residue poly(A) tail and showed that regulation of poly(A) length is controlled independently by two cis-acting poly(A)-limiting elements (PLE A and PLE B) located in the terminal exon. The present study sought to determine the generality of this regulatory mechanism. Transferrin mRNA also has a discrete <20-nt poly(A) tail, and deletion mapping experiments identified an element homologous to the albumin gene PLE B within the terminal exon of the transferrin gene that conferred poly(A) length regulation on a globin reporter mRNA. Based on this similarity the PLE B sequence was used in a database search to identify candidate mRNA targets for regulated polyadenylation. Of the several hundred sequences identified in this manner we focused on HIV-EP2/Schnurri-2, a member of a family of genes encoding related zinc finger transcription factors. A striking feature of the PLE-like element in these genes is its location 10–33 bp upstream of the translation stop codon. We demonstrate that HIV-EP2 mRNA has a <20-nt poly(A) tail, for which the identified PLE-like sequence is responsible. These results indicate that the presence of a PLE can predict mRNAs with <20-nt poly(A) tails, and that nuclear regulation of poly(A) tail length is a feature of many mRNAs.
The 3′ processing of nuclear pre-mRNAs begins with the ordered assembly of cleavage and polyadenylation specificity factor, cleavage stimulatory factor, two cleavage factors (CF Im, CF IIm), and poly(A) polymerase on conserved sequence elements flanking the site of poly(A) addition (reviewed in refs. 1 and 2). Poly(A) is added onto the upstream fragment after cleavage by an as-yet-unidentified endonuclease. The process of poly(A) polymerization occurs in two steps: a slow, distributive addition of 10+ residues followed by the rapid, processive addition of 200–250 residues (3). Size limitation is imposed on the growing polymer by poly(A) binding protein II (PAB II) (4), which acts to limit the overall length of the newly synthesized chain to 200–250 residues (5).
A number of sequence elements have been found to affect the efficiency of pre-mRNA 3′ processing. Upstream sequence elements that increase the efficiency of pre-mRNA 3′ processing have been identified in adenovirus (6), simian virus 40 (7–9), HIV-1 proviral RNA (10, 11), and the C2 complement pre-mRNA (12). Inhibitory upstream sequence elements also have been identified in HIV proviral RNA, where the 5′ major splice site of HIV inhibits cleavage at the polyadenylation site within the 5′ long terminal repeat (13); bovine papilloma virus, where late gene expression also is repressed by the presence of an upstream 5′ splice site (14); and U1 small nuclear ribonucleoprotein A protein mRNA, where U1A binding to duplicated U1A binding sites upstream of AAUAAA inhibits poly(A) addition onto the cleaved mRNA (15).
Controlling the length of added poly(A) represents a third level of regulated pre-mRNA 3′ processing. Cytoplasmic control of poly(A) length plays a key role in activating or repressing gene expression during meiosis and early embryonic development where transcription is silent (reviewed in ref. 16). Although there are numerous examples of changes in poly(A) length of somatic mRNAs (reviewed in ref. 17), most of these either represent deadenylation linked to decay or increases in poly(A) length caused by unknown mechanisms. Several years ago we identified an unusually short, discrete 17-nt poly(A) tail on the mRNA encoding Xenopus serum albumin (18). By using a reverse transcription–PCR (RT-PCR) protocol (19) adapted to measure poly(A) length on pre-mRNA, we determined that this short, discrete poly(A) tail was present on albumin pre-mRNA before splicing of the terminal intron (20). Because other mRNAs in Xenopus liver had the expected long, heterogeneous poly(A) tails, this result indicated that albumin mRNA had a specific cis-acting element that regulated either poly(A) addition or rapid deadenylation before splicing. Two homologous poly(A)-limiting elements, or PLEs, subsequently were identified in the terminal exon of albumin pre-mRNA (21). The first element (PLE A) comprises the first 24 bp of exon 15, beginning 128 bp upstream of the cleavage site, and a second downstream element (PLE B) was identified in the sequence spanning 53–80 bp upstream of the cleavage site. A striking feature of these sequence elements was their ability to function in the context of the strong synthetic polyadenylation element SPA (22).
