Abstract
A central question in lignin biosynthesis is how guaiacyl intermediates are hydroxylated and methylated to the syringyl monolignol in angiosperms. To address this question, we cloned cDNAs encoding a cytochrome P450 monooxygenase (LsM88) and a caffeate O-methyltransferase (COMT) from sweetgum (Liquidambar styraciflua) xylem. Mass spectrometry-based functional analysis of LsM88 in yeast identified it as coniferyl aldehyde 5-hydroxylase (CAld5H). COMT expressed in Escherichia coli methylated 5-hydroxyconiferyl aldehyde to sinapyl aldehyde. Together, CAld5H and COMT converted coniferyl aldehyde to sinapyl aldehyde, suggesting a CAld5H/COMT-mediated pathway from guaiacyl to syringyl monolignol biosynthesis via coniferyl aldehyde that contrasts with the generally accepted route to sinapate via ferulate. Although the CAld5H/COMT enzyme system can mediate the biosynthesis of syringyl monolignol intermediates through either route, kcat/Km of CAld5H for coniferyl aldehyde was ≈140 times greater than that for ferulate. More significantly, when coniferyl aldehyde and ferulate were present together, coniferyl aldehyde was a noncompetitive inhibitor (Ki = 0.59 μM) of ferulate 5-hydroxylation, thereby eliminating the entire reaction sequence from ferulate to sinapate. In contrast, ferulate had no effect on coniferyl aldehyde 5-hydroxylation. 5-Hydroxylation also could not be detected for feruloyl-CoA or coniferyl alcohol. Therefore, in the presence of coniferyl aldehyde, ferulate 5-hydroxylation does not occur, and the syringyl monolignol can be synthesized only from coniferyl aldehyde. Endogenous coniferyl, 5-hydroxyconiferyl, and sinapyl aldehydes were detected, consistent with in vivo operation of the CAld5H/COMT pathway from coniferyl to sinapyl aldehydes via 5-hydroxyconiferyl aldehyde for syringyl monolignol biosynthesis.
Lignin in angiosperms is composed of guaiacyl and syringyl monomers, whereas gymnosperm lignin consists almost entirely of guaiacyl moieties (1). The importance of the syringyl constituent in facilitating overall lignin degradation for more efficient materials and energy production from angiosperm than from gymnosperm wood has long been established (2–4). In contrast, the biosynthesis of syringyl lignin is not well understood.
It has been thought that syringyl lignin biosynthesis involves a cytochrome P450 ferulate 5-hydroxylase (F5H)-catalyzed conversion of guaiacyl intermediate ferulate to 5-hydroxyferulate followed by a caffeate O-methyltransferase (COMT)-mediated reaction to sinapate (1, 5–9). Current understanding of F5H function in plants is based solely on the in vitro enzymatic activity of proteins from Populus euramericana (8). Ferulate was believed to be converted into a product that was “tentatively proposed” as 5-hydroxyferulate (8), but the structure of this product must yet be corroborated to demonstrate F5H activity. In fact, 5-hydroxyferulate as an intermediate for monolignol biosynthesis has not been reported in planta. Although a putative F5H cDNA was recently cloned from Arabidopsis, its biochemical function remains unknown (10, 11). Overexpression of this cDNA in Arabidopsis mutant (fah1) lines deficient in syringyl lignin (12) restored the accumulation of syringyl-enriched lignin, but did not result in detectable F5H enzymatic activity (11). It is also known that, based on in vitro studies, proteins from various angiosperm species cannot activate sinapate into its CoA derivative for syringyl lignin biosynthesis (13–15). Taken together, these results challenge the conventional concept of a ferulate 5-hydroxylation/methylation-regulated biosynthesis of syringyl lignin in angiosperms. Based on this and on the general lack of evidence that ferulate 5-hydroxylation is involved in syringyl monolignol biosynthesis, we hypothesize that F5H may encode an enzyme that catalyzes 5-hydroxylation of guaiacyl intermediates other than ferulate to initiate the biosynthesis of the syringyl monolignol. We therefore examined the 5-hydroxylation and methylation reactions in lignifying xylem of an angiosperm tree species, sweetgum (Liquidambar styraciflua), to investigate the entrance pathways to syringyl lignin and thereby test the validity of the traditionally accepted pathway.
MATERIALS AND METHODS
Plant Material.
Differentiating stem xylem was collected from vegetatively propagated 3-yr-old sweetgum trees (Forest Experimental Station of International Paper Co., Bainbridge, GA) and stored in liquid nitrogen before protein, RNA, and DNA isolation, as described (15).
cDNA Cloning of Sweetgum Monooxygenases and COMT, and Northern and Southern Blotting.
Poly(A)+ RNA isolated from xylem was used to construct a cDNA library in λZAP II vector (Stratagene), as described (16). A pool of phagemid cDNAs rescued from the cDNA library was used as a template for PCR amplification of P450 cDNAs. A degenerate primer (5′-CTAGTCTAGACCATTCGGNDCNGGNMGNNMG-3′) for the conserved P450 heme-binding domain (PFGXGRR) with an introduced 5′ XbaI site and an oligo-dT primer were used for PCR. The amplified cDNAs were cloned into a pCRII vector (Invitrogen) and sequenced. Sequencing of 24 independent clones classified them into five separate groups, of which three, namely LsM4, LsM7, and LsM8, could be assigned to the CYP73, CYP98, and CYP84 gene families, respectively. LsM8 was used to screen the cDNA library to obtain a full length cDNA, designated LsM88, and sequenced (GenBank AF139532). By using an aspen (Populus tremuloides) COMT-encoding cDNA (PtOMT1) (17) to screen the same sweetgum cDNA library, a full-length cDNA, designated LsCOMT, was cloned and sequenced (GenBank AF139533), and exhibited 80% amino acid sequence identity to PtOMT1. The gcg software package (Genetics Computer Group, Madison, WI) was used for sequence analysis. Northern and Southern blotting were performed according to Tsai et al. (18).
