Abstract
Aims: To evaluate a polymerase chain reaction (PCR) based assay to detect fungi in scrapings from infected corneas.
Methods: A PCR assay was developed to amplify a portion of the fungal 18S ribosome gene. Corneal scrapings from 30 patients with presumed infectious keratitis were evaluated using this assay, as well as by standard microbiological techniques, and the results were compared. Conjunctival swabs from each patient's healthy, fellow eye were also evaluated by PCR.
Results: PCR and fungal culture results matched (were both positive or both negative for fungi) in 22 (74%) of 30 scrapings from infected corneas. Three (10%) of 30 samples were PCR positive but fungal culture negative; two of these appeared clinically to represent fungal infections, and the third was clinically indeterminate. Four (13%) scrapings were positive by PCR but also by bacterial and not fungal culture. One specimen (3%) was PCR negative but fungal culture positive. Of the conjunctival swabs from each patient's healthy fellow eye, five (17%) of 30 were positive by PCR, and the opposite, infected eye of all five of these harboured a fungal infection.
Conclusions: PCR is promising as a means to diagnose fungal keratitis and offers some advantages over culture methods, including rapid analysis and the ability to analyse specimens far from where they are collected.
Keywords: molecular diagnostics, India, keratomycosis, field studies
Infectious keratitis causes extensive ocular morbidity worldwide. The true extent of visual impairment from this condition is thought to far exceed the recognised prevalence, particularly among agricultural workers in the developing world, where a “silent epidemic” of corneal blindness has been postulated.1 Fungal corneal infections in particular occur most frequently in individuals who work in agriculture.2,3 This condition is also associated with diabetes mellitus4 and the acquired immune deficiency syndrome (AIDS).5
Standard microbiological techniques for diagnosing microbial keratitis rely on culturing the organisms in nutrient media. The frequency of apparent diagnostic failure (that is, no organism is isolated though an infection is clinically evident) ranges from 20%6 to 60%.7 An additional problem is that such techniques require days to weeks for complete results, which can significantly delay appropriate treatment.
The potential utility of polymerase chain reaction (PCR) based techniques for improving the diagnosis of ocular infection is well recognised,8,9 and the use of PCR for this purpose is expanding.10–27 Assays targeting Candida, Aspergillus, and Fusarium have been tested preliminarily in vitreous specimens,28,29 and recently an assay using panfungal primers has been used more extensively in India.30 A PCR assay has also been applied to detect Fusarium in rabbit corneal infections.31
The current study aims to (1) use PCR to detect fungal DNA in corneal scrapings from patients clinically suspected to have fungal keratitis; (2) compare the diagnostic accuracy of PCR analysis and standard microbiological techniques for diagnosing keratomycosis; (3) evaluate the practicality of PCR as a diagnostic method in field or epidemiological studies of ocular surface infections.
MATERIALS AND METHODS
Clinical specimen collection and processing
Patients who presented to the LV Prasad Eye Institute (LVPEI) in Hyderabad, India, with eye findings suspicious for fungal keratitis, were eligible to contribute cornea samples for this study. Thirty such patients submitted samples between March and June 2000. Patients with bilateral disease were excluded. No patients acknowledged using antibiotic or antifungal eye drops before presentation. Scrapings from an affected area of each infected cornea were obtained with a flame sterilised platinum spatula, and were streaked onto an agar plate. The platinum spatula was then rinsed in 250 μl of 1X magnesium free PCR buffer (Promega, Madison, WI, USA) in a 1.5 ml vial, flame sterilised, cooled, passed again across the cornea, streaked across another agar plate, rinsed again in the same 1.5 ml vial, and again sterilised and cooled. This process was repeated for each of three agar plates and one nutrient broth vial used as part of the LVPEI standard microbiological investigation. The 1.5 ml vial was stored at −70°C for up to 3 months until being sent to the Yale Eye Center for PCR analysis. Specimens were stored at ambient temperature during transport. Upon arrival, specimens were frozen at −20°C for up to 4 weeks until testing.
