Abstract
Glucocorticoids exert their effects on gene transcription through ubiquitous receptors that bind to regulatory sequences present in many genes. These glucocorticoid receptors are present in all cell types, yet glucocorticoid action is controlled in a tissue-specific way. One mechanism for this control relies on tissue-specific transcriptional activators that bind in the vicinity of the glucocorticoid receptor and are required for receptor action. We now describe a gene-specific and tissue-specific inhibitory mechanism through which glucocorticoid action is repressed by a tissue-restricted transcription factor, hepatocyte nuclear factor-6 (HNF-6). HNF-6 inhibits the glucocorticoid-induced stimulation of two genes coding for enzymes of liver glucose metabolism, namely 6-phosphofructo-2-kinase and phosphoenolpyruvate carboxykinase. Binding of HNF-6 to DNA is required for inhibition of glucocorticoid receptor activity. In vitro and in vivo experiments suggest that this inhibition is mediated by a direct HNF-6/glucocorticoid receptor interaction involving the amino-terminal domain of HNF-6 and the DNA-binding domain of the receptor. Thus, in addition to its known property of stimulating transcription of liver-expressed genes, HNF-6 can antagonize glucocorticoid-stimulated gene transcription.
Keywords: liver, gene transcription, glucocorticoid receptor, 6-phosphofructo-2-kinase, phosphoenolpyruvate carboxykinase
Glucocorticoids, an important class of steroid hormones secreted by the adrenal gland, are essential for life (1–3). Besides their role in the suppression of immune and inflammatory responses, glucocorticoids regulate carbohydrate, protein, and lipid metabolism. They increase the level of circulating glucose by stimulating the expression of rate-limiting enzymes of the gluconeogenic pathway and by decreasing glucose uptake and use (3). Glucocorticoids act via a ubiquitous intracellular receptor, called the glucocorticoid receptor (GR). The GR contains several functional domains, among which are a constitutive amino-terminal activation domain (τ1), a central DNA-binding domain (DBD), and a carboxyl-terminal ligand-binding domain that includes a ligand-dependent activation function (τ2). On steroid binding, the GR becomes activated and migrates from the cytoplasm into the nucleus where it regulates gene transcription. This regulation results from binding of the GR to glucocorticoid response elements (GRE) in specific genes or from an interaction of the GR with DNA-bound regulatory factors (4). The GR is present in all cell types, and GREs are present in many genes; however, glucocorticoid action is controlled in a tissue-specific way. Investigations into the mechanisms that mediate this specificity led to the description of glucocorticoid response units (GRU). These are gene regulatory regions in which the GRE is associated with other binding sites for ubiquitous or tissue-specific transcription factors that, through synergistic activation, enable the GR to exert its function (5). One such factor is hepatocyte nuclear factor (HNF)-3. HNF-3 cooperates with the GR to activate genes that code for proteins involved in glucose homeostasis, namely phosphoenolpyruvate carboxykinase (PEPCK), tyrosine aminotransferase, insulin-like growth factor-binding protein-1, and 6-phosphofructo-2-kinase (PFK-2; EC 2.7.1.105; refs. 6–10). No inhibitory mechanism has been described for the tissue-specific control of glucocorticoid action. Nuclear factor κB, c-jun, Nur77, and signal transducer and activator of transcription factor-5 reportedly interfere with the GR-induced stimulation of GRE-containing promoters (11–15). However, these factors exert this inhibition without binding to DNA and without tissue-specificity.
We (16) and others (17) have studied a GRU located in the first intron of the gene coding for hepatic PFK-2. This bifunctional enzyme catalyzes the synthesis and degradation of fructose 2,6-bisphosphate, a potent allosteric regulator of glycolysis and gluconeogenesis (18). We previously identified the transcription factors that bind to the pfk-2 GRU and showed that nuclear factor-I, which is ubiquitous, as well as HNF-3 and members of the CAAT/enhancer-binding protein family, which are liver-enriched, cooperate with the GR to stimulate pfk-2 gene transcription. This GRU contains a consensus binding sequence, 100 bp downstream from the GRE, for another liver-enriched transcription factor called HNF-6 (10). HNF-6 possesses a bipartite DBD consisting of a cut domain and an atypical homeodomain (Hd; ref. 19). It is the prototype of the recently defined ONECUT class of Hd proteins, which are conserved from Caenorhabditis elegans to humans (20). The aim of this work was to investigate the role of HNF-6 in the function of the pfk-2 GRU. Although HNF-6 has been identified as a transcriptional activator (19, 21, 22), we now show that HNF-6 antagonizes glucocorticoid action when bound to the GRU of the pfk-2 gene. We also show that this action of HNF-6 extends to the pepck gene where HNF-6 binding again interferes with glucocorticoid-stimulated gene transcription.