Expression of Recombinant LsM88 and LsCOMT in Escherichia coli, Preparation of Anti-LsM88 Polyclonal Antibodies, and Western Blotting.
The plasmid vector pQE30 (Qiagen, Chatsworth, CA) was used for the expression of LsM88 cDNA in E. coli. The first 30 amino acids of LsM88 consisting of many hydrophobic residues involved in endoplasmic reticulum targeting were replaced by an alanine codon (GCT) for high expression of the transgene (19) in E. coli. This modification was achieved by PCR mutagenesis by using a pair of LsM88-specific primers that introduced BamHI sites at the 5′ and 3′ ends of the PCR product that was cloned at the BamHI site of the pQE30 vector to give the expression plasmid pQEΔLsM88. This expression plasmid, in which the LsM88 cDNA was sequenced and confirmed to have no PCR errors, was used to transform E. coli strain M15 for expression according to Li et al. (20). The truncated LsM88 protein was harvested from bacterial cells and affinity purified by using a Ni2+-NTA-agarose column (Qiagen) according to the manufacturer’s protocol. Anti-LsM88 polyclonal antibodies were raised in rabbits (Alpha Diagnostic, San Antonio, TX) and used for Western blotting, as described (20). E. coli-expressed LsCOMT was prepared according to Li et al. (20).
Coexpression of Sweetgum LsM88 with Arabidopsis NADPH-Cytochrome P450 Reductase (CPR) in Yeast.
To use adenine as a selection marker, we altered the ADE 2 gene (21) of the INVSc1 host strain of yeast Saccharomyces cerevisiae (Invitrogen) and designated the mutated form as INVSc2. The Arabidopsis CPR cDNA (EST clone G8A6, ABRC) driven by a GAL promoter was then integrated into the INVSc2 genome by homologous recombination, giving rise to the INVSc2(CPR) strain. LsM88 cDNA driven by a GAL promoter was placed into the autonomously replicating vector pYAL by cloning the ADE2 gene into the pYX243 vector (Novagen/R & D Systems) and then selected by using adenine and leucine as the markers. This LsM88 expression vector (pYAL-LsM88) was transferred into INVSc2(CPR) to create the INVSc2(CPR)/pYAL-LsM88 yeast strain for coexpressing CPR and LsM88 cDNAs. The expression of INVSc2(CPR)/pYAL-LsM88 and control cells transformed with pYAL alone [INVSc2(CPR)/pYAL], and the preparation of microsomal fractions from these cells was carried out as described (22). P450 was measured from the reduced-CO difference spectrum (23). Microsomal NADPH-cytochrome c reductase activity was determined as described (24). Protein concentrations were determined by using the Bradford dye-binding reagent (Bio-Rad) with BSA as the standard.
Chemicals.
5-Hydroxyferulate, feruloyl-CoA, and 5-hydroxyferuloyl-CoA thioesters were synthesized as described (20, 25). 5-Hydroxyconiferyl aldehyde was synthesized from 5-hydroxyvanillin by first condensing it with monoethyl malonate to give ethyl 5-hydroxyferulate, which was ethoxyethylated with ethyl vinyl ether and DL-10-camphorsulfonic acid in CH2Cl2/tetrahydrofuran (10/1) to yield ethyl 5-hydroxyferulate diethoxyethyl ether. This ether was reduced by diisobutylaluminum hydride in CH2Cl2 to give 5-hydroxyconiferyl alcohol diethoxyethyl ether, followed by oxidation with activated MnO2 in CH2Cl2 to afford 5-hydroxyconiferyl aldehyde diethoxyethyl ether, of which the ethoxyethyl groups were hydrolyzed by HCl in acetone to produce 5-hydroxyconiferyl aldehyde, and its structure was confirmed by 1H- and 13C-NMR, C,H-correlation spectroscopy, and heteronuclear multiple bond connectivity, and MS. NMR spectra were recorded with a JNM-LA400MK FT-NMR System (JEOL). Electron impact mass spectrometry (70eV) was recorded with a JMS-DX303HF mass spectrometer equipped with a JMA-DA5000 Mass Data System (JEOL). 5-Hydroxyconiferyl aldehyde: 1H-NMR (acetone-d6, carbon numbers are shown in Fig. 2A), δ 3.88 (3H, s, OCH3), 6.60 (1H, dd, J = 15.6, J = 7.8, C8H), 6.88 (1H, d, J = 1.7, C6H), 6.95 (1H, d, J = 1.7, C2H), 7.50 (1H, d, J = 15.6, C7H), 9.61 (1H, d, J = 7.8, C9H); 13C-NMR (acetone-d6), δ 56.6 (OCH3), 104.7 (C2), 111.2 (C6), 126.5 (C1), 127.2 (C8), 138.3 (C4), 146.5 (C5), 149.3 (C3), 154.4 (C7), 193.8 (C9); MS m/z (%), 194 (M+, 100), 177 (10.8), 166 (24.4), 151 (53.9), 133 (8.0), 123 (21.1), 105 (7.0). All other chemicals used were obtained from Sigma/Aldrich.