For comparison purposes, a specimen was obtained from the fellow eye of each patient by rubbing a nylon swab along the inferior conjunctival fornix after instilling a topical anaesthetic. This swab was then immersed in a separate vial containing 250 μl of 1X PCR buffer. This vial was stored and transported to the Yale Eye Center together with that from the fellow infected eye.
Standard microbiological testing of scrapings from infected corneas at the LVPEI included fungal smears and solid and liquid media that support the growth of fungi, bacteria, and acanthamoeba. These techniques have been reported elsewhere.32–35 Briefly, corneal scrapings were plated on blood, chocolate, potato dextrose, and Sabouraud's dextrose agar, and incubated at appropriate temperature and conditions for 7–14 days. Gram, Giemsa, 10% potassium hydroxide, and calcofluor white stained slides were also prepared and examined by light microscopy. Fungal isolates were considered positive if (1) growth was consistent with smear results, (2) a fungus was grown in two or more media in the absence of fungus on smears, or (3) a fungus was grown in at least one medium in the presence of fungus on smears.
All patients over the age of 18 years gave verbal consent for participation in this study, and parents gave consent for younger patients. This study was approved by the institutional review boards of the LV Prasad Eye Institute and Yale University School of Medicine. Corneal scrapings were transported to the United States with permission from the Centers for Disease Control.
PCR amplification strategy
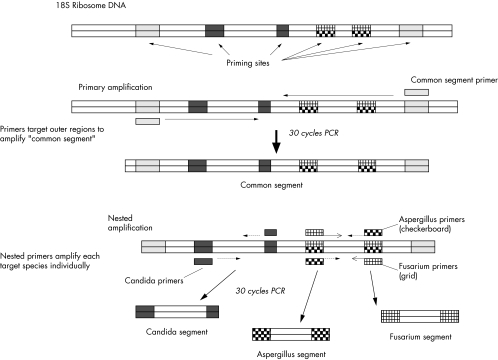
The PCR reaction utilises two rounds of DNA amplification. Primers for primary amplification were selected based on an alignment of published DNA sequences of Candida albicans (Genbank accession No AF 114470), Aspergillus fumigatus (AF 138288), and Fusarium oxysporum (AF 141951). Alignments and all primer selection were performed using the Lasergene sequence analysis software (Dnastar Inc, Madison, WI, USA). These primers amplify a portion of the 18S RNA gene that was found on alignment to be similar among these three target fungal species. This amplified segment is between 800 and 900 base pairs (bp) long for all three species, and is hereafter referred to as the “common segment” (see Fig 1). The sequence of these primers is shown in Table 1.
Figure 1.
Diagram of polymerase chain reaction amplification scheme.
Table 1.
Primer sequences
| Primer name | Sequence | Product length* |
| Primary amplification | ||
| Common segment upper | CAGGGGAGGTAGTGACAATAAATA | ∼870 bp† |
| Common segment lower | ACAAATCACTCCACCAACTAAGAA | |
| Nested amplification | ||
| Candida albicans upper | CAGCCGAGCCTTTCCTTCTGGT | 423 bp |
| Candida albicans lower | CCATACTCCCCCCAGAACCCAAAG | |
| Aspergillus fumigatus upper | TAGTCGGGGGCGTCAGTATTCAGC | 214 bp |
| Aspergillus fumigatus lower | GTAAGGTGCCGAGCGGGTCATCAT | |
| Fusarium oxysporum upper | GACAGTCGGGGGCATCAGTATTCAAT | 214 bp |
| Fusarium oxysporum lower | GTAAGGTGCCGAACGGGTCAAAAAAT |
*Predicted product length is based on an alignment of the published 18S RNA gene sequences of the fungal species used in primer design.
†Base pairs, actual number varies between species.
For the second round of PCR, each specimen was amplified with three different primer pairs separately. These nested primers pairs were chosen within variable regions in the common segment (see Fig 1). The nested primers for A fumigatus and F solani target the same variable region, and the C albicans nested primers target a separate DNA variable segment. Based on the aligned DNA sequences, the nested primers for each species exclude the other two (for example, A fumigatus nested primers do not match the C albicans or F solani DNA sequences, etc).