MATERIALS AND METHODS
Electrophoretic Mobility-Shift Assays (EMSAs).
Liver nuclear extracts were prepared as described (23), and wheat germ extracts were programmed according to the supplier’s instructions (Promega). EMSAs were performed as described (20) with a rat pfk-2 GRU probe (bp 114 to 137, 5′-TGAAAGTTATGGATTTTTTTTGTT-3′) or a pepck probe (bp −267 to −244, 5′-CAAAGTTTAGTCAATCAAACGTTG-3′) (the consensus HNF-6-binding sites are underlined) radiolabeled with [γ-32P]ATP (Amersham Pharmacia) by T4 polynucleotide kinase (New England Biolabs) and purified with the Quick Spin Columns from Boehringer Mannheim. The HNF-6 antiserum used was generated by immunizing a rabbit with a bacterially produced glutathione S-transferase (GST)–HNF-6 fusion protein as described (21). Immunoglobulins were precipitated from preimmune and immune serum by addition of ammonium sulfate.
Plasmids.
The luciferase reporter vectors GRU and GRUH6 were constructed by inserting the RsaI–RsaI fragment of the rat pfk-2 GRU (17) into the HindIII site of pPLLuc138 (24), located upstream of the minimal (138-bp) pfk-2 L promoter (10, 16). GRU17 and GRUG or GRUGH6 were constructed from GRU or GRUH6 by replacing the HNF-6-binding site region (bp 121 to 137, 5′-TATGGATTTTTTTTGTT-3′) or the GRE (bp 18 to 34, 5′-CAGAACTATCTGTTCCT-3′) with the 17-bp (5′-CGGAGTACTGTCCTCCG-3′) GAL4-binding site (25). The luciferase reporter vector PEPCK was constructed by inserting the rat pepck promoter (bp −600 to +69) into the BglII–HindIII sites of pXP2 (26). The expression vectors for wild-type or mutated rat HNF-6 (20), human GR (27), or human androgen receptor (pSVAR0; ref 28), have been described. A simian virus 40-driven GAL4–DBD (residues 1 to 95) expression plasmid was constructed to obtain the fusion proteins GAL4–DBD/HNF-6α, GAL4–DBD/HNF-6α Cut-Hd, and GAL4–DBD/HNF-6 ΔCut-Hd. Rat cDNAs coding for full-length HNF-6α, HNF-6α Cut-Hd, or HNF-6 ΔCut-Hd were cloned in frame with the GAL4–DBD sequence in the expression vector.
Cell Culture and Transient Transfections.
Rat hepatoma FTO-2B cells were grown as monolayers in a humidified atmosphere (5% CO2/95% air, vol/vol) in a 1:1 mixture of DMEM and Ham’s F-12 medium (GIBCO/BRL) supplemented with 10% (vol/vol) FCS. Human hepatoma HepG2 cells were grown under the same conditions in DMEM containing 10% (vol/vol) FCS. Cotransfections in these cells were carried out with LipofectAMINE PLUS (GIBCO/BRL) by using 300 ng of luciferase reporter construct, 15 ng of renilla luciferase control vector, and 15 ng of expression vector in 17.6-mm dishes for 4 h. After a 24-h treatment with dexamethasone (1 μM), dihydrotestosterone (0.1 μM), or ethanol (0.01%), cell extracts were prepared, and luciferase reporter activities were measured with a Lumac or a TD20/20 (Promega) luminometer and normalized for renilla luciferase (dual-luciferase reporter assay system, Promega). Transfection of rat hepatoma H4IIE cells has been described (29, 30). Cos-7 cells were transfected in 6-well tissue culture plates with LipofectAMINE PLUS by using 1 μg of expression vector for 4 h in DMEM. After 48 h, cells were scraped and allowed to lyse for 20 min on ice in 100 μl of lysis buffer (50 mM Tris, pH 7.4/150 mM NaCl/5 mM EDTA/1% Nonidet P-40/0.2% deoxycholate/1 mM PMSF/1 μg/ml aprotinin/1 μg/ml leupeptin). Cell extracts were cleared by centrifugation at 13,100 × g for 3 min in a refrigerated centrifuge. Protein concentrations were measured by using a Bio-Rad protein assay kit, and 20 μg of the extract was resolved on a SDS/10% PAGE gel for immunoblotting. HNF-6 proteins that were expressed by transfection were detected by chemiluminescence by using a rabbit antipeptide (amino acids 266 to 277 of HNF-6) antibody.