We wondered whether other mRNAs might be subject to the same type of regulated polyadenylation as albumin mRNA. A previous report showed that the mRNA for another highly abundant liver protein product, transferrin, also has a <20-nt poly(A) tail (23). In the first portion of this study we demonstrate that the <20-nt poly(A) tail on transferrin mRNA results from regulated polyadenylation of its nuclear pre-mRNA and identify a PLE related to the sequence of the albumin PLE B in the terminal exon of the transferrin gene. The conserved sequence then was used to search sequence databases to identify PLE-like elements in other genes. This search produced an unexpectedly large number of such genes. We focused here on the HIV-EP2/Schnurri-2 gene, which encodes a T cell-specific member of a family of zinc finger transcription factors that bind NF-κB sites in a number of enhancers. The PLE-like sequence in this gene begins 33 bp upstream of the translation stop codon and lies 1,675 bp upstream of the poly(A) addition site. Nevertheless, our results show that HIV-EP2 mRNA has a <20-nt poly(A) tail, and poly(A) length regulation is controlled by the identified PLE. These findings demonstrate that regulation of poly(A) tail length on nuclear pre-mRNAs is a more general phenomenon than previously appreciated.
MATERIALS AND METHODS
Plasmid Constructions.
The 3′ end of the transferrin gene (GenBank accession no. AF139169) was isolated from a genomic library that was prepared from DNA of a homozygous frog (a gift from Tom Sargent, National Institutes of Health). Portions of the transferrin gene were cloned into the expression vector CMV-glo-SPA (21) as follows. DNA primers 5′-TAAAGGTACCGCAGCACCACAAAATCTGG and 5′-CAGATCTAGAGCCAGGAGCCCTGTTTAGG were used to PCR-amplify the entire terminal intron and 13 bp of the 5′ and 3′ flanking exon sequence of the transferrin gene. 5′-TAAAGGTACCAGGGCTCCTGGCTTCCTGC and 5′-CAGATCTAGAACAAAAACAACAGAAGACTA were used to PCR-amplify all but the last 29 bp of the transferrin terminal exon. The numbering convention for primers used here corresponds to the nucleotide positions upstream of the poly(A) addition sites. For the 5′ deletions of the terminal exon (−122/−22 and −78/−22), 5′-TAACGGTACCTGGTAGATTGTCCTGTTTCC and 5′-TAACGGTACCTATTCTCAGATGTGGGAGGG were used as the upstream primers and 5′-CAGATCTAGAGGATAAACAAAAACAACAGAAGA was the downstream primer in the PCR amplification. Fragment −51/−22 was obtained by annealing primers 5′-TAAAGGTACCGTGTTAGTCTTCTGTTGTTTTTGTTTATCC and 5′-CAGATCTAGAGGATAAACAAAAACAACAGAAGA, and end-filling with Klenow fragment of DNA polymerase. For the 3′-deletions (−171/−52, −171/−92, and −171/−132), 5′-CAGATCTAGAAAATACCCCCTCCCACATCT, 5′-CAGATCTAGAGGTTATATGATGGAAACAGG, and 5′-CAGATCTAGATGTTCAGCAGCTGTGGAATG were used as the downstream primers, and 5′-TAAAGGTACCAGGGCTCCTGGCTTCCTGC was the upstream primer in the PCR amplification. All of the above DNA fragments were digested with KpnI and XbaI, gel-purified, and cloned into KpnI + XbaI digested CMV-glo-SPA. A plasmid bearing the transferrin PLE (see Fig. 2D, CMV-glo-tf PLE-SPA) was prepared by annealing the primer 5′- AGUUUAUUCUCAGAUGUGGGAGG with its complement bearing appropriate overhanging sequence for cloning into KpnI + XbaI-digested CMV-glo-SPA as described above.
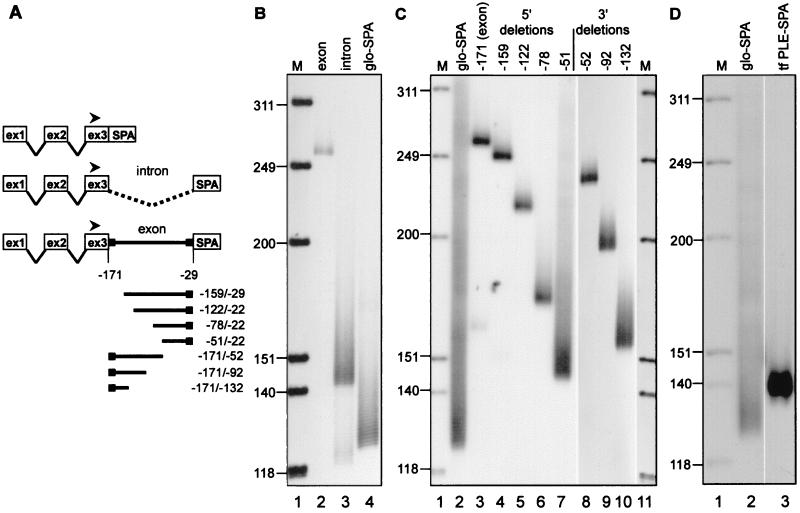
Figure 2.
