Abstract
The structure of a stable recombinant ectodomain of influenza hemagglutinin HA2 subunit, EHA2 (23–185), defined by proteolysis studies of the intact bacterial-expressed ectodomain, was determined to 1.9-Å resolution by using x-ray crystallography. The structure reveals a domain composed of N- and C-terminal residues that form an N cap terminating both the N-terminal α-helix and the central coiled coil. The N cap is formed by a conserved sequence, and part of it is found in the neutral pH conformation of HA. The C-terminal 23 residues of the ectodomain form a 72-Å long nonhelical structure ordered to within 7 residues of the transmembrane anchor. The structure implies that continuous α helices are not required for membrane fusion at either the N or C termini. The difference in stability between recombinant molecules with and without the N cap sequences suggests that additional free energy for membrane fusion may become available after the formation of the central triple-stranded coiled coil and insertion of the fusion peptide into the target membrane.
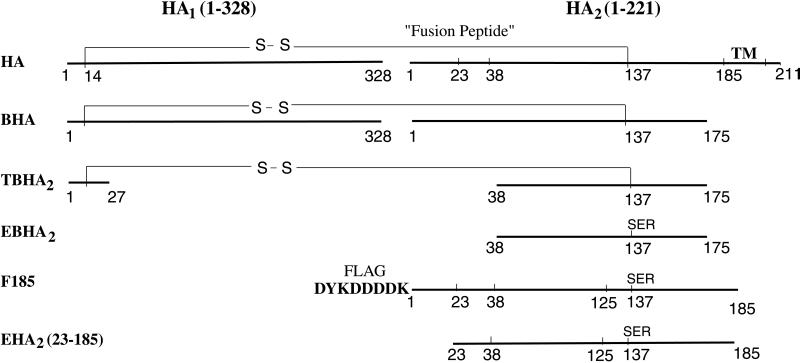
Influenza virus hemagglutinin (HA) is synthesized as a single polypeptide of ≈550 aa with a membrane-anchor sequence near its C terminus. It undergoes posttranslational cleavage into two disulfide-linked chains, HA1 and HA2 (Fig. 1). The cleavage generates a hydrophobic N terminus of HA2 (the fusion peptide) (1) and is essential for virus infection (2, 3). Soluble neutral pH HA (BHA, Fig. 1) can be released from virus by bromelain digestion, which removes HA2 residues 176–221, including the transmembrane anchor (residues 186–211) (4). In 1981, the structure of BHA was determined by using x-ray crystallography (5, 6). At the low pH of endosomes (pH 5.0), the HA undergoes a major conformational change (7) that activates membrane fusion required during viral entry. BHA incubated at pH 5.0 irreversibly acquires hydrophobic properties (7, 8). It can be solubilized by trypsin and thermolysin cleavage, generating a trimeric fragment (TBHA2, Fig. 1) containing residues 1–27 of HA1 disulfide linked to residues 38–175 of HA2 (9, 10).
Figure 1.
Schematic of influenza virus HA and HA2 sequences. EHA2 (E. coli-expressed HA2 residues 23–185) is the protein structure determined here. TM, transmembrane anchor; S—S, interchain disulfide bond from HA1 Cys-14 to HA2 Cys-137; Flag octapeptide (in F185), bold.
The x-ray crystal structure of TBHA2 (11) defined the details of a conformational rearrangement throughout the length of the HA2 chain, one part of which had been proposed from studies of model peptides (12). Three major refolding events occurred. (i) An extended loop and short α-helix became an extension of the central triple-stranded coiled coil found in native HA, relocating the fusion peptide over 100 Å from its previously buried position (11, 12); (ii) The middle of the long α-helix of the native HA2 coiled coil unfolded to form a reverse turn, and the second half of the long α-helix jackknifed back to lie antiparallel against the first half (11), also relocating residues by over 100 Å in relation to coiled-coil residues 76–105, which form a common structure in both conformations. (iii) Residues 141–175 C-terminal to a small β-sheet hairpin that accompanied the jackknifed α-helix appeared to be extruded from their compact association in native HA2 to become a mostly extended structure that reached about three-fourths of the way down the central coiled coil packing antiparallel within the groove between adjacent α-helices (11). The overall effect of this refolding is apparently to deliver the fusion peptide toward the target membrane and to bend the molecule in half so that the fusion peptide and the viral membrane anchor are near the same end of the rod-shaped molecule (11, 13, 14).