Figure 2.
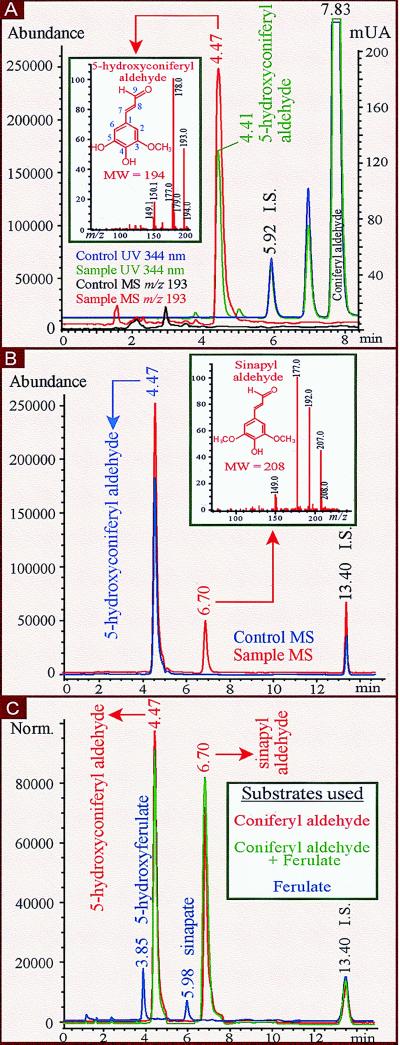
HPLC separation of products from enzymatic reactions with recombinant proteins. (A) HPLC-UV (344 nm) and HPLC-MS [Selected Ion Monitoring (SIM), 70 V; m/z 193] chromatograms of reaction mixture after incubating 0.5 mM coniferaldehyde (Rt 7.83 min/UV) with 100 pmol LsM88 (CAld5H)-containing (Sample) or 500 μg vector-only (Control) yeast microsomes. Internal standard (I.S.) was sinapate. Negative ion electrospray (NI-ES) mass spectrum (Inset, scanning mode at 150 V) of the reaction product 5-hydroxy coniferyl aldehyde (Rt 4.47 min/MS, 4.41 min/UV) is identical to that of the authentic standard. (B) HPLC-MS [SIM, 70 V; m/z 193 (0–5.50 min), m/z 207 (5.50–10.00 min), m/z 163 (10.00–15.00 min)] chromatograms of reaction mixture after incubating 0.05 mM 5-hydroxyconiferyl aldehyde with 9.5 μg protein extract from LsCOMT-containing (Sample) E. coli cells or with boiled (Control) LsCOMT recombinant proteins. NI-ES mass spectrum (Inset, scanning mode at 150 V) of product sinapyl aldehyde (Rt 6.70 min) is identical to that of the authentic standard. (C) HPLC-MS [SIM, 70 V; m/z 209 (0–4.10 min), m/z 193 (4.10–5.20 min), m/z 223 (5.20–6.30 min), m/z 207 (6.30–10.00 min), m/z 163 (10.00–15.00 min)] chromatograms of reaction mixture after incubating either 0.5 mM coniferyl aldehyde, 0.5 mM ferulate, or 0.5 mM of both with a mixture of 100 pmol CAld5H-containing yeast microsomes and 9.5 μg protein extract from LsCOMT-containing E. coli cells. 5-Hydroxyconiferyl aldehyde and sinapyl aldehyde were products (red or green) from coniferyl aldehyde or a mixture of coniferyl aldehyde and ferulate. 5-Hydroxyferulate (Rt 3.85 min) and sinapate (Rt 5.98 min) were products (blue) from ferulate. I.S. was o-coumarate in B and C.
Hydroxylase and O-Methyltransferase Enzyme Assays and HPLC-UV/Mass Spectrometer Detector (MSD) Analysis of Reaction Products.