Identification of the PLE in the transferrin gene. (A) The constructs that were transfected into LM(tk−) cells for analysis of poly(A) tail length on expressed mRNAs in B and C are diagrammed with the expression plasmid CMV-glo-SPA shown on top. The location of the radiolabeled globin exon 3 primer used for RT-PCR of all samples is indicated with a filled arrowhead. The numerical positions indicated on the diagram refer to the nucleotide locations within transferrin mRNA relative to the site of poly(A) addition. (B) Poly(A) tail lengths were determined for mRNAs expressed from constructs bearing the terminal transferrin exon (lane 2, exon), the terminal intron plus 13 bp of flanking exon sequence to retain functional splice sites (lane 3, intron), and vector control with no inserted transferrin DNA (lane 4, glo-SPA). M, molecular size marker HinfI-digested OX174 DNA. (C) Poly(A) tail lengths were determined for mRNAs expressed from constructs bearing nested 5′ and 3′ deletions of the terminal transferrin exon. The numbering at the top refers to the positions of the 5′ or 3′ ends relative to the site of poly(A) addition on transferrin mRNA. The poly(A) tail length of mRNA expressed from the vector control is shown in lane 2 (glo-SPA). M, molecular size marker HinfI-digested OX174 DNA. (D) Poly(A) tail lengths were determined for mRNA expressed from vector control (lane 2, glo-SPA) or vector carrying a fragment spanning nucleotides −82/−60 of transferrin mRNA inserted upstream of SPA (lane 3, tf PLE-SPA). M, molecular size marker HinfI-digested OX174 DNA.
The fragments EP99 and EP62 from the HIV-EP2 gene were obtained by PCR amplification of Jurkat cell DNA. The upstream primers consisted of 5′-AAAGGTACCAAAGATCCTTCATCAGAAAAG, which corresponds to position −1675/−1654, and 5′-AAAGGTACCCTATGATGCATGGAGACTTTC, which corresponds to position −1638/−1617, and the downstream primer was 5′-CCGTCGA- CAAAGAACACTCCTTAATGTA, which corresponds to position −34/−53. The resultant products were digested with Asp-718 and XbaI and inserted into similarly digested CMV-glo-SPA. To construct CMV-glo-EP/PLE-SPA the primers 5′-GTGGTACCAGAGGAAAGCAAAGATCCTTCATCAGAAAA and 5′-GGTCTAGATGTAGCTGACTCTTTTCTGATGAAGGATCT were annealed, end-filled with Klenow fragment of DNA polymerase, and digested with Asp-718 and XbaI for cloning into CMV-glo-SPA.
Manipulation of Cells.
LM(tk−) and Jurkat T cells were obtained from the American Type Culture Collection. Transient transfection, RNA extraction, and RT-PCR analysis of poly(A) tail length using a globin intron 2 primer (see Fig. 4C) or a globin exon 3 primer were performed as described (21). Jurkat cells were maintained in RPMI medium supplemented with 10% FCS. Induction of HIV-EP2 and IL-2 mRNA was done by treating 2 × 106 cells in 60-mm plates with 1 μg/ml of phytohemagglutinin and 50 ng/ml of phorbol-12-myristate-13-acetate (PMA). Nuclear RNA was extracted from cells harvested at the time points indicated in Fig. 3.
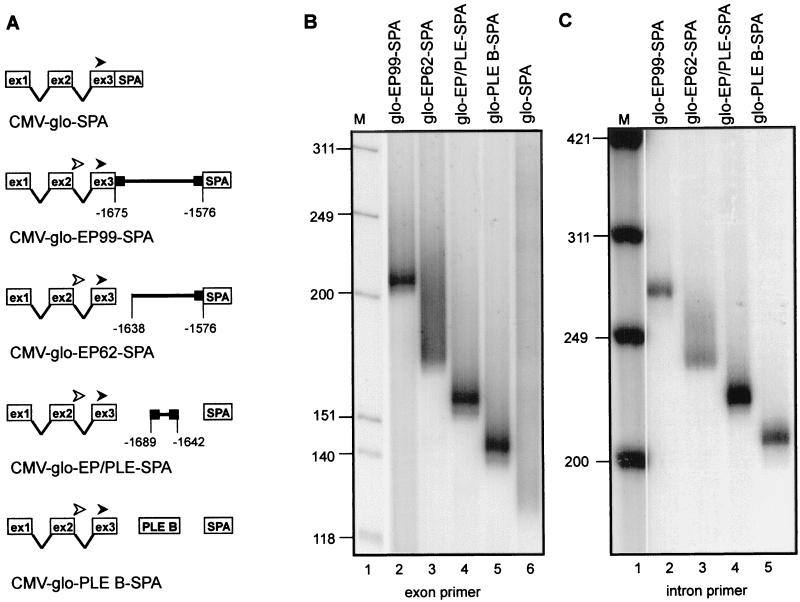
Figure 4.