Similarities to the fusion-pH induced conformation of TBHA2 have been found in recombinant ectodomains or peptide fragments of the fusion proteins of other viruses, including the retroviruses Moloney murine leukemia virus, HIV-1, SIV-1, and human T-lymphotrophic virus type 1, the filovirus, Ebola virus, and the paramyxovirus, SV5 (14–22). A related rod-shaped structure has been found for the v- and t-SNARE complex involved in synaptic fusion (23–25). In each case, the termini expected to be embedded in the two prefusion membranes are found at one end of a rod-shaped molecule containing an α-helical coiled coil. The formation of these rod-like structures has been suggested to provide both a mechanism for the close apposition of the prefusion membranes and a driving force for the fusion of the bilayers (14, 25, 26).
In TBHA2 crystals (HA2 residues 38–175 in the fusion-pH conformation), the C terminus of one monomer is disordered beyond residue 153, and the other two monomers are disordered beyond residue 162. The absence of HA2 residues 1–37 from this fragment also limits our knowledge of the locations of the N-terminal fusion peptides. The present structure determination addresses both of these regions.
Previously, we described (27) the spontaneous folding and trimerization of the HA2 ectodomain, EBHA2 (residues 38–175) (Fig. 1), which is expressed in bacteria in the absence of HA1 and structurally similar to TBHA2. [Shorter fragments have also been reported and their fusion activity studied (28, 29).] More recently, we described a soluble chimera of the complete HA2 ectodomain (residues 1–185), F185, with the highly polar Flag octapeptide at its N terminus (30) that data also indicated has the same coiled-coil structure as the fusion-pH conformation of viral HA.
Here, we have identified a core structure of the ectodomain of HA2 (EHA2) expressed in bacteria (Fig. 1) by incubating F185 with a number of proteases. Residues corresponding to this stable domain (23–185) were then expressed in Escherichia coli, and the structure was determined to 1.9-Å resolution. An N capping domain was found that suggests that α-helices are not required at the linking regions to either prefusion membrane. The N cap domain also presents a potential source of free energy for driving the apposition and fusion of membranes available after the insertion of the fusion peptide by the extended coiled coil.
MATERIALS AND METHODS
Proteolysis, Expression, and Crystallization.
Thermolysin, trypsin, α-chymotrypsin, elastase, and V8-protease digestions (Fig. 1) were performed (37°C) at protease/F185 ratios of 1:20 (wt/wt) in TBS with 2 mM CaCl2. Aliquots were inactivated by boiling in SDS sample buffer after 30 min and 2.5 hr incubation and analyzed by SDS/ 15% PAGE. Control F185 was incubated without protease at 37°C for 2.5 hr.
DNA encoding HA2 residues 23–185 was cloned (PLM1 vector) and expressed, and the protein was purified by using a heat treatment as described (27) and an added affinity column of protein A Sepharose (Sigma) coupled with mAb IIF4 (31). Concentrated protein (Centricon-30; Amicon) at 10 mg/ml in 10 mM Hepes (pH 7.5) was crystallized by hanging drop vapor diffusion. Droplets were made by 1:1 dilution of the protein with the well solution (2% PEG 4000, pH 4.4, 30–150 mM NaCl).
Crystals were harvested into 8% PEG 4000 (pH 4.4) and serially transferred into cryobuffer containing 40% PEG 400 (pH 4.4) (five steps soaking, 30 min per step, 4°C).
X-Ray Crystallography.
Two crystal forms with space group C2 were grown under the same conditions, one with a c axis length twice the other (Table 1). X-ray data were collected at 100 K on PhosphorImage plates at the CHESS F2 beamline and integrated by using the program denzo (HKL Research). Residues 40–162 of HA2 from the structure of TBHA2 (11) and data from 10.0–3.5 Å were used to locate the molecule in the form I crystal by using amore (32). One trimer was located with a translation solution 5 σ higher than the next peak (correlation coefficient = 0.54, r = 0.48). Model refinement with cns (33), three-fold ncs phase improvement with dm (34), and model building in o (35) yielded a model containing residues 45–166 (Rwork = 0.36 and Rfree = 0.37) with no clear electron density for the remaining residues.
Table 1.