Protein–Protein Interaction Assays.
For the single-hybrid assays, Rat-1 cells (3 × 105 cells per 6-cm dish) were transfected in DMEM without FCS by lipofection with N-[1-(2, 3-dioleoyloxy)propyl]-N, N,N-triethylammonium methylsulfate (DOTAP, Boehringer Mannheim). After 6 h, the cells were washed with PBS and further incubated for 45 h in DMEM plus 10% (vol/vol) FCS before measuring luciferase activities. Luciferase values were normalized for protein concentration in the cell extracts. The cells were transfected with 3 μg of pHNF-6/HNF-3β(6×)-TATA-luc, 100 ng of vector coding for HNF-6α (pCMV-HNF-6α), and/or 100 ng of a vector coding for a GR–VP16 (pCMV-GR(1–565)-VP16) or a GAL4–VP16 fusion protein. pHNF-6/HNF-3β(6×)-TATA-luc contains six HNF-6-binding sites from the hnf-3β promoter (bp −141 to −127) upstream of a TATA box and the luciferase coding sequence. The amount of cytomegalovirus promoter-containing plasmid was kept identical in each transfection by addition of empty expression vector (pCMV-NH) where needed, and the total amount of plasmid (5 μg) was adjusted by addition of pGEM-3 (Promega). For the GST pull-down experiments, HNF-6α and hGRα were produced in Escherichia coli as GST fusion proteins by addition of 1 mM isopropyl-β-d-thiogalactopyranoside at 30°C for 3 h. The cells were lysed with a French press in a solution containing 150 mM NaCl, 16 mM Na2HPO4, and 4 mM NaH2PO4 (pH 7.3) and cleared by centrifugation at 6,300 × g for 10 min. Cleared lysates were incubated at 4°C on a rocking platform for 1 h with glutathione-Sepharose beads; 14C-labeled full-length recombinant HNF-6α and hGRα synthesized in vitro by using the TNT-coupled wheat germ extract and reticulocyte lysate, respectively (Promega), were incubated in buffer (20 mM Hepes, pH 7.6/150 mM KCl/0.1 mM EDTA/2.5 mM MgCl2/1 mM DTT/0.05% Nonidet P-40) at 4°C for 2 h with the immobilized fusion proteins. After extensive washing in the same buffer, the beads were boiled, and the eluate was loaded on a SDS/8% PAGE gel, which was dried and subjected to autoradiography.
RESULTS AND DISCUSSION
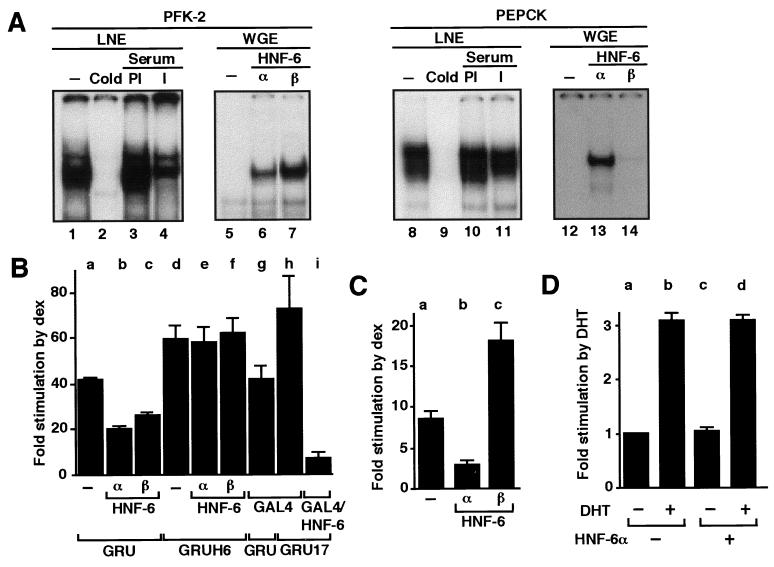
To evaluate the possible influence of HNF-6 on the expression of the pfk-2 gene, we first determined whether HNF-6 binds to the PFK-2 GRU. In EMSAs, liver nuclear extracts produced a complex, the formation of which was prevented by an excess of unlabeled probe and by an antiserum (21) raised against HNF-6 (Fig. 1A, lanes 1–4). There are two hepatic isoforms of HNF-6, α (465 residues) and β (491 residues), which differ in the length of the linker between the cut domain and the Hd and in their DNA-binding properties (20). Both HNF-6 isoforms bound to the pfk-2 GRU (Fig. 1A, lanes 5–7).