Characterization of the PLE in the HIV-EP2 gene. (A) The constructs used for transfection into Jurkat T cells are diagrammed, with the location of each fragment with respect to the site of poly(A) addition on HIV-EP2 mRNA indicated numerically. The position of the globin exon 3 primer used in B is indicated with a filled arrowhead, and the position of the globin intron 2 primer used in C is indicated with an open arrowhead. (B) Poly(A) tail lengths were determined on the total population of each mRNA expressed in transfected Jurkat T cells by using a globin exon 3 primer for RT-PCR amplification. These constructs carried a 99-bp fragment of the HIV-EP2 gene with the PLE-like sequence located at the very 5′ end (glo-EP299-SPA, lane 2), a 62-bp fragment with the PLE-like sequence deleted (glo-EP62-SPA, lane 3), a 47-bp fragment spanning the PLE-like sequence (glo-EP/PLE-SPA, lane 4), the albumin gene PLE B (glo-PLE B-SPA, lane 5), or no inserted DNA (glo-SPA, lane 6). M, molecular size marker HinfI-digested OX174 DNA. (C) Poly(A) tail lengths were determined on pre-mRNAs present in the population analyzed in B by using a globin intron 2 primer for RT-PCR analysis rather than the exon 3 primer used in B. M, molecular size marker HinfI-digested OX174 DNA.
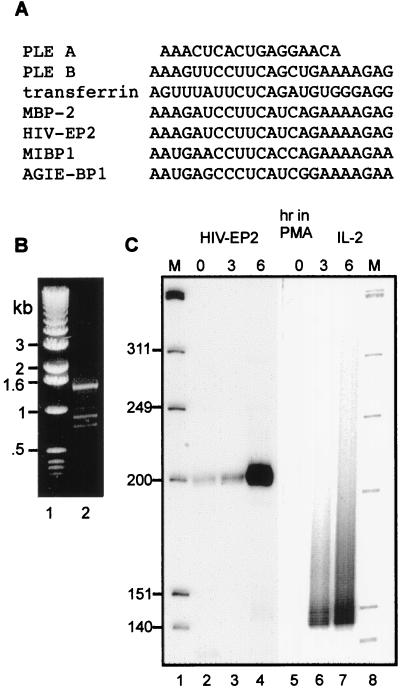
Figure 3.
Identification of PLE-like elements in other genes. (A) PLEs A and B of the albumin gene and the transferrin gene PLE are aligned with PLE-like sequences in mRNAs from several members of a family of related zinc finger transcription factors. See the text for the full names of each transcription factor. (B) PCR was performed on DNA extracted from Jurkat T cells to determine whether the PLE-like element identified in the HIV-EP2 gene is within the terminal exon. Primers were designed to encompass a 1,516-bp region of the cDNA sequence from the PLE-like element to a site 158 bp 5′ to the poly(A) addition site. The PCR products were separated on a 1% agarose gel and visualized by ethidium bromide staining. (C) Poly(A) tail length was determined for HIV-EP2 and IL-2 mRNAs in PMA-treated Jurkat T cells. Nuclear RNA was harvested from Jurkat T cells treated with phytohemagglutinin plus PMA for the indicated times. The products of the RT-PCR assay for poly(A) tail length were separated on a 6% polyacrylamide/urea gel and visualized by autoradiography. M, molecular size marker HinfI-digested OX174 DNA.
PCR Assays for Transferrin, HIV-EP2, and IL-2 mRNAs.
To determine whether the PLE-like element is in the terminal exon of the HIV-EP2 gene PCR was performed on DNA isolated from Jurkat cells by using primers 5′-AAAGATCCTTCATCAGAAAAGAGT (spanning −1674/−1650) and 5′-TTCGTGCACCCCTCCCTCTC (spanning −158/−178). The products of this reaction were analyzed by electrophoresis on a nondenaturing 1% agarose gel. The RT-PCR assay for poly(A) length on HIV-EP2 mRNA used the primer 5′-GAGAGGGAGGGGTGCAGCAA corresponding to nucleotides −178/−159, and the assay for IL-2 mRNA used the primer 5′-GTGCCCTAGGGGCTCTAAAATGG corresponding to nucleotides −123/−100 upstream of the poly(A) addition site. RT-PCR analysis of poly(A) length on transferrin pre-mRNA used the primer 5′-CATGTTGGTCTAGGAACAGC within the terminal intron corresponding to nucleotides −225/−205, and 5′-TTCCTGCACAATTCCACA, located within the terminal exon, corresponding to nucleotides −159/−143.
RESULTS
Identification of a PLE in the Transferrin Gene.