Data collection, structural, and refinement statistics
| Form I | Form II | |||
| Unit cell dimensions | ||||
| a, b, c, Å | 113.54, 49.7, 111.53 | 113.24, 48.88, 221.30 | ||
| α, β, γ, ° | 90.0, 103.91, 90.0 | 90.0, 103.26, 90.0 | ||
| Resolution | 35.0–2.5 Å | 2.59–2.5 Å | 35.0–1.9 Å | 1.97–1.90 Å |
| Rmerge, %* | 0.065 | 0.243 | 0.09 | 0.277 |
| Completeness, % | 94.2 | 61.3 | 94.2 | 83.9 |
| >3σI | 77.0 | 24.5 | 66.8 | 21.9 |
| Number of reflections | 20,013 | 1,292 | 87,961 | 7,801 |
| Average redundancy | 3.8 | 1.7 | 2.6 | 1.5 |
| Refinement, Form II | ||||
| Number of reflections | 82,895 | |||
| Rcryst,†% | 0.223 (0.332)‡ | |||
| Rfree,†% | 0.248 (0.337)‡ | |||
| Average B, Å2 | 46.7 | |||
| rms deviations from ideal | ||||
| Bond lengths, Å | 0.014 | |||
| Bond angles, ° | 1.47 | |||
| % in most favored region§ | 93.0 | |||
| % in additional allowed regions§ | 6.7 | |||
Rmerge = ΣhΣi|Ii(h) − 〈I(h〉)|/ΣhΣiIi(h), where Ii(h) is the ith measurement, and 〈I(h)〉 is the weighted mean of all measurements of I(h).
Rcryst and Rfree = Σh||F(h)obs|−|F(h)calc||/Σh|F(h)obs| for reflections in the working and test sets, respectively. Test set is 6% of the data selected at random.
Numbers in parentheses are for final shell 1.97–1.90 Å.
As defined in procheck (41).
The refined partial model from crystal form I (residues 45–166) and data from 10.0–3.5 Å were used to locate the molecules in the form II crystal by using amore (32). A translational search located the first trimer (solution 4 σ above the next peak, correlation coefficient = 0.23, r = 0.54). A large peak at (−0.023, 0.0, 0.503) (fractional coordinates) in the native Patterson function suggested that the two trimers in the asymmetric unit are related by a simple translation. PC refinement of the two trimers and a translational search with xplor (36) refined the location of both trimers (second solution 5 σ above next highest; r = 0.44).
The phases were improved by density modification and ncs averaging with dm (34) by using the threefold rotational symmetry of the two trimers in crystal form II. The sixfold averaged monomer electron density allowed the model to be extended to residues 37–168. Because the two crystal forms were related, form II having a noncrystallographic translation along the c axis approximately identical to the crystallographic translation in form I, all three unique trimers were packed in almost identical environments. Phase improvement by density modification and threefold translational averaging from both crystals of the three unique trimers revealed more of the model, including elements of the trimers effected by crystal packing and not conforming to the molecular threefold symmetry axis [using mave (37)]. Cycles of model building and refinement using torsion angle dynamics and a maximum-likelihood target were performed with form II data from 35.0- to 1.9-Å resolution, with a bulk solvent correction and group and individual B factor refinement [with cns (36) and o (35)]. The final model contains 883 residues, trimer 1: residues 33–178; 33–178; 31–185; trimer 2: residues 34–175; 33–176; 36–185, and 782 solvent molecules (Table 1).
RESULTS
Defining, Producing, and Crystallizing EHA2 (23–185), a Recombinant HA2 Ectodomain.
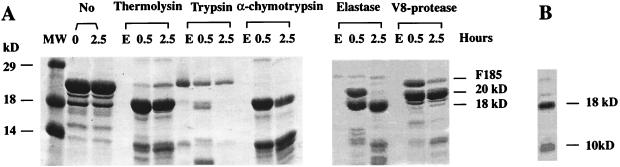
A stable ectodomain of HA2 was first defined by proteolysis of a Flag-solubilized, intact ectodomain and then produced by expression in bacteria. Five proteases with a broad range of specificities were incubated with the bacterially expressed complete ectodomain (including fusion peptide) of HA2 solubilized by the N-terminal addition of the very polar Flag octapeptide (MDYKDDDDK), F185 (Fig. 1) (30). SDS/PAGE analysis of the proteolytic products showed that F185 is sensitive to digestion by all five enzymes (Fig. 2A). Digestion of F185 with thermolysin, α-chymotrypsin, or elastase all yielded fragments of ≈18 and ≈10 kDa. Trypsin completely degraded the protein in 2.5 hr. Digestion with V8-protease yielded a ≈20-kDa fragment. Degradation of F185 to two fragments of ≈18 kDa and ≈10 kDa was also observed on storage at 4°C for 60 days, presumably resulting from digestion by a contaminating bacterial protease (Fig. 2B). All of these results suggested that the ≈18-kDa proteolysis product is a stable, well folded domain potentially suitable for crystallization. The thermolytic digestion product was identified as residues 24–185 by mass spectrometry and N-terminal sequencing (data not shown), indicating that the molecule had only been cleaved at the N terminus with the removal of the Flag octapeptide and residues 1–23, which includes the nonpolar fusion peptide. Mild thermolysin digestion of viral HA incubated at fusion-pH, pH 5.0, also generated a fragment with this N terminus (10).