Figure 1.
Binding of HNF-6 to the GRU inhibits glucocorticoid stimulation of the pfk-2 and pepck genes. (A) Radiolabeled oligonucleotide probes containing the HNF-6-binding sites from the pfk-2 or pepck GRU were incubated with liver nuclear extracts (LNE) or with wheat germ extracts (WGE) programmed or not to synthesize HNF-6α or β. A 50-fold excess of cold probe, the serum of a rabbit immunized (I) against recombinant HNF-6, or preimmune serum (PI) was added as indicated. For lanes 1 and 8, no competitor or serum was added; for lanes 5 and 12, unprogrammed extracts were used. (B) FTO-2B cells were transiently cotransfected with expression vectors for the recombinant proteins indicated and with a luciferase reporter construct driven by the pfk-2 promoter linked to the intact pfk-2 GRU (GRU) or to the GRU in which the HNF-6-binding site has been disrupted (GRUH6) or replaced by a GAL4-binding site (GRU17). After transfection, the cells were treated for 24 h with dexamethasone and assayed for luciferase activity as described (10, 15). (C) H4IIE cells were treated as in B, except that the reporter construct was driven by the pepck promoter and the cells were exposed for 16 h to dexamethasone. (D) FTO-2B cells were transiently transfected with a luciferase reporter construct driven by the pfk-2 promoter linked to the intact pfk-2 GRU and with expression vectors for the androgen receptor and for HNF-6α as indicated. After transfection, the cells were treated for 24 h with dihydrotestosterone (DHT) as indicated. Values are means ± SEM from three to five experiments.
The functional consequence of HNF-6 binding to the GRU was studied by transfection as described in the legend to Fig. 1B. Dexamethasone, a GR agonist, produced the expected GRU-dependent stimulation of the pfk-2 promoter (Fig. 1B, bar a). Disruption of the HNF-6-binding site in the GRU (GRUH6), verified by in vitro footprinting (data not shown), enhanced the glucocorticoid effect (Fig. 1B, bar d). This result suggested that endogenous HNF-6 is inhibitory. Overexpression of HNF-6α or β reduced the glucocorticoid-stimulated expression of the wild-type reporter (Fig. 1B, bars b and c) and was without effect on the construct that cannot bind HNF-6 (Fig. 1B, bars e and f). This result indicated that HNF-6 must bind to the GRU to exert its inhibitory effect. If so, the same inhibition was predicted to occur when exogenous HNF-6 is anchored to the GRU. We therefore replaced the HNF-6-binding site in the GRU with a binding site for the DBD of the yeast GAL4 transcription factor. The glucocorticoid-stimulated expression of this construct, designated GRU17, was increased by the HNF-6/GAL4 site substitution (Fig. 1B, bar h), in the same manner observed after disruption of the HNF-6-binding site (Fig. 1B, bar d). Consistent with the prediction, expression of a GAL4–DBD/HNF-6 fusion protein markedly reduced the glucocorticoid-stimulated expression of the GRU17 pfk-2 promoter construct (Fig. 1B, bar i), whereas expression of the GAL4–DBD alone had no effect (Fig. 1B, bar g).
The GRU located in the promoter of the gene coding for PEPCK (31), a rate-controlling enzyme of gluconeogenesis, also contains an HNF-6-binding site located 100 bp downstream from the GRE. Consistent with earlier work (32), several specific complexes were observed in EMSA with liver nuclear extracts by using a probe corresponding to the pepck HNF-6 site (ref. 19; Fig. 1A, lane 8). This site is localized in the P3II footprint region previously shown to bind CAAT/enhancer-binding protein family members and activator protein-1 (33). We identified the lower band as HNF-6, based on the use of the anti-HNF-6 serum (Fig. 1A, lanes 10 and 11). Recombinant HNF-6α, but not β, bound to this site (Fig. 1A, lanes 12–14). As the sequence of this site differs from the HNF-6 site in the pfk-2 GRU, this result was not surprising in view of the differences in DNA recognition properties of HNF-6α and β (20). We therefore predicted that overexpressed HNF-6α, but not β, would inhibit the stimulation of the pepck promoter by dexamethasone. This prediction held true (Fig. 1C). In fact, HNF-6β amplified the glucocorticoid response by an unknown mechanism.
To test whether the inhibitory effect of HNF-6 was specific to the GR, we verified that androgen receptor action was not inhibited by HNF-6α. Indeed, the androgen receptor stimulated the pfk-2 GRU in the presence of dihydrotestosterone (Fig. 1D, bars a and b), consistent with earlier work (34). However, this androgen-mediated stimulation was not inhibited by overexpression of HNF-6α (Fig. 1D, bars c and d).