For hydroxylase activity, 500 μl of reaction mixture (saturated with oxygen) containing 50 mM NaH2PO4 (pH 7.5), 1 mM β-mercaptoethanol, 200 nM P450 from transformed yeast cells or 720 μg microsomal proteins from xylem, 0.5 mM substrate, and 1 mM NADPH was incubated at 30°C for 15 min followed by the addition of 20 μl 6N HCl to terminate the reaction and 1 μg sinapate as internal standard. For kinetic analyses, the reaction time was 5 min with 15 nM P450 from transformed yeast and varying concentrations of coniferyl aldehyde (1 to 32 μM) or ferulate (100 to 3,200 μM) to measure the Km, Vmax, and kcat. To measure the Ki for coniferyl aldehyde, hydroxylation of ferulate (100 to 3,200 μM) was assayed in the presence of coniferyl aldehyde at various concentrations (0.25 to 5 μM). O-methyltransferase activity was assayed according to Li et al. (20), except nonradioactive S-adenosyl-l-methionine was used and 1 μg o-coumarate as internal standard. The ethyl acetate extracted and dried reaction mixtures were dissolved in 30 μl of HPLC mobile phase (20% acetonitrile in 10 mM formic acid, pH 2.7). Samples of 15 μl were injected automatically onto a Supelcosil LC-ABZ column (15 cm × 4.6 mm × 5 μm, Supelco), and compounds were separated isocratically at 40°C and a flow rate of 1 ml/min with an HP 1100 LC system and detected by an HP 1100 diode array detector and an HP 1100 LC-MSD with an Atmospheric Pressure Ionization-electrospray source in negative ion mode (Hewlett Packard). The reaction products were identified and quantified based on the authentic standards. 5-Hydroxyconiferyl aldehyde; UV (HPLC mobile phase) λ max I 244 nm, λmax II 344 nm, λ min 273 nm; MS (150 V) m/z (%), 194 (5.9), 193 ([M-H]−, 55), 178 (100), 150 (18.1); retention time (Rt) 4.41 min/UV, 4.47 min/MS. 5-Hydroxyferulate; UV, λ max I 236 nm, λ max II 322 nm, λ min 263 nm; MS (150 V) m/z (%), 210 (9.8), 209 ([M-H]−, 71.9), 194 (100), 150 (79.3); Rt 3.79 min/UV, 3.85 min/MS. Sinapyl aldehyde; UV, λ max I 244 nm, λ max II 344 nm, λmin 275 nm; MS (150 V) m/z (%), 208 (7.9), 207 ([M-H]−, 44), 192 (77), 177(100), 149 (13); Rt 6.64 min/UV, 6.70 min/MS. Sinapate; UV, λmax I 238 nm, λ max II 324 nm, λ min 264 nm; MS (150 V) m/z (%), 224 (11.3), 223 ([M-H]−, 100), 208 (44.5), 193 (59); Rt 5.92 min/UV, 5.98 min/MS.
Measurement of Kinetic Constants.
Km and Vmax values were determined from Lineweaver–Burk plots, and kcat values by dividing Vmax by the enzyme concentration, based on three to four independent assays. Ki was derived from a Dixon plot.
RESULTS AND DISCUSSION
Cloning of a Sweetgum Cytochrome P450 Monooxygenase LsM88 and Coexpression with Arabidopsis CPR cDNA.
We focused on the 5-hydroxylation of guaiacyl monolignol precursors by first cloning cytochrome P450 monooxygenases from lignifying stem xylem of sweetgum. A full-length cDNA LsM88 (GenBank AF139532) was isolated and was 1,883-bp long, encoding an ORF of 511 amino acids with a calculated Mr of 57,503 and a pI of 5.94. LsM88 has a 75% amino acid sequence identity (82% similarity) to the Arabidopsis putative F5H (10), suggesting that LsM88 encodes a plant CYP84. However, the N-terminal 34-aa sequence of LsM88 is highly divergent from Arabidopsis F5H and contains the hydrophobic region typical of the uncleavable signal peptide for anchoring P450 protein to the endoplasmic reticulum membrane (26). A proline-rich region following the putative signal peptide and a cytochrome P450 heme-binding signature (PFGXGRR) toward the C terminus, which is typical of plant P450 proteins, were also identified in LsM88. Northern blot analysis of sweetgum xylem mRNA showed a 1.9-Kb transcript matching well with the length of the LsM88 clone, and Southern blot analysis indicated that a small LsM88 gene family of two to three members may be present in the sweetgum genome (data not shown). Western blotting showed that LsM88 protein is present in syringyl lignin-forming secondary growth tissue, including developing stem xylem, but not in leaves or the guaiacyl lignin-enriched primary growth tissue (15, 18, 27) (Fig. 1), suggesting a role for LsM88 enzyme in the biosynthesis of the syringyl monolignol.
Figure 1.
Western blot analysis of 1 μg of purified E. coli-expressed LsM88 recombinant protein (lane 1) and 10 μg microsomal proteins each from leaves (without midveins, lane 2), stem internodes 1 to 4 (primary growth tissue, lane 3), internodes 6 to 10 (secondary growth tissue, lane 4), and stem xylem (lane 5) of sweetgum.
To investigate LsM88 enzyme function, we used a yeast expression system. Yeast strain INVSc2 (CPR) exhibited a 5-fold higher microsomal NADPH-cytochrome c reductase activity (500 nmol/minute per mg) than INVSc2 without the CPR transgene. INVSc2(CPR)/pYAL-LsM88 yeast cells expressed a ≈58-kDa protein that localized to the microsome fraction and crossreacted strongly with the anti-LsM88 antibody, whereas no such crossreaction could be detected in microsomes of control INVSc2(CPR)/pYAL cells (data not shown). P450 expression levels in INVSc2(CPR)/pYAL-LsM88 were typically ≈300 pmol P450 per mg of microsomal proteins. No P450 protein was detectable in control INVSc2(CPR)/pYAL.
LsM88 Is a Coniferyl Aldehyde 5-Hydroxylase (CAld5H).