Figure 2.
Proteolysis of the F185 ectodomain of HA2 defined a stable EHA2 (23–185) ectodomain. (A) SDS/PAGE of digestion products of F185 (Fig. 1) with thermolysin, trypsin, α-chymotrypsin, elastase, and V8-protease. The first two lanes are controls without protease. Lanes labeled E are the proteases alone. (B) SDS/PAGE analysis of degradation products of F185 stored at 4°C for 60 days. The full length protein (22 kDa) was completely degraded to ≈18 kDa and ≈10 kDa bands. Fig. prepared with molscript (42).
To obtain a large quantity of HA2 (23–185) for crystallographic study, a protein consisting of these residues was expressed in E. coli and purified by using methods previously described for recombinant EBHA2 (38–175) (Fig. 1) (27).
Refinement of the crystallization conditions at 12°C yielded large single crystals with average dimensions of 400 × 150 × 100 μm from 2% PEG 4000, 50 mM NaCl, and 50 mM Na acetate (pH 4.4).
X-Ray Crystal Structure Determination.
The x-ray crystal structure of EHA2(23–185) was determined by molecular replacement and iterative real-space phase averaging by using two crystal forms and refined to 1.9-Å resolution (Rwork = 0.223; Rfree = 0.248). Trimers of EHA2 were packed very similarly in two crystal forms, with one trimer per asymmetric unit in one crystal and two trimers related by an approximate translation in the second crystal form. The molecular-replacement phases were first improved by iterative sixfold real space averaging of the monomer in the two trimers in crystal form I, within an envelope encompassing residues 40–162, which had been defined earlier in the viral TBHA2 (Fig. 1) structure. Iterative threefold translational averaging of the three trimers, two in one crystal and one in the other revealed the new parts of the structure. Because the crystal packing of each of the three unique trimers was almost identical, the translational averaging defined crystal contacts and other deviations from the molecular threefold rotational symmetry of the trimer.
X-ray diffraction data from the crystal with two trimers per asymmetric unit were measured to 1.9 Å for least-squares refinement. The final model of two trimers contains 883 residues: trimer 1 (33–178, 33–178, 31–185) and trimer 2 (34–175, 33–176, 36–185).
Tertiary and Quaternary Structure.
Four of the six copies of the EHA2 (23–185) monomer (Fig. 3A) in the asymmetric unit of the crystal make no or very minor crystal-packing contacts, and they are only ordered from residues 33 or 34 to 176 or 178, whereas two monomers, one in each trimer, make an extensive crystal-packing contact at one end and are, as a result ordered from residue 31 to the C terminus at residue 185 (Fig. 3C). As the structures of residues 31–33 and 178–185, ordered in only one monomer per trimer (Fig. 3C), appear to depend on this crystal contact, they are unlikely to have the same structure in solution and will not be discussed further.
Figure 3.
Ribbon diagrams of monomeric and trimeric HA2 ectodomains. (A) Stereodiagram of the EHA2 (23–185) monomer (blue) superimposed on HA2 (red) from neutral pH BHA (5). Residues 1–33 not visible in EHA2 (23–185) are colored white in the HA2. (B) Trimer of HA2 (blue, red, yellow monomers) from neutral pH BHA (5). Residues 1–33 not visible in EHA2 (23–185) are colored white in HA2. (C) Trimer of the EHA2 (23–185) structure (colored as in B). Figure prepared with ribbons (43).
The monomer structure is a 110-Å long hairpin, beginning and ending at one extreme end (blue in Fig. 3A). A long α-helix extends 68 residues from residue 38 to residue 105. After a 180° bend (residues 106–109), a short 18-residue α-helix (residues 110–128) packs antiparallel to the long helix. A short β-sheet hairpin then precedes a two-turn α-helix that crosses the long α-helix almost perpendicular to its axis. This short helix (146–153) has the only disulfide bond in the molecule at its N terminus from Cys-144 to Cys-148. After this α-helix, the 23 residues from 154 to 178 extend 72 Å antiparallel along the N-terminal α-helix in an approximately polyproline type II conformation (extended with side chains emerging about every 120° around the axis