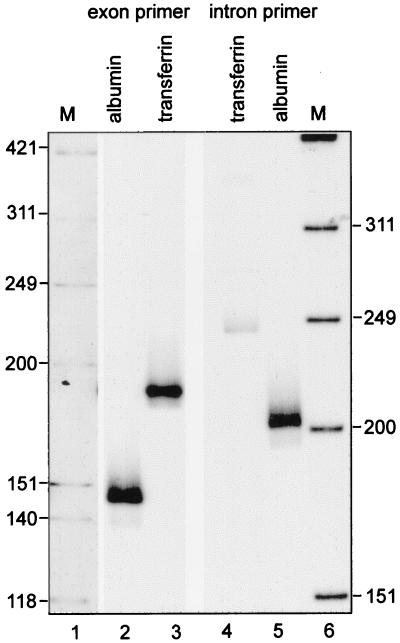
Previous work showed that, like albumin mRNA, transferrin mRNA has a <20-nt poly(A) tail (24), which made the transferrin gene a good target to search for sequence elements involved in regulating poly(A) tail length. Before embarking on this study we first confirmed that the general pathway regulating poly(A) tail length was similar for both genes. The <20-nt poly(A) tail on albumin pre-mRNA was determined by an RT-PCR assay for poly(A) tail length in which the upstream primer was located in the terminal intron of the albumin gene (20). The sequence of the terminal transferrin gene intron and the location of the boundary with the terminal exon were determined by a clone for the 3′ end of the transferrin gene (GenBank accession no. AF139169). Radiolabeled primers to the terminal intron and exon then were used for poly(A) length analysis by RT-PCR, with the resulting products analyzed by electrophoresis on high-resolution polyacrylamide/urea gels. Based on our previous work (21), a <20-nt poly(A) tail on transferrin mRNA would generate a discrete 183-bp product using a primer located 159 bp upstream of the cleavage site in the terminal exon (159 + 12 A + 12 bp of GC adapter sequence), and transferrin pre-mRNA would generate a 249-bp product using a primer located 225 bp upstream of the cleavage site in the terminal intron (225 + 12 A + 12 bp of GC adapter). The data in Fig. 1, lanes 3 and 4 confirm this result, indicating that transferrin transcripts undergo the same nuclear regulation of poly(A) tail length as albumin transcripts.
Figure 1.
Identification of a <20-nt poly(A) tail on transferrin pre-mRNA. Poly(A) tail lengths on transferrin mRNA and pre-mRNA were determined by RT-PCR poly(A) analysis of a preparation of total Xenopus liver RNA. RT was primed with an oligo(dT) primer-adapter as described (21), and poly(A) tail lengths on transferrin mRNA and pre-mRNA were determined by PCR amplification using 5′ 32P-labeled primers for the terminal exon (lane 3) or the terminal intron (lane 4), respectively. For comparison poly(A) length was determined on albumin mRNA (lane 2) and pre-mRNA (lane 5) (21). The products of the PCRs were visualized by autoradiography after electrophoresis on 6% polyacrylamide/urea gels. M, molecular size marker HinfI-digested OX174 DNA.
The location of the PLE in the 3′ end of the transferrin gene was determined in a manner similar to that used to identify the PLEs in albumin pre-mRNA (21). Portions of the transferrin gene were inserted as shown in Fig. 2A between a human β-globin gene truncated at the EcoRI site in exon 3 and the highly efficient 3′ processing element SPA (22), in the vector CMV-glo-SPA (21). The length of poly(A) on hybrid globin mRNAs extracted from nuclei of transfected murine fibroblasts was determined by RT-PCR using a primer for globin exon 3 (arrowhead in Fig. 2A). The results in Fig. 2B compare poly(A) tail lengths on mRNAs expressed from constructs bearing 152 bp of the terminal transferrin exon (lane 2) versus the terminal transferrin intron (lane 3). The portion of the terminal transferrin exon used here is complete except for the last 29 bp (containing AAUAAA) to insure that 3′ processing was strictly controlled by the SPA element. The discrete product seen in lane 2 indicates that the transferrin PLE is located within this 152 bp of exon sequence. The correctly spliced product from the plasmid bearing the terminal transferrin intron (lane 3) yielded mRNA with a heterogeneous poly(A) tail similar to that of the vector without inserted transferrin DNA (lane 4). Thus, like the albumin gene, the PLE of the transferrin gene is located in the terminal exon.
Mapping the PLE Within the Terminal Exon of the Transferrin Gene.