Having found that HNF-6 can inhibit glucocorticoid action when it binds in the vicinity of the GRE, we next delineated the region of HNF-6 that mediates this effect. Deletion mutants of HNF-6 were tested in transfected cells for their ability to inhibit the GRU-dependent, dexamethasone-induced, transcriptional stimulation of the PFK-2 promoter-luciferase reporter construct. These mutants (Fig. 2A) were all expressed at similar levels, as determined by immunoblotting and EMSA (Fig. 2 B and C). Fig. 2 D and E show that the domain responsible for the antiglucocorticoid activity resides in the amino-terminal half of HNF-6. Indeed, deletion of this portion of the protein abolished the inhibitory effect of HNF-6 (Fig. 2D, bar g). Furthermore, when fused to a GAL4–DBD and targeted to the GAL4 site in the GRU17 reporter construct, the amino-terminal half of HNF-6 inhibited the effect of dexamethasone as well as intact HNF-6 (Fig. 2E, bars j and k). Deletion of the Hd, the cut domain, or both also abolished the antiglucocorticoid effect of HNF-6 (Fig. 2D, bars e, f, and h). However, this observation is probably explained by the fact that deletion of the cut domain abolished binding of HNF-6 to DNA (Fig. 2C, lane f) and that deletion of the Hd markedly reduced DNA binding (Fig. 2C, lane e). Moreover, these domains were not inhibitory when fused to the GAL4–DBD (Fig. 2E, bar l), suggesting that the DBD of HNF-6 is required only for anchoring the amino-terminal inhibitory region to DNA.
Figure 2.
The amino-terminal segment of HNF-6 inhibits glucocorticoid action. (A) Structure of the recombinant proteins tested. The lowercase letters refer to the same proteins in all panels. (B) Immunoblot of the recombinant proteins with an anti-HNF-6 antibody. Recombinant protein g was not detected, because it lacks the epitopes recognized by this antibody; however, it was detected by EMSA (see C). (C) EMSA of the recombinant proteins with the HNF-6-binding site in the pfk-2 GRU as a probe. Asterisks indicate specific complexes. No specific band was seen with the empty vector or with recombinant proteins f and h, because they lack the DBD(s). Recombinant proteins f and h were detected by immunoblotting (see B). Recombinant protein g gave a fast-migrating complex because of its much smaller size. (D) Effect of the overexpressed recombinant proteins on the glucocorticoid response of the GRU luciferase reporter construct measured as described for Fig. 1B. Tests were performed in transfected FTO-2B cells. For recombinant protein a, stimulation by dexamethasone was 40-fold (see Fig. 1B). Values are means ± SEM from three or four experiments. (E) Effect of GAL4–DBD (bar i) or the indicated fusion proteins (bars j, k, and l) on the glucocorticoid response of the GRU17 luciferase reporter constructs in transfected FTO-2B cells. Stimulation by dexamethasone in the presence of GAL4–DBD was 40-fold. Values are means ± SEM from three to five experiments.
To investigate whether HNF-6 inhibits glucocorticoid action by interacting with the GR, we used an in vivo single-hybrid test. We determined, by transfection, whether HNF-6α could recruit a GR–VP16 fusion protein to a reporter gene that contains HNF-6-binding sites but not GR-binding sites. The GR–VP16 protein contains the activation domain of the viral transcription factor VP16 (amino acids 412 to 490) fused downstream of amino acids 1 to 565 of the GR (GR devoid of ligand-binding domain). Transfection of HNF-6 in Rat-1 fibroblasts, which have no endogenous HNF-6, increased reporter gene activity (Fig. 3A), consistent with earlier results (20). Transfection of GR–VP16 alone did not affect the activity of the reporter gene significantly. In contrast, cotransfection of HNF-6 with GR–VP16 reproducibly stimulated transcription much more than HNF-6 alone (Fig. 3A Left), suggesting that HNF-6 interacts with the GR and recruits it to the reporter gene. To determine whether HNF-6 recruits GR–VP16 via the GR portion of the fusion protein and not via the VP16 activation domain, the GR portion was replaced by the GAL4–DBD (residues 1 to 148) to make a GAL4–VP16 fusion protein. GAL4–VP16 was not recruited by HNF-6 to the reporter construct, as determined by the fact that it did not amplify the effect of HNF-6 in the same experiments (Fig. 3A Right). This result showed that the interaction between HNF-6 and GR–VP16 is GR-specific. To confirm an interaction between HNF-6 and the GR further, we performed in vitro GST pull-down experiments (Fig. 3B). These experiments showed that matrix-immobilized HNF-6 can interact with GR and vice versa.