We focused on monolignol biosynthetic pathways starting from ferulate. After HPLC separation, all enzymatic reaction products were confirmed for their purity and identity and were quantified based on the authentic compounds. In preliminary experiments, plant protein extracts were used to test the enzymatic 5-hydroxylation of ferulate and its downstream derivatives, feruloyl-CoA, coniferyl aldehyde, and coniferyl alcohol. To mimic an in vivo situation, a mixture of these four monolignol intermediates was incubated with microsomal proteins from lignifying stem xylem of sweetgum. This experiment should result in the formation of 5-hydroxyferulate according to the traditional ferulate 5-hydroxylation pathway for syringyl monolignol biosynthesis. But no 5-hydroxyferulate was detected. Instead, the single product formed exhibited the UV and MS spectral profiles of 5-hydroxyconiferyl aldehyde. To verify this reaction and to test whether it is mediated by LsM88, the assays were repeated by using microsomes from xylem and LsM88-transformed yeast cells with coniferyl aldehyde as the lone substrate. In both cases, the single product formed coeluted with authentic 5-hydroxyconiferyl aldehyde and exhibited UV and MS spectral properties identical to those of authentic 5-hydroxyconiferyl aldehyde (Fig. 2A). This identifies LsM88 as a protein that hydroxylates coniferyl aldehyde, instead of ferulate, to initiate the biosynthesis of the syringyl monolignol from the guaiacyl pathway. These results confirm a 5-hydroxylation function involved in monolignol metabolism (Table 1). We therefore designated LsM88 as CAld5H and propose that CYP84 genes also encode a coniferyl aldehyde 5-hydroxylase.
Table 1.
Substrate specificity of recombinant sweetgum LsM88 (CAld5H) and LsCOMT (COMT) proteins and of sweetgum xylem proteins
| Substrate | Substrate specificity (pmol/min per mg protein)
|
|||||
|---|---|---|---|---|---|---|
| Recombinant proteins
|
Xylem proteins
|
|||||
| CAld5H | COMT | CAld5H + COMT | Microsome | Soluble | Microsome + soluble | |
| Coniferyl aldehyde | 723.9 ± 24.0 | 382.2 ± 15.2 | 114.0 ± 3.6 | 13.8 ± 1.2 | ||
| (5-OH-CAld) | (SAld) | (5-OH-CAld) | (SAld) | |||
| Ferulate | 60.7 ± 3.2 | 32.2 ± 2.4 | 26.3 ± 1.2 | 12.0 ± 1.4 | ||
| (5-OH-FA) | (SA) | (5-OH-FA) | (SA) | |||
| 5-Hydroxyconiferyl aldehyde | 14,370 ± 200 | 5,900 ± 35 | ||||
| (SAld) | (SAld) | |||||
| 5-Hydroxyferulate | 12,710 ± 70 | 5,700 ± 90 | ||||
| (SA) | (SA) | |||||
| 526.5 ± 30.0 | 247.5 ± 17.2 | 8.8 ± 1.4 | 4.8 ± 1.9 | |||
| Coniferyl aldehyde + ferulate | (5-OH-CAld) | (SAld) | (5-OH-CAld) | (SAld) | ||
| 0 | 0 | 0 | 0 | |||
| (5-OH-FA) | (SA) | (5-OH-FA) | (SA) | |||
Substrate and recombinant protein concentrations and the control experiments were the same as described in Fig. 2. For assaying xylem proteins, 9.5 and 720 μg of soluble and microsome proteins were used, respectively, with the same substrate concentrations as for recombinant proteins and boiled plant proteins as control. Specific activities were mean ± SD (n = two to three independent assays). Reaction products: 5-OH-CAld, 5-hydroxyconiferyl aldehyde; 5-OH-FA, 5-hydroxyferulate; SAld, sinapyl aldehyde; SA, sinapate.
Coniferyl Aldehyde Is 5-Hydroxylated and Methylated in Series by CAld5H and COMT to Form Sinapyl Aldehyde.
To support the proposed model of coniferyl aldehyde-initiated syringyl monolignol biosynthesis, methylation of 5-hydroxyconiferyl aldehyde to sinapyl aldehyde, as suggested by Higuchi (28), must be demonstrated. The results from two sets of experiments attested to such a model. First, 5-hydroxyconiferyl aldehyde was converted exclusively into sinapyl aldehyde when incubated with E. coli-expressed LsCOMT (Fig. 2B and Table 1), providing the first evidence that the aldehyde precursor is methylated during monolignol biosynthesis and that COMT can catalyze this reaction. Second, when coniferyl aldehyde was incubated with a mixture of CAld5H-containing yeast P450 and E. coli-expressed COMT, it was converted into sinapyl aldehyde via 5-hydroxyconiferyl aldehyde (Fig. 2C, in red, and Table 1). Thus, CAld5H catalyzes 5-hydroxylation of coniferyl aldehyde into 5-hydroxyconiferyl aldehyde, which in turn is methylated by COMT to sinapyl aldehyde, supporting the idea of a hydroxylation/methylation flux in vivo from guaiacyl to syringyl monolignol biosynthesis via coniferyl aldehyde. This finding stands in sharp contrast to the generally accepted idea that regulation of syringyl and guaiacyl lignin composition occurs upstream at the ferulate 5-hydroxylation step (1, 9, 10).
Coniferyl Aldehyde Inhibits Ferulate 5-Hydroxylation, Eliminating the Reaction Sequence from Ferulate to Sinapate.