The deletion constructs shown in the bottom of Fig. 2A were used to map the location of the transferrin PLE within the terminal exon in Fig. 2C. Length regulation decreased when 93 bp were removed from the 5′ end of exon 15 (−78/−22) and was lost completely when 120 bp were removed (−51/−22). Deleting 52 bp from the 3′ end of the exon yielded mRNA with <20-nt poly(A) (−171/−52); however, deleting an additional 40 bp (−171/−92) yielded mRNA with heterogeneous poly(A). This defined a region of the exon containing a sequence shown in Fig. 3A resembling albumin PLE B. To test whether this sequence was indeed the transferrin PLE it was inserted into CMV-glo-SPA, and poly(A) tail length was determined on mRNA expressed in transfected cells. The results in Fig. 2D confirm that the identified element was sufficient to impart poly(A) length regulation on the expressed product, thus demonstrating that the length of the poly(A) tail on transferrin mRNA is regulated in the same manner as on albumin mRNA by an element homologous to the albumin PLE B in the terminal exon of the transferrin gene.
Identification of PLEs in Other Genes.
The high degree of sequence similarity between the transferrin PLE and albumin PLE B suggested that this motif might be conserved in other genes. A database search using the albumin PLE B as query sequence revealed hundreds of genes and expressed sequence tags from Caenorhabditis elegans to human bearing PLE-like sequence elements. A particularly high degree of homology was found to a sequence element present in genes for members of an extended family of zinc finger transcription factors. The products of these genes include AT-BP, which binds to the enhancer for α1-antitrypsin; MBP-2, which binds to the enhancer for class I MHC genes; MIBP1, which binds a 9-bp sequence in rat c-myc intron 1 and is homologous to PRDII (a protein that binds the positive regulatory domain of the human INF-β gene promoter); AGIE-BP1, which is a cytokine-induced transcription factor that binds to the angiotensinogen enhancer, and HIV-EP2/Schnurri 2, a T cell-specific human homolog of the Drosophila Schnurri-2 transcription factor that stimulates HIV-1 provirus transcription. The alignment of the PLEs from the albumin and transferrin genes with these PLE-like elements is shown in Fig. 3A. The mRNAs of this family of transcription factors are unusually long, ranging from 8 to 10 kb, and they have 3′ untranslated regions ranging in length from 1 to several kb. However, in all of the genes listed here, the PLE-like element resides within the coding region, spanning the sequence from 10 to 33 bp upstream of the translation stop codon.
We chose HIV-EP2/Schnurri-2 mRNA for further study to test whether the element identified within this gene acts as a PLE to regulate poly(A) tail length. As noted above, both the albumin and transferrin PLEs are located in the terminal exons of their respective genes, and recent experiments indicate the PLE must be in the terminal exon to regulate poly(A) tail length (J.D.G., unpublished work). Because the sequence of the HIV-EP2 gene is not available, we used PCR to determine whether the PLE-like sequence is in the terminal exon. PCR was performed on DNA extracted from Jurkat cells by using primers spanning a 1,516-bp region of the cDNA, beginning with the PLE-like element and ending 158 bp upstream of the site of poly(A) addition. A predominant PCR product of ≈1,500 bp was obtained (Fig. 3B), and there was no evidence of larger products indicative of intervening sequences. Therefore, the PLE-like sequence of the HIV-EP2 gene resides in a large terminal exon.
The experiment in Fig. 3C examined the length of poly(A) on HIV-EP2 mRNA. In this experiment IL-2 mRNA was used as a control for an mRNA with a long, heterogeneous poly(A) tail. Because these mRNAs are significantly induced by stimulation of Jurkat T cells with phytohemagglutinin (PHA) and PMA (25) both untreated and PHA+PMA-treated cells were examined to determine whether induction is accompanied by changes in poly(A) tail length. The results indicate that HIV-EP2 mRNA has a poly(A) tail <20 nt (lanes 2–4), whereas IL-2 mRNA has a 100- to 200-nt poly(A) tail (lanes 6 and 7). Both mRNAs were induced after 3 and 6 hr of PHA+PMA treatment and there was no observed effect of this treatment of poly(A) length on either mRNA.
Functional Analysis of the PLE in HIV-EP2 mRNA.