Figure 3.
Interaction between HNF-6 and the GR. (A) Rat-1 fibroblasts were transiently transfected with a reporter containing the luciferase gene driven by a TATA box and six HNF-6-binding sites, in the presence or absence of expression vectors coding for HNF-6α or for the GR–VP16 and GAL4–VP16 fusion proteins as indicated. Representative experiments performed in duplicate are shown. (B) Bacterially expressed GST and the GST–HNF-6α and GST–GR fusion proteins were bound to glutathione-Sepharose beads; 14C-labeled GR or 14C-labeled HNF-6α produced in a wheat germ extract was incubated as indicated for 2 h at 4°C with the beads, which were then washed and processed for SDS/PAGE followed by autoradiography. An aliquot (1/10 of the input in the incubation mixture) of the radioactive proteins was run as a control (lanes 1 and 8).
Because HNF-6 binds to the GR, we next sought to delineate the GR domain necessary for this interaction. Mutants of GR were used to define the region required for the repression of the glucocorticoid-stimulated GRU reporter construct by endogenous HNF-6. In cells transfected with GR mutants, the glucocorticoid response of the wild-type pfk-2 promoter construct was compared with that of a construct that lacks the HNF-6 site (GRUH6). We reasoned that a glucocorticoid-mediated increase of reporter gene expression seen with the GRUH6 construct, as compared with the wild-type GRU construct, would indicate that the transfected GR is still inhibited by endogenous HNF-6. HepG2 cells were used for these experiments, because the induction of reporter gene expression by glucocorticoids is lower than in FTO-2B cells in the absence of cotransfected GR, thus allowing an easier detection of the effect of exogenous GR. Fig. 4A shows the structure of the transfected GR mutants and of the GRU contained in the reporter constructs. The GR mutants had not lost their ability to stimulate the pfk-2 GRU (Fig. 4B), an observation consistent with earlier reports in which other reporter constructs were employed (27). Fig. 4B shows that, in the absence of exogenous GR, the GRUH6 reporter responded better than the wild-type GRU to dexamethasone (Fig. 4B, bars a and b), as expected from the loss of the DNA-dependent inhibitory interaction between HNF-6 and the endogenous GR. This phenomenon was amplified after cotransfection with exogenous GR (Fig. 4B, bars c and d). Similarly, the response caused by transfection of a GR lacking the carboxyl-terminal half (Fig. 4B, bars e and f) or the amino-terminal half of the molecule (Fig. 4B, bars g and h) was inhibited by HNF-6 binding to the GRU. Both of these two GR mutants retain the DBD; thus, we hypothesized that this domain is the target of HNF-6 action. It is impossible to confirm this hypothesis by simply deleting the DBD from the transfected GR, because this mutant would be unable to bind DNA and stimulate transcription. We therefore replaced the GRE in the wild-type GRU and GRUH6 reporters with a GAL4-binding site and then cotransfected a GR mutant in which the DBD has been replaced by the GAL4–DBD (27). This GR mutant activated these substituted (GRUG and GRUGH6) reporters but was insensitive to HNF-6 (Fig. 4B, bars i and j), thereby confirming the requirement of the GR DBD for the antiglucocorticoid activity of HNF-6.
Figure 4.
The target of HNF-6 is the DBD of the GR. (A) Structure of the GR and GRU constructs. LBD, ligand-binding domain; τ1 and τ2, transactivation domains. In GR–GAL4–GR, the DBD of the GR was replaced by the DBD of GAL4. (B) Activity of wild-type and mutated GR on reporter genes driven by a wild-type or mutated GRU. After transient transfection, HepG2 cells were treated with dexamethasone (except after cotransfection with GR1–488, which lacks the ligand-binding domain and is constitutively active) before determination of luciferase activity as described in Fig. 1B. Data are shown as relative values in the graph, and the absolute values corresponding to 100% are given on the right of the graph. Values are means ± SEM from three to six experiments.