Consistent with the reaction observed in xylem microsomes, recombinant CAld5H also selectively mediated the conversion of coniferyl aldehyde into 5-hydroxyconiferyl aldehyde from a substrate mixture of ferulate, feruloyl-CoA, coniferyl aldehyde, and coniferyl alcohol. Interestingly, when these substrates were incubated individually with either recombinant CAld5H or xylem microsomes, both coniferyl aldehyde and ferulate were 5-hydroxylated (Table 1). Although the catalytic efficiency with ferulate is considerably lower than that with coniferyl aldehyde (Table 1), a bifunctional 5-hydroxylase activity is implied. This bifunctional activity, however, is inconsistent with the exclusive detection of 5-hydroxyconiferyl aldehyde in mixed substrate reactions mediated either by xylem or CAld5H-containing yeast microsomes. This surprising discovery prompted us to determine whether coniferyl aldehyde inhibits the 5-hydroxylation of ferulate. Equal molar coniferyl aldehyde and ferulate were incubated with recombinant CAld5H. This mixed substrate reaction resulted in a complete inhibition of ferulate 5-hydroxylation, but the conversion of coniferyl aldehyde into 5-hydroxyconiferyl aldehyde was conserved (Table 1). Similarly, incubation of such mixed substrates with a mixture of yeast CAld5H and E. coli LsCOMT recombinant proteins resulted in the production of 5-hydroxyconiferyl aldehyde and sinapyl aldehyde, but not of 5-hydroxyferulate and sinapate (Fig. 2C, in green). Elimination of coniferyl aldehyde from the substrate mixture restored the production of 5-hydroxyferulate and sinapate (Fig. 2C, in blue). Thus, these results provide unambiguous evidence that coniferyl aldehyde inhibits ferulate 5-hydroxylation, thereby eliminating the entire reaction sequence from ferulate to sinapate, the traditionally accepted branch to syringyl lignin biosynthesis (1, 9).
Coniferyl Aldehyde Is Both a Noncompetitive Inhibitor of Ferulate 5-Hydroxylation and the Substrate for 5-Hydroxylation Initiating Syringyl Monolignol Biosynthesis.
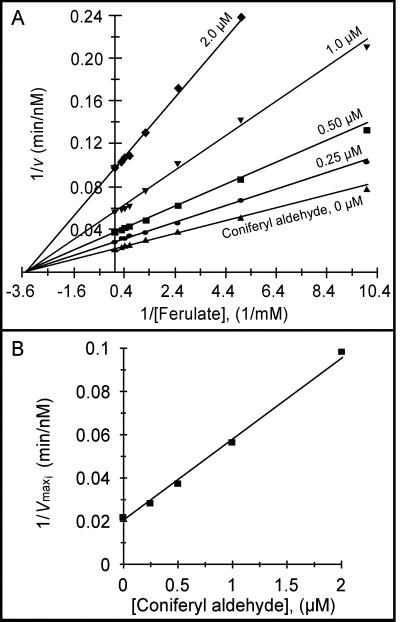
To understand how coniferyl aldehyde may inhibit ferulate 5-hydroxylation in vivo, we studied the kinetics of CAld5H reaction. The specificity constant (kcat/Km) values indicated that coniferyl aldehyde 5-hydroxylation is ≈140 times more efficient than ferulate 5-hydroxylation (Table 2), suggesting that coniferyl aldehyde 5-hydroxylation would be the dominant reaction leading to syringyl monolignol biosynthesis. In fact, when coniferyl aldehyde and ferulate are used as substrates together, coniferyl aldehyde is both the favored substrate for 5-hydroxylation and a noncompetitive inhibitor of ferulate 5-hydroxylation (Fig. 3A), with a Ki of 0.59 ± 0.01 μM (Fig. 3B). Consistent with the predicted degree of inhibition based on the Ki value, no ferulate 5-hydroxylation activity could be detected at coniferyl aldehyde concentrations ≥4 μM, regardless of ferulate concentrations (100 to 3,200 μM). Various concentrations (0.02 to 2 μM) of product 5-hydroxyconiferyl aldehyde had no effect on the CAld5H reaction with ferulate, confirming that substrate coniferyl aldehyde is solely responsible for the observed inhibition. In contrast, ferulate did not inhibit coniferyl aldehyde 5-hydroxylation at any concentrations tested. Based on the Ki for coniferyl aldehyde and kcat/Km values for coniferyl aldehyde and ferulate, we concluded that ferulate 5-hydroxylation is unlikely to take place in the presence of coniferyl aldehyde in vivo.
Table 2.
Kinetic constants for CAld5H enzyme
| Substrate | Km, μM | Vmax, nM⋅min−1 | kcat, min−1 | kcat/Km, min−1⋅μM−1 |
|---|---|---|---|---|
| Coniferyl aldehyde | 2.77 ± 0.04 | 64.58 ± 2.08 | 4.31 ± 0.14 | 1.56 |
| Ferulate | 286.05 ± 0.35 | 46.50 ± 1.10 | 3.10 ± 0.07 | 0.0108 |
Values were mean ± SD (n = three or four independent assays).
Figure 3.
(A) Lineweaver–Burk plot of CAld5H-catalyzed ferulate 5-hydroxylation in the presence of coniferyl aldehyde at different concentrations (as shown). Ferulate concentrations: 100 to 3,200 μM. (B) A replot of 1/Vmaxi for each reciprocal plot (in A) vs. the corresponding coniferyl aldehyde (inhibitor) concentration at which it was obtained. Slope = 1/Vmax Ki.