PCR optimisation
The PCR assay was optimised using dilute suspensions of fungal isolates in sterile, deionised water. Fungal isolates were obtained from the clinical laboratories of Yale New Haven Hospital and the LVPEI. The suspensions were overlaid with mineral oil and heated in thin walled PCR tubes for 20 minutes at 94°C to lyse the fungi. Reagent mixtures containing the appropriate primers were added after heat extraction of the sample DNA. The final reaction mixture contained 0.8 μM of each primer, 2.5 units of taq polymerase (Buffer B, Promega) 0.25 mM deoxyribonucleoside triphosphates (Boehringer Mannhein, Germany), 2.0 mM magnesium chloride (Promega), and 1X PCR buffer (Promega). For optimisation, PCR reactions were run in 50 μl volumes using thin walled 500 μl PCR tubes. Thermocycling was performed in a Stratagene Robocycler; 30 rounds were used for both primary and nested amplification. Reaction tubes were heated for 3 minutes at 94°C, followed by DNA melting for 30 seconds at 94°C, annealing for 40 seconds at 57°C, and extension for 1 minute at 72°C. The same annealing temperature was used for all primers.
For the second round of amplification, the amplified common segment was diluted 1/500 in sterile water, and 25 μl of this diluted product were used as template DNA for each of three nested reactions, each of which contained a species directed primer pair. The reaction product was resolved by electrophoresis using 2% agarose gels containing ethidium bromide, 0.375 μg/ml. A PCR result was considered positive if a DNA band of the predicted length for the primers used was present.
Determining the sensitivity of the PCR assay
The lower limit of detection of the PCR assay using 30 rounds each of primary and nested amplification was determined using serial dilutions of quantified fungal suspensions. Fungal suspensions were quantified microscopically using a haemocytometer. When quantifying filamentous fungi, any hyphal element, conidiospore, or particle thereof was counted as a fungal element.
Determining the specificity of the PCR assay
Each set of primer pairs was used to assay the following fungal species: C albicans; C parapsilosus; Torulopsis glabrata; C krusei; C tropicalis; A fumigatus; A flavus; A niger; A versicolor; F solani; Corvularia species (spp); Alternaria spp; Penicillium spp; Cladosporium spp; Saccharomyces cerevesiae, and Cryptococcus spp. The PCR assay was also tested using reference strains of Staphylococcus aureus, group B Streptococcus, Enterococcus faecalis, Klebsiella spp, Pseudomonas aeruginosa, Bacillus cereus, and Escherichia coli, and clinical isolates of Nocardia spp.
Clinical specimen analysis
In preparation for PCR analysis, specimens from LVPEI were thawed at room temperature, and centrifuged for 6 minutes at 13 000 rpm. Fifteen μl were removed from the bottom of the centrifuged vial, and used for PCR analysis. PCR reactions were run in 25 μl volumes using 200 μl thin walled PCR tubes. Fungal DNA extraction, two rounds of nested PCR amplification, and target product identification were performed as described above for test isolates.
RESULTS
PCR sensitivity
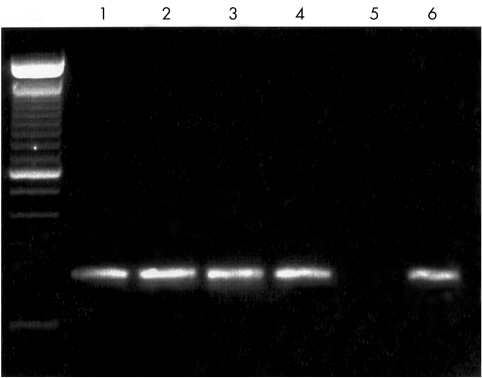
Clear DNA signals were produced from as few as 38 C albicans organisms, and five A fumigatus (see Fig 2) or 10 F solani elements. The PCR products of amplification from each species were sequenced, and found to match the predicted target sequence. The primers for each of these three designated species always yielded a target band when sufficient organisms of that species were known to be present.
Figure 2.
Agarose gel demonstrating sensitivity of PCR assay. Serial dilutions of quantified Aspergillus fumigatus suspensions were assayed with 30 cycles of primary and 30 cycles of species directed PCR amplification. Far left lane shows 100 bp DNA ladder, (200 bp fragment obscured by loading dye). Lane 1: Aspergillus fumigatus 500 fungal elements. Target 214 bp product DNA band visible, as predicted for A fumigatus directed primers. Lane 2: 100 elements, target DNA band present. Lane 3: 25 elements, target DNA band present. Lane 4: 5 elements, target DNA band present. Lane 5: 0 elements, no target DNA. Lane 6: template A fumigatus DNA, positive control.
PCR specificity
The primer pairs were variably cross reactive among filamentous fungi. Primers designed to amplify A fumigatus generated product DNA bands of the predicted length from several filamentous species tested, including most Aspergillus and some Fusarium species. This cross reactivity varied among isolates of the same species. Primers designed to amplify F solani also showed cross reactivity within and among various filamentous fungal species. Cross reactivity of these primers with yeast was not observed.
Primers based on C albicans generated a target band from multiple fungal species of all types, and this cross reactivity also varied between isolates of the same species. No bacteria sustained DNA amplification by this PCR assay. As such, this PCR assay appears to differentiate filamentous fungi from yeast, without further taxonomic specificity.
Clinical specimens
Thirty clinical specimens were evaluated by PCR and by culture techniques. The results are shown in Table 2. Of the 30 specimens analysed, fungal keratitis was definitively diagnosed by culture in 16 (53%). Fifteen (94%) of these 16 specimens were PCR positive. One specimen (3% of the 30 total) was fungal culture positive but PCR negative—an apparent “false negative” PCR result. Fourteen (47%) of 30 specimens were fungal culture negative, and seven (50%) of these 14 were also PCR negative.
Table 2.
Results of culture and polymerase chain reaction (PCR) analysis of corneal scrapings from patients with presumed infectious keratitis
| N = 30* | Culture positive for fungi | Culture negative for fungi |
| PCR positive for fungi† | 15 (50%) | 7 (23%) |
| Culture results: | Clinical impression: | |
| Fusarium: 5 (17%) | Fungal keratitis: 2 (7%) | |
| Aspergillus 2 (7%) | Bacterial keratitis: 4 (13%) | |
| Unidentified: 8 (27%)‡ | Uncertain: 1 (3%) | |
| PCR negative for fungi | 1 (3%) | 7 (23%) |
| Culture result: unidentified‡ | Culture results: | |
| Bacteria: 4 (13%) | ||
| Clinical impression: fungal keratitis | Acanthamoeba: 2 (7%) | |
| No growth: 1 (3%) | ||
| 16 (54%) | 14 (46%) |
*Percentages of the total of 30 specimens are shown in parentheses
†Specimens were considered to be “PCR positive” if a positive DNA product was obtained using any 1 of the 3 primer sets described.
‡These fungal culture isolates were examined microscopically and recorded as “hyaline” or “dematiaceous.” They were not speciated.
Seven patients' corneal scrapings were PCR positive but fungal culture negative (see Table 2); their clinical charts were reviewed. Based on the result of fungal staining and their response to antimicrobial treatment, two patients appeared clinically to have fungal keratitis despite negative fungal culture results. Four patients were judged clinically to have bacterial infections, and one patient was lost to follow up with an uncertain clinical course.
Among the 16 culture positive specimens (see Table 2), five harboured Fusarium in culture, two had Aspergillus, and eight culture isolates were not speciated. No specimen was found in culture to harbour yeast, and no specimen was positive with only C albicans primers.
Of the seven specimens negative by both PCR and fungal culture (see Table 2), four showed bacterial growth, two grew Acanthamoeba, and one had no growth.
All 30 specimens were examined by light microscopy with fungal staining, and 19 (63%) showed fungi. Three (16%) of these 19 specimens showed fungi on smear but were fungal culture negative, and two of these three specimens were PCR positive.
Conjunctival swabs from the fellow eyes of all 30 patients were analysed by PCR. Five (17%) swabs were PCR positive, and the opposite, infected eye in all five cases was found to harbour fungus. In one of these five patients, the corneal scraping from the opposite, infected eye was PCR negative but fungal culture positive (the apparent false negative PCR result).