It is known that HNF-6 is a transcriptional activator when bound to certain gene promoters (19–21, 35). Here, we describe an inhibitory action of HNF-6 that results from its binding to a GRU. This effect involves the amino-terminal portion of HNF-6 and the DBD of the GR. Our data suggest that a direct interaction between HNF-6 and the GR plays a role in this inhibitory action. HNF-6 expression is controlled by tissue-specific transcription factors (21, 36), by developmental cues (21, 35), and by growth hormone in liver (37). This tissue-specific, developmental and hormonal regulation of HNF-6 expression could indirectly modulate glucocorticoid action via the mechanism described here. Glucocorticoids are potent antiinflammatory drugs, but their clinical use is plagued by complications that include perturbations of glucose metabolism. The discovery of the antiglucocorticoid activity of HNF-6 might help in the design of modulators of the pharmacological actions of glucocorticoids that would limit their impact in the tissues that express HNF-6, e.g., the liver, while allowing their therapeutic effect in other tissues.
Acknowledgments
We thank S. Neou, S. Durviaux, and Y. Peignois for help, as well as M. Müller, G. Verhoeven, J. Trapman, and K. R. Yamamoto for plasmids. C.E.P and D.D. hold a fellowship from the Fonds pour la Formation à la Recherche dans l’Industrie et l’Agriculture (Belgium), and F.P.L. is Senior Research Associate of the Fonds National de la Recherche Scientifique (Belgium). This work was supported by grants from the Poles d’Attraction Interuniversitaires, the Délégation Générale Higher Education and Scientific Research, the Fonds National de la Recherche Scientifique (Belgium), and the Fonds de la Recherche Scientifique Médicale (Belgium), as well as by National Institutes of Health Grants DK35107 and DK20593 (to The Vanderbilt Diabetes Research and Training Center).
ABBREVIATIONS
- GR
glucocorticoid receptor
- DBD
DNA-binding domain
- GRE
glucocorticoid response element
- GRU
glucocorticoid response unit
- HNF
hepatocyte nuclear factor
- PEPCK
phosphoenolpyruvate carboxykinase
- PFK-2
6-phosphofructo-2-kinase
- EMSA
electrophoretic mobility-shift assay
- Hd
homeodomain
- GST
glutathione S-transferase
References
- 1.Pearce D, Yamamoto K R. Science. 1993;259:1161–1165. doi: 10.1126/science.8382376. [DOI] [PubMed] [Google Scholar]
- 2.Beato M, Herrlich P, Schütz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 3.Karin M. Cell. 1998;93:487–490. doi: 10.1016/s0092-8674(00)81177-0. [DOI] [PubMed] [Google Scholar]
- 4.Reichardt H M, Kaestner K H, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, et al. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 5.Lucas P C, Granner D K. Annu Rev Biochem. 1992;61:1131–1173. doi: 10.1146/annurev.bi.61.070192.005411. [DOI] [PubMed] [Google Scholar]
- 6.Ganss R, Weih F, Schütz G. Mol Endocrinol. 1994;8:895–903. doi: 10.1210/mend.8.7.7984151. [DOI] [PubMed] [Google Scholar]
- 7.Suwanichkul A, Allander S V, Morris S L, Powell D R. J Biol Chem. 1994;269:30835–30841. [PubMed] [Google Scholar]
- 8.Sassi H, Fromont-Racine M, Grange T, Pictet R. Proc Natl Acad Sci USA. 1995;92:7197–7201. doi: 10.1073/pnas.92.16.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J-C, Stromstedt P-E, O’Brien R M, Granner D K. Mol Endocrinol. 1996;10:794–800. doi: 10.1210/mend.10.7.8813720. [DOI] [PubMed] [Google Scholar]
- 10.Pierreux C E, Urso B, De Meyts P, Rousseau G G, Lemaigre F P. Mol Endocrinol. 1998;12:1343–1354. doi: 10.1210/mend.12.9.0172. [DOI] [PubMed] [Google Scholar]