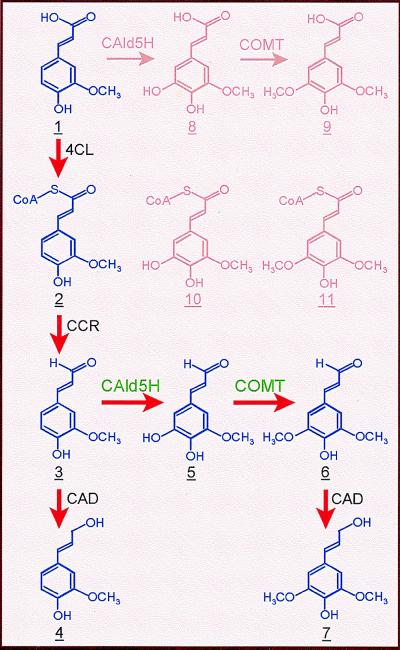
Enzymatic reactions analogous to those of CAld5H and COMT recombinant proteins, including the coniferyl aldehyde-induced elimination of ferulate 5-hydroxylation and methylation, were obtained by using xylem proteins (Table 1). Furthermore, by using HPLC-mass spectrometer detector analysis of methanol extracts from sweetgum xylem cells, we report detection of endogenous coniferyl, 5-hydroxyconiferyl, and sinapyl aldehydes, at 12, 4, and 110 ng/g fresh weight, respectively. The detection of these aldehydes in xylem cells and the kinetic results are consistent with a reaction sequence in planta from coniferyl aldehyde (3; in Fig. 4) to sinapyl aldehyde (6) via 5-hydroxyconiferyl aldehyde (5) catalyzed by CAld5H and COMT that diverts guaiacyl intermediates into syringyl monolignol biosynthesis. Proteins from aspen stem xylem catalyzed reactions (data not shown) similar to those catalyzed by sweetgum proteins, suggesting that coniferyl aldehyde-modulated CAld5H function for mediating syringyl monolignol biosynthesis may be common to angiosperm tree species.
Figure 4.
Biosynthetic pathway from ferulate to coniferyl alcohol and sinapyl alcohol for the formation of guaiacyl-syringyl lignin in angiosperms. 4CL, 4-coumarate:CoA ligase; CCR, cinnamoyl-CoA reductase; CAld5H, coniferyl aldehyde 5-hydroxylase; COMT, caffeate O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase.
To our knowledge, this is the first observation that noncompetitive inhibition among substrates may modulate a plant P450 activity. Our current results indicate a coniferyl aldehyde-sensitive 5-hydroxylation activity that regulates ferulate metabolism in angiosperms. In the absence of coniferyl aldehyde, CAld5H-catalyzed conversion of ferulate is possible, leading to the formation of 5-hydroxyferulate (8, Fig. 4) and sinapate (9) as the intermediates, for instance, for sinapoyl malate for UV protection in leaf epidermis (12, 29), or as donors of acyl groups for esterification to the cell wall (30). In its presence because of lignification in tissues such as developing xylem, coniferyl aldehyde, which interrupts ferulate 5-hydroxylation/methylation to divert the carbon flow toward biosynthesis of lignin, becomes 5-hydroxylated to initiate syringyl monolignol biosynthesis. This finding may also suggest that 5-hydroxylation of ferulate and coniferyl aldehyde are developmentally regulated and that coniferyl aldehyde 5-hydroxylation is specific to lignifying tissues.
CONCLUSION
Our finding that coniferyl aldehyde in cooperation with CAld5H can block the ferulate 5-hydroxylation/methylation reaction sequence challenges those ferulate-initiated branch pathways placing 8–11 (in Fig. 4) as the intermediates for monolignol biosynthesis. Another proposed branch from feruloyl CoA (2) to 5-hydroxyferuloyl CoA (10) for syringyl monolignol (31) also could not be substantiated, because there was no detectable hydroxylation of feruloyl CoA by either soluble or microsomal proteins from sweetgum lignifying xylem. Our results, supported by chemical, biochemical, and enzyme kinetic evidence, led us to conclude that CAld5H/COMT regulates the diversion of guaiacyl intermediates toward synthesis of syringyl monolignol. The CAld5H/COMT pathway (3 to 6) also provides clarification as to (i) why syringyl lignin is formed when sinapate is not activated to its CoA ester by 4CL (13–15); (ii) why enrichment of syringyl lignin biosynthesis in transgenic Arabidopsis is not associated with detectable ferulate 5-hydroxylation activity (11); and (iii) why abnormal 5-hydroxyconiferyl aldehyde-derived monolignols accumulate in lignin of COMT-suppressed transgenic trees (18, 32). Thus, based on the previously identified substrate preference of 4CL (13–15), CCR (33, 34), and CAD (35), these proteins, together with CAld5H and COMT, constitute a dynamic enzyme system (red arrows in Fig. 4) that efficiently mediates conversion of ferulate into the guaiacyl and syringyl monolignols, coniferyl alcohol (4), and sinapyl alcohol (7), respectively, for the biosynthesis of guaiacyl-syringyl lignin in angiosperms.
Acknowledgments
We thank C. J. Tsai and S. A. Harding for valuable comments on the manuscript. This research was supported by grants from the United States Department of Agriculture NRICGP (95-37103-2061), the United States Department of Agriculture McIntire–Stennis Forestry Research Program, and International Paper Company.
ABBREVIATIONS
- F5H
ferulate 5-hydroxylase
- CPR
cytochrome P450 reductase
- CAld5H
coniferyl aldehyde 5-hydroxylase
- COMT
caffeate O-methyltransferase
Footnotes
References
- 1.Higuchi T. Biochemistry and Molecular Biology of Wood. New York: Springer; 1997. pp. 131–233. [Google Scholar]
- 2.Sarkanen K V. In: Lignins: Occurrence, Formation, Structure and Reaction. Sarkanen K V, Ludwig C H, editors. New York: Wiley Interscience; 1971. pp. 639–694. [Google Scholar]
- 3.Chang H M, Sarkanen K V. Tappi. 1973;56:132–136. [Google Scholar]
- 4.Chiang V L, Funaoka M. Holzforschung. 1990;44:309–313. [Google Scholar]
- 5.Higuchi T, Brown S A. Can J Biochem Physiol. 1963;41:65–76. [PubMed] [Google Scholar]
- 6.Higuchi T, Brown S A. Can J Biochem Physiol. 1963;41:613–620. [PubMed] [Google Scholar]
- 7.Higuchi T, Brown S A. Can J Biochem Physiol. 1963;41:621–627. [PubMed] [Google Scholar]
- 8.Grand C. FEBS Lett. 1984;169:7–11. [Google Scholar]
- 9.Whetten R W, MacKay J J, Sederoff R R. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:585–609. doi: 10.1146/annurev.arplant.49.1.585. [DOI] [PubMed] [Google Scholar]
- 10.Meyer K, Cusumano J C, Somerville C, Chapple C C S. Proc Natl Acad Sci USA. 1996;93:6869–6874. doi: 10.1073/pnas.93.14.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer K, Shirley A M, Cusumano J C, Bell-Lelong D A, Chapple C C S. Proc Natl Acad Sci USA. 1998;95:6619–6623. doi: 10.1073/pnas.95.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapple C C S, Vogt T, Ellis B E, Somerville C. Plant Cell. 1992;4:1413–1424. doi: 10.1105/tpc.4.11.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kutsuki H, Shimada M, Higuchi T. Phytochemistry. 1982;21:267–271. [Google Scholar]
- 14.Gross G G, Mansell R L, Zenk M H. Biochem Physiol Pflanz. 1975;168:41–51. [Google Scholar]
- 15.Hu W J, Kawaoka A, Tsai C J, Lung J, Osakabe K, Ebinuma H, Chiang V L. Proc Natl Acad Sci USA. 1998;95:5407–5412. doi: 10.1073/pnas.95.9.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X-H, Chiang V L. Plant Physiol. 1997;113:65–74. doi: 10.1104/pp.113.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bugos R C, Chiang V L, Campbell W H. Plant Mol Biol. 1991;17:1203–1215. doi: 10.1007/BF00028736. [DOI] [PubMed] [Google Scholar]
- 18.Tsai C J, Popko J L, Mielke M R, Hu W J, Podila G K, Chiang V L. Plant Physiol. 1998;117:101–112. doi: 10.1104/pp.117.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Looman A C, Bodlaender J, Comstock L J, Eaton D, Thurani R, deBoer H R, Van Knippenberg P H. EMBO J. 1987;6:2489–2492. doi: 10.1002/j.1460-2075.1987.tb02530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Popko J L, Zhang X-H, Osakabe K, Tsai C J, Joshi C P, Chiang V L. Proc Natl Acad Sci USA. 1997;94:5461–5466. doi: 10.1073/pnas.94.10.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stotz A, Linder P. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- 22.Pompon D, Louerat B, Bronine A, Urban P. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- 23.Omura T, Sato R. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 24.Yasukochi Y, Masters B S S. J Biol Chem. 1976;251:5337–5344. [PubMed] [Google Scholar]
- 25.Stockigt J, Zenk M H. Z Naturforsch. 1975;30:352–358. doi: 10.1515/znc-1975-5-609. [DOI] [PubMed] [Google Scholar]
- 26.Mizutani M, Ohta D. Plant Physiol. 1998;116:357–367. doi: 10.1104/pp.116.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bland D E. Holzforschung. 1966;20:12–18. [Google Scholar]
- 28.Higuchi T. In: Biosynthesis and Biodegradation of Wood Components. Higuchi T, editor. New York: Academic; 1985. pp. 141–160. [Google Scholar]
- 29.Ruegger M, Meyer K, Cusumano J C, Chapple C C S. Plant Physiol. 1999;119:101–110. doi: 10.1104/pp.119.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mock H P, Strack D. Phytochemistry. 1993;32:575–579. [Google Scholar]
- 31.Ye Z H, Kneusel R E, Matern U, Varner J E. Plant Cell. 1994;6:1427–1439. doi: 10.1105/tpc.6.10.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Doorsselaere L, Baucher M, Chognot E, Chabbert B, Tollier M T, Petit-Conil M, Leple J C, Pilate G, Cornu D, Monties B, et al. Plant J. 1995;8:855–864. [Google Scholar]
- 33.Sarni F, Grand C, Boudet A M. Eur J Biochem. 1984;139:259–265. doi: 10.1111/j.1432-1033.1984.tb08002.x. [DOI] [PubMed] [Google Scholar]
- 34.Lacombe E, Hawkins S, Van Doorsselaere J, Piquemal J, Goffner D, Poeydomenge O, Boudet A M, Grima-Pettenati J. Plant J. 1997;11:429–441. doi: 10.1046/j.1365-313x.1997.11030429.x. [DOI] [PubMed] [Google Scholar]
- 35.Kutsuki H, Shimada M, Higuchi T. Phytochemistry. 1982;21:19–23. [Google Scholar]