To determine whether the identified PLE-like sequence in the HIV-EP2 gene functions to regulate poly(A) tail length a 99-bp fragment beginning at the 5′ end of the element [position −1675 with respect to the poly(A) addition site] was inserted into CMV-glo-SPA to make the plasmid CMV-glo-EP99-SPA (Fig. 4A). This plasmid was transfected into Jurkat cells and the length of the poly(A) tail on expressed mRNA was determined by RT-PCR. The 99-bp element directed the addition of a discrete poly(A) tail of <20 nt (Fig. 4B, lane 2), indicating the presence of a PLE within this sequence. Deleting the PLE-like sequence in the plasmid CMV-glo-EP62-SPA yielded mRNA with a heterogeneous poly(A) tail (Fig. 4B, lane 3), supporting the identification of this PLE-like element in regulating poly(A) tail length. Final confirmation that the sequence identified in the HIV-EP2 gene is a PLE was accomplished by transfecting a construct carrying just the PLE-like element from the HIV-EP2 gene (CMV-glo-EP/PLE-SPA, Fig. 4B, lane 4). Both this plasmid and a plasmid carrying the albumin gene PLE B (CMV-glo-PLE B-SPA, Fig. 4B, lane 5) produced mRNAs with discrete <20-nt poly(A) tails, indicating that the homologous sequence identified in the HIV-EP2 gene is an authentic PLE. As noted above, the albumin gene PLEs regulate the length of poly(A) on intron-containing nuclear pre-mRNA. To confirm that the same phenomenon was at work here, RT-PCR assays were repeated by using an upstream primer located in globin intron 2. The results in Fig. 4C are identical to those in Fig. 4B, confirming that the HIV-EP2 PLE acts like the albumin gene PLEs in regulating the length of the poly(A) tail on nuclear pre-mRNAs.
DISCUSSION
It has been widely accepted that, with the exception of histone mRNAs, the 3′ processing of vertebrate mRNAs involves the polymerization of a 200+-residue poly(A) tail. Cytoplasmic deadenylation then yields a population of mRNAs bearing heterologous lengths of poly(A), perhaps through the general pathway of mRNA degradation, which begins with poly(A) shortening before the degradation of the body of the mRNA. The first hint that all mRNAs might not receive a 200+-residue poly(A) tail came from results showing that albumin mRNA extracted from Xenopus liver had a 17-nt poly(A) tail whether or not it was undergoing estrogen-induced destabilization (18). Subsequent work showed that transferrin mRNA had a similarly short poly(A) tail (24). The <20-nt poly(A) tail on albumin mRNA results from the presence of a cis-acting element, the PLE, in the terminal exon, which acts in an as-yet-unknown manner to either restrict the action of poly(A) polymerase or to effect rapid deadenylation of pre-mRNA before splicing of the terminal intron (21).
The purpose of the present study was to determine whether regulation of poly(A) tail length by the PLE was unique to albumin pre-mRNA, or whether it might represent a more general phenomenon. We began by searching for a PLE in the transferrin gene, because previous work had shown that transferrin mRNA has a <20-nt poly(A) tail. The results in Figs. 1 and 2 demonstrated that transferrin pre-mRNA undergoes the same type of regulation of poly(A) tail length as albumin pre-mRNA, because of the presence of a PLE similar in sequence to the albumin PLE B in the terminal exon. More surprising were the results from a database search for PLE-like sequences. Such sequences were found in hundreds of known and as-yet-unidentified genes. Our attention was drawn to one family of genes that encodes related zinc finger transcription factors because the cDNA sequences of each of these had a PLE-like element in the region 10–33 bp upstream of the translation stop codon (Fig. 3). It should be noted that the PLEs of the albumin and transferrin genes are downstream of the stop codon. The predictive value of this search was confirmed by the results in Figs. 3 and 4 showing that HIV-EP2 mRNA has a <20-nt poly(A) tail and a functional PLE within the terminal exon. This latter observation is significant, as all of the PLEs identified to date have been found in the terminal exon, and moving the PLE to different locations within the gene results in the production of mRNAs with heterogeneous poly(A) tails (J.D.G. and D.R.S., unpublished work). Our results draw into question the generally accepted view that all mRNAs exit the nucleus with a 200+-residue poly(A) tail. Instead, it appears that there are two categories of nuclear polyadenylated mRNAs: those that have a long, heterogeneous poly(A) tail, and those that have a discrete poly(A) tail of <20 residues.
Poly(A) length has functional implications for mRNA turnover, translation, and, perhaps, mRNA export. The role for deadenylation in mRNA decay has been noted above. A positive relationship between poly(A) tail length and translation efficiency has been demonstrated in vitro in translationally active yeast extract (26, 27) and rabbit reticulocyte lysate (28). These results complement findings in yeast (29–31) and mammalian cell systems (32) showing cap-dependent translation is stimulated by interaction of the poly(A) tail with the 5′ end of the mRNA through the binding of poly(A) binding protein (PABP) and eukaryotic initiation factor (eIF)-4E to eIF-4G. One PABP molecule binds to every 20–25 nt of poly(A) (33), so a typical mRNA with a 100- to 200-nt poly(A) tail will have 5–10 molecules of poly(A)-bound PABP. Although it has not been proven, the implication of these results is that the stimulation of translation initiation with increasing length of poly(A) results from the presence of multimers of bound PABP on the 3′ end of the mRNA. In spite of its mRNA having a <20-nt poly(A) tail albumin is readily translated to produce one of the most abundant hepatic protein products. This finding raises the fascinating possibility that the PLE, perhaps through interaction with a PLE-binding protein, might functionally replace the elongated poly(A) tail to stimulate translation initiation on mRNAs with short poly(A) tails.