- 11.Schüle R, Rangarajan P, Kliewer S, Ransone L J, Bolado J, Yang N, Verma I M, Evans R M. Cell. 1990;62:1217–1226. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- 12.Yang-Yen H-F, Chambart J-C, Sun Y-L, Smeal T, Schmidt T J, Drouin J, Karin M. Cell. 1990;62:1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- 13.Philips A, Maira M, Mullick A, Chamberland M, Lesage S, Hugo P, Drouin J. Mol Cell Biol. 1997;17:5952–5959. doi: 10.1128/mcb.17.10.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stöcklin E, Wissler M, Gouilleux F, Groner B. Nature (London) 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 15.McKay L I, Cidlowski J A. Mol Endocrinol. 1998;12:45–56. doi: 10.1210/mend.12.1.0044. [DOI] [PubMed] [Google Scholar]
- 16.Zimmermann P L, Pierreux C E, Rigaud G, Rousseau G G, Lemaigre F P. DNA Cell Biol. 1997;16:713–723. doi: 10.1089/dna.1997.16.713. [DOI] [PubMed] [Google Scholar]
- 17.Lange A J, Espinet C, Hall R, El-Maghrabi M R, Vargos A M, Miksicek R J, Granner D K, Pilkis S J. J Biol Chem. 1992;267:15673–15680. [PubMed] [Google Scholar]
- 18.Rousseau G G, Hue L. Prog Nucleic Acid Res Mol Biol. 1993;45:99–127. doi: 10.1016/s0079-6603(08)60868-5. [DOI] [PubMed] [Google Scholar]
- 19.Lemaigre F P, Durviaux S M, Truong O, Lannoy V L, Hsuan J J, Rousseau G G. Proc Natl Acad Sci USA. 1996;93:9460–9464. doi: 10.1073/pnas.93.18.9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lannoy V, Bürglin T, Rousseau G G, Lemaigre F P. J Biol Chem. 1998;273:13552–13562. doi: 10.1074/jbc.273.22.13552. [DOI] [PubMed] [Google Scholar]
- 21.Landry C, Clotman F, Hioki T, Oda H, Picard J J, Lemaigre F P, Rousseau G G. Dev Biol. 1997;192:247–257. doi: 10.1006/dbio.1997.8757. [DOI] [PubMed] [Google Scholar]
- 22.Spek C A, Lannoy V J, Lemaigre F P, Rousseau G G, Bertina R M, Reitsma P H. J Biol Chem. 1998;273:10168–10173. doi: 10.1074/jbc.273.17.10168. [DOI] [PubMed] [Google Scholar]
- 23.Hattori M, Tugores A, Veloz L, Karin M, Brenner D A. DNA Cell Biol. 1990;9:777–781. doi: 10.1089/dna.1990.9.777. [DOI] [PubMed] [Google Scholar]
- 24.Lemaigre F P, Durviaux S M, Rousseau G G. Mol Cell Biol. 1991;11:1099–1106. doi: 10.1128/mcb.11.2.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webster N, Jin J R, Green S, Hollis M, Chambon P. Cell. 1988;52:169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- 26.Nordeen S K. BioTechniques. 1988;6:454–457. [PubMed] [Google Scholar]
- 27.Müller M, Baniahmad C, Kaltschmidt C, Renkawitz R. Mol Endocrinol. 1991;5:1498–1503. doi: 10.1210/mend-5-10-1498. [DOI] [PubMed] [Google Scholar]
- 28.Brinkmann A O, Faber P W, van Rooij H C J, Kuiper G G J M, Ris C, Klaassen P, van der Korput J A G M, Voorhorst M M, van Laar J H, Mulder E, et al. J Steroid Biochem. 1989;34:307–310. doi: 10.1016/0022-4731(89)90098-8. [DOI] [PubMed] [Google Scholar]
- 29.Peterson D D, Magnuson M A, Granner D K. Mol Cell Biol. 1988;8:96–104. doi: 10.1128/mcb.8.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall R K, Scott D K, Noisin E L, Lucas P C, Granner D K. Mol Cell Biol. 1992;12:5527–5535. doi: 10.1128/mcb.12.12.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imai E, Stromstedt P-E, Quinn P G, Carlstedt-Duke J, Gustafsson J-A, Granner D K. Mol Cell Biol. 1990;10:4712–4719. doi: 10.1128/mcb.10.9.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roesler W J, Vandenbark G R, Hanson R W. J Biol Chem. 1989;264:9657–9664. [PubMed] [Google Scholar]
- 33.Roesler W J, Simard J, Graham J G, McFie P. J Biol Chem. 1994;269:14276–14283. [PubMed] [Google Scholar]
- 34.McFarlan S C, Zhang Q, Miksicek R J, Lange A. Mol Cell Endocrinol. 1997;129:219–227. doi: 10.1016/s0303-7207(97)00069-5. [DOI] [PubMed] [Google Scholar]
- 35.Rausa F, Samadani U, Ye H, Lim L, Fletcher C F, Jenkins N A, Copeland N G, Costa R H. Dev Biol. 1997;192:228–246. doi: 10.1006/dbio.1997.8744. [DOI] [PubMed] [Google Scholar]
- 36.Rastegar M, Szpirer C, Rousseau G G, Lemaigre F P. Biochem J. 1998;334:565–569. doi: 10.1042/bj3340565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lahuna O, Fernandez L, Karlsson H, Maiter D, Lemaigre F P, Rousseau G G, Gustafsson J-A, Mode A. Proc Natl Acad Sci USA. 1997;94:12309–12313. doi: 10.1073/pnas.94.23.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]