DISCUSSION
This study demonstrates that fungi can be detected in infected corneas using PCR techniques. Advantages of PCR as shown here include greater speed than culture methods, and the ability to analyse specimens far from where they are collected. Limitations to the PCR assay used in this study include suboptimal specificity and the inability to identify and test the drug sensitivities of fungal pathogens.
The PCR assay used in this study requires 4 hours to generate results, significantly faster than the 2 days to 2 weeks required by any fungal culture technique. While fungal smears can be analysed by light microscopy in minutes, the effectiveness of this technique is more variable, and the results are not definitive. The ability of PCR based assays to detect or rule out the presence of fungi in less time would represent an advance in the management of ocular infections, and may also facilitate efforts to recognise and study fungal keratitis.
PCR allows investigators to analyse specimens far from where they are collected, and thus offers a significant advantage for those conducting field or epidemiological studies of fungal keratitis. PCR in this study enabled collaboration between centres in India and the United States; corneal surface samples were sent across the world for PCR analysis, without special packaging or shipping arrangements. Analysis of these specimens by culture techniques after shipment would have been impossible. While clinical samples of larger size (for example, stool samples) are occasionally shipped long distances on ice for culture analysis,36 the minute quantities of micro-organisms present in ocular surface scrapings would generally not survive long range shipment.
The sensitivity of PCR approximated that of standard culture methods for detecting fungi in ocular surface scrapings in this study. PCR and fungal culture results were either both positive or both negative in 22 (73%) of 30 case specimens. In two (7%) case specimens, PCR detected fungi where no organism was found in culture. Both of these patients appeared clinically to have fungal infections, and fungi were present on KOH smear for both. As such, PCR in this study did not increase diagnostic sensitivity to the extent shown in other comparisons with standard culture methods.37,38 However, given the paucity of precedent for using PCR to diagnose infectious keratitis in a clinical setting, and allowing for refinement and optimisation of multiple aspects of the PCR system, this technique holds promise as a diagnostic method for ocular surface infections.
The specificity of the PCR assay in this study is probably not adequate for clinical use. PCR appears to have yielded falsely positive results in at least four (14%) case specimens that grew bacteria in culture. Although our laboratory protocol did not enable us to run PCR on these bacterial isolates to rule out non-specific primer targeting, we consider it quite unlikely, based on pretrial testing, that this assay would generate positive results on non-fungal organisms. Clearly, the ocular surface is not germ free, and a high non-specific yield may be due to innocuous flora on the cornea or in the tear film, though laboratory contamination is always a possibility. Previous studies have suggested that antibiotic use or the mere presence of ocular inflammation could increase the level at which fungi are detected on the ocular surface.39,40 We expect that increasing the stringency of the current PCR assay would result in detecting fewer colonising organisms, and increased specificity.
The observation that fungal DNA was found by PCR in the healthy fellow eyes of five patients with fungal keratitis further highlights this issue. This interesting finding also suggests that fungal infection in one eye may increase the level of fungal colonisation of the fellow eye. It is noteworthy that in one of these five cases, the infected eye was culture positive but PCR negative, suggesting possible error (by switching vials) during specimen collection.
The PCR assay used here does not enable precise identification of fungal pathogens, or the ability to test isolates for drug sensitivity. From a practical standpoint, this would probably be an acceptable limitation in an otherwise highly sensitive and fungi specific assay. Particularly in the tropics, where the vast majority of keratomycosis occurs, almost all infections result from filamentous organisms,41–43 for which treatment is nearly uniform. In non-tropical areas, the differentiation between yeast and filaments is necessary to direct therapy. The current PCR assay appeared to make this differentiation in initial testing on known specimens, although no yeast infections occurred among the clinical cases to test this. Nevertheless, advancing understanding of fungal molecular genetics offers the potential for molecular diagnostic assays to precisely identify fungal pathogens and genes that code for antifungal drug resistance. The assay used here is but a start in that direction.
The cost of PCR to diagnose infections generally exceeds that of conventional culture methods, a factor that currently limits its widespread use.44,45 The added expenditure may be merited in certain research settings such as those in which specimens must be analysed far from where they are collected, as shown in this study, or in studying the epidemiology of certain infections where culture techniques are known to lack sensitivity. For clinical purposes, the cost-benefit assessment of PCR may improve as the technology becomes more widely available, the technique more automated, and the decreased morbidity—and hence cost—afforded by its use more evident.
Jaeger et al have used a PCR assay similar to that shown here in testing endophthalmitis specimens.29 The PCR primers used in their study also target the 18S ribosome, though the primer sequences are different. At the time that our PCR primers were developed, alignment of available fungal DNA sequences suggested that ribosome genes were logical targets for PCR amplification of DNA segments common to multiple fungal genuses but containing genus specific intervening variable regions. Currently, the continually expanding database of fungal gene sequences offers the opportunity for improved primer targeting and design.
PCR has been shown in non-ophthalmic settings to enable the detection of infectious pathogens in patients who had already received antimicrobial treatment or were cultured late in their illness.46 We were not able to address this issue in our study, since all of our patients presented acutely and none had used antifungal agents before presentation. None the less, we feel that PCR may prove particularly useful for this purpose in the setting of ocular surface infections, where previous topical antimicrobial therapy frequently complicates diagnosis by culture.
Our findings suggest that PCR is a potentially valuable tool for diagnosing keratomycosis. A variety of modifications in the PCR protocol, including specimen collection, primer design, thermocycling parameters, and resolution of the product DNA merit modification. Optimisation will also require ongoing evaluations in multiple clinical settings with more rigorous control specimens for comparison. Eventually, PCR might solidly complement the current “gold standard” diagnostic techniques for guiding management or supporting research studies of fungal keratitis.
Funding sources: NEI grant R01EY 08362.
The authors have no proprietary interests in this study.
REFERENCES
- 1.Whitcher JP, Srinivasan M. Corneal ulceration in the developing world–a silent epidemic. Br J Ophthalmol 1997;81:622–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deshpande SD, Koppikar GV. A study of mycotic keratitis in Mumbai. Ind J Pathol Microbiol 1999;42:81–7. [PubMed] [Google Scholar]
- 3.Johnson GJ, Minassian DC, Weale R. The epidemiology of eye disease. 1st ed. New York: Chapman and Hall Medical, 1998:143.
- 4.Ormerod LD, Hertzmark E, Gomez DS, et al. Epidemiology of microbial keratitis in southern California. A multivariate analysis. Ophthalmology 1987;94:1322–33. [DOI] [PubMed] [Google Scholar]
- 5.Mselle J. Fungal keratitis as an indicator of HIV infection in Africa. Tropical Doctor 1999;29:133–5. [DOI] [PubMed] [Google Scholar]
- 6.Upadhyay MP, Karmacharya PC, Koirala S, et al. Epidemiologic characteristics, predisposing factors, and etiologic diagnosis of corneal ulceration in Nepal. Am J Ophthalmol 1991;111:92–9. [DOI] [PubMed] [Google Scholar]
- 7.Wahl JC, Katz HR, Abrams DA. Infectious keratitis in Baltimore. Ann Ophthalmol 1991;23:234–7. [PubMed] [Google Scholar]
- 8.Gordon YJ. Rapid diagnostic tests for infectious ocular disease. Int Ophthalmol Clin 1993;33:153–61. [DOI] [PubMed] [Google Scholar]
- 9.Reischl U, Lohmann CP. [Polymerase chain reaction (PCR) and its possible applications in diagnosis of infection in ophthalmology.] Klin Monatsbl Augenheilkd 1997;211:227–34. [DOI] [PubMed] [Google Scholar]
- 10.Bersudsky V, Rehany U, Tendler Y, et al. Diagnosis of chlamydial infection by direct enzyme-linked immunoassay and polymerase chain reaction in patients with acute follicular conjunctivitis. Graefes Arch Clin Exp Ophthalmol 1999;237:617–20. [DOI] [PubMed] [Google Scholar]
- 11.Biswas J, Therese L, Madhavan HN. Use of polymerase chain reaction in detection of Mycobacterium tuberculosis complex DNA from vitreous sample of Eales' disease [letter]. Br J Ophthalmol 1999;83:994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bou G, Figueroa MS, Marti-Belda P, et al. Value of PCR for detection of Toxoplasma gondii in aqueous humor and blood samples from immunocompetent patients with ocular toxoplasmosis. J Clin Microbiol 1999;37:3465–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowen EF. Cytomegalovirus reactivation in patients infected with HIV: the use of polymerase chain reaction in prediction and management. Drugs 1999;57:735–41. [DOI] [PubMed] [Google Scholar]
- 14.Bowyer JD, Gormley PD, Seth R, et al. Choroidal tuberculosis diagnosed by polymerase chain reaction. A clinicopathologic case report. Ophthalmology 1999;106:290–4. [DOI] [PubMed] [Google Scholar]
- 15.Danise A, Cinque P, Vergani S, et al. Use of polymerase chain reaction assays of aqueous humor in the differential diagnosis of retinitis in patients infected with human immunodeficiency virus. Clin Infect Dis 1997;24:1100–6. [DOI] [PubMed] [Google Scholar]
- 16.Dondey JC, Sullivan TJ, Robson JM, et al. Application of polymerase chain reaction assay in the diagnosis of orbital granuloma complicating atypical oculoglandular cat scratch disease. Ophthalmology 1997;104:1174–8. [DOI] [PubMed] [Google Scholar]
- 17.Elnifro EM, Storey CC, Morris DJ, et al. Polymerase chain reaction for detection of Chlamydia trachomatis in conjunctival swabs. Br J Ophthalmol 1997;81:497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feinberg AS, Spraul CW, Holden JT, et al. Conjunctival lymphocytic infiltrates associated with Epstein-Barr virus. Ophthalmology 2000;107:159–63. [DOI] [PubMed] [Google Scholar]
- 19.Hidalgo F, Melon S, de Ona M, et al. Diagnosis of herpetic keratoconjunctivitis by nested polymerase chain reaction in human tear film. Eur J Clin Microbiol Infect Dis 1998;17:120–3. [DOI] [PubMed] [Google Scholar]
- 20.Kerkhoff FT, Bergmans AM, van Der Zee A, et al. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol 1999;37:4034–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knox CM, Cevallos V, Margolis TP, et al. Identification of bacterial pathogens in patients with endophthalmitis by 16S ribosomal DNA typing. Am J Ophthalmol 1999;128:511–2. [DOI] [PubMed] [Google Scholar]
- 22.Lee SM, Choi YJ, Ryu HW, et al. Species identification and molecular characterization of Acanthamoeba isolated from contact lens paraphernalia. Korean J Ophthalmol 1997;11:39–50. [DOI] [PubMed] [Google Scholar]
- 23.Lohmann CP, Heeb M, Linde HJ, et al. Diagnosis of infectious endophthalmitis after cataract surgery by polymerase chain reaction. J Cataract Refract Surg 1998;24:821–6. [DOI] [PubMed] [Google Scholar]
- 24.Lohmann CP, Gabel VP, Heep M, et al. Listeria monocytogenes-induced endogenous endophthalmitis in an otherwise healthy individual: rapid PCR-diagnosis as the basis for effective treatment. Eur J Ophthalmol 1999;9:53–7. [PubMed] [Google Scholar]
- 25.Messmer EM, Raizman MB, Foster CS. Lepromatous uveitis diagnosed by iris biopsy. Graefes Arch Clin Exp Ophthalmol 1998;236:717–9. [DOI] [PubMed] [Google Scholar]
- 26.Uchio E, Matsuura N, Takeuchi S, et al. Acute follicular conjunctivitis caused by adenovirus type 34. Am J Ophthalmol 1999;128:680–6. [DOI] [PubMed] [Google Scholar]
- 27.Lehmann OJ, Green SM, Morlet N, et al. Polymerase chain reaction analysis of corneal epithelial and tear samples in the diagnosis of Acanthamoeba keratitis. Invest Ophthalmol Vis Sci 1998;39:1261–5. [PubMed] [Google Scholar]
- 28.Okhravi N, Adamson P, Mant R, et al. Polymerase chain reaction and restriction fragment length polymorphism mediated detection and speciation of Candida spp causing intraocular infection. Invest Ophthalmol Vis Sci 1998;39:859–66. [PubMed] [Google Scholar]
- 29.Jaeger EE, Carroll NM, Choudhury S, et al. Rapid detection and identification of Candida, Aspergillus, and Fusarium species in ocular samples using nested PCR. J Clin Microbiol 2000;38:2902–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anand AR, Madhavan HN, Neelam V, et al. Use of polymerase chain reaction in the diagnosis of fungal endophthalmitis. Ophthalmology 2001;108:326–30. [DOI] [PubMed] [Google Scholar]
- 31.Alexandrakis G, Jalali S, Gloor P. Diagnosis of Fusarium keratitis in an animal model using the polymerase chain reaction. Br J Ophthalmol 1998;82:306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao NA. A laboratory approach to rapid diagnosis of ocular infections and prospects for the future. Am J Ophthalmol 1989;107:283–91. [DOI] [PubMed] [Google Scholar]
- 33.Reddy PS, Satyendran OM, Satapathy M, et al. Mycotic keratitis. Ind J Ophthalmol 1972;20:101–8. [PubMed] [Google Scholar]
- 34.Liesegang TJ, Forster RK. Spectrum of microbial keratitis in South Florida. Am J Ophthalmol 1980;90:38–47. [DOI] [PubMed] [Google Scholar]
- 35.Foster CS. Fungal keratitis. Infect Dis Clin N Am 1992;6:851–7. [PubMed] [Google Scholar]
- 36.Sethabutr O, Venkatesan M, Yam S, et al. Detection of PCR products of the ipaH gene from Shigella and enteroinvasive Escherichia coli by enzyme linked immunosorbent assay. Diag Microbiol Infect Dis 2000;37:11–6. [DOI] [PubMed] [Google Scholar]
- 37.Therese KL, Anand AR, Madhavan HN. Polymerase chain reaction in the diagnosis of bacterial endophthalmitis [see comments]. Br J Ophthalmol 1998;82:1078–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arora SK, Gupta V, Gupta A, et al. Diagnostic efficacy of polymerase chain reaction in granulomatous uveitis. Tubercle Lung Dis 1999;79:229–33. [DOI] [PubMed] [Google Scholar]
- 39.Nema HV, Ahuja OP, Bal A, et al. Effects of topical corticosteroids and antibiotics on mycotic flora of conjunctiva. Am J Ophthalmol 1968;65:747–50. [DOI] [PubMed] [Google Scholar]
- 40.Ando N, Takatori K. Fungal flora of the conjunctival sac. Am J Ophthalmol 1982;94:67–74. [DOI] [PubMed] [Google Scholar]
- 41.Dunlop AA, Wright ED, Howlader SA, et al. Suppurative corneal ulceration in Bangladesh. A study of 142 cases examining the microbiological diagnosis, clinical and epidemiological features of bacterial and fungal keratitis. Aust NZ J Ophthalmol 1994;22:105–10. [DOI] [PubMed] [Google Scholar]
- 42.Gonawardena SA, Ranasinghe KP, Arseculeratne SN, et al. Survey of mycotic and bacterial keratitis in Sri Lanka. Mycopathologia 1994;127:77–81. [DOI] [PubMed] [Google Scholar]
- 43.Srinivasan M, Gonzales CA, George C, et al. Epidemiology and aetiological diagnosis of corneal ulceration in Madurai, south India. Br J Ophthalmol 1997;81:965–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Press N, Romney M, Takaya S, et al. Molecular diagnosis of infections in the new millennium. Can Med Ass J 1999;161:1294. [PMC free article] [PubMed] [Google Scholar]
- 45.Louie M, Louie L, Simor AE. The role of DNA amplification technology in the diagnosis of infectious diseases.Can Med Ass J 2000;163:301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sethabutr O, Echeverria P, Hoge CW, et al. Detection of Shigella and enteroinvasive Escherichia coli by PCR in the stools of patients with dysentery in Thailand. J Diarrhoeal Dis Res 1994;12:265–9. [PubMed] [Google Scholar]