Acknowledgments
We thank Tom Sargent for his gift of the Xenopus genomic library. This work was supported by National Institutes of Health Grant GM55407 from the National Institute of General Medical Sciences to D.R.S. and Center Grant P30 CA16058 from the National Cancer Institute to The Ohio State University Comprehensive Cancer Center.
ABBREVIATIONS
- PLE
poly(A)-limiting element
- PAB II
poly(A) binding protein II
- PABP
poly(A) binding protein
- RT-PCR
reverse transcription–PCR
- PMA
phorbol-12-myristate-13-acetate
Footnotes
Data deposition: The transferrin gene sequence reported in this paper has been deposited in the GenBank database (accession no. AF139169).
References
- 1.Colgan D F, Manley J L. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 2.Wahle E, Keller W. Trends Biochem Sci. 1996;21:247–250. [PubMed] [Google Scholar]
- 3.Sheets M D, Wickens M. Genes Dev. 1990;3:1401–1412. doi: 10.1101/gad.3.9.1401. [DOI] [PubMed] [Google Scholar]
- 4.Wahle E. Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- 5.Wahle E. J Biol Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 6.DeZazzo J D, Imperiale M J. Mol Cell Biol. 1989;9:4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutz C S, Alwine J C. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 8.Lutz C S, Murthy K G K, Schek N, O’Connor J P, Manley T L, Alwine J C. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 9.Schek N, Cooke C, Alwine J C. Mol Cell Biol. 1992;12:5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilmartin G M, Fleming E S, Oetjen J. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmartin G M, Fleming E S, Oetjen J, Graveley B R. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 12.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley J L, Proudfoot N J. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashe M P, Pearson L H, Proudfoot N J. EMBO J. 1997;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunderson S I, Polycarpou-Schwarz M, Mattaj I W. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 15.Boelens W C, Jansen E J R, Vanvenrooij W J, Stripecke R, Mattaj I W, Gunderson S I. Cell. 1993;72:881–892. doi: 10.1016/0092-8674(93)90577-d. [DOI] [PubMed] [Google Scholar]
- 16.Stebbins-Boaz B, Richter J D. Crit Rev Eukaryot Gene Exp. 1997;7:73–94. doi: 10.1615/critreveukargeneexpr.v7.i1-2.50. [DOI] [PubMed] [Google Scholar]
- 17.Baker E J. In: Control of Messenger RNA Stability. Belasco J, Brawerman G, editors. San Diego: Academic; 1993. pp. 367–415. [Google Scholar]
- 18.Schoenberg D R, Moskaitis J E, Smith J H, Jr, Pastori R L. Mol Endocrinol. 1989;3:805–814. doi: 10.1210/mend-3-5-805. [DOI] [PubMed] [Google Scholar]
- 19.Salles F J, Richards W G, Strickland S. Methods. 1999;17:38–45. doi: 10.1006/meth.1998.0705. [DOI] [PubMed] [Google Scholar]
- 20.Rao M N, Chernokalskaya E, Schoenberg D R. Nucleic Acids Res. 1996;24:4078–4083. doi: 10.1093/nar/24.20.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das Gupta J, Gu H, Chernokalskaya E, Gao X, Schoenberg D R. RNA. 1998;4:766–776. doi: 10.1017/s1355838298971837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levitt N, Briggs D, Gil A, Proudfoot N J. Genes Dev. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- 23.Pastori R L, Moskaitis J E, Buzek S W, Schoenberg D R. Mol Endocrinol. 1991;5:461–468. doi: 10.1210/mend-5-4-461. [DOI] [PubMed] [Google Scholar]
- 24.Pastori R L, Moskaitis J E, Buzek S W, Schoenberg D R. J Steroid Biochem Mol Biol. 1992;42:649–657. doi: 10.1016/0960-0760(92)90105-r. [DOI] [PubMed] [Google Scholar]
- 25.Nomura N, Zhao M J, Nagase T, Maekawa T, Ishizaki R, Tabata S, Ishii S. J Biol Chem. 1991;266:8590–8594. [PubMed] [Google Scholar]
- 26.Preiss T, Hentze M W. Nature (London) 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 27.Preiss T, Muckenthaler M, Hentze M W. RNA. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munroe D, Jacobson A. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wells S E, Hillner P E, Vale R D, Sachs A B. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 30.Tarun S Z, Jr, Wells S E, Deardorff J A, Sachs A B. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarun S Z, Jr, Sachs A B. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 32.Imataka H, Gradi A, Sonenberg N. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sachs A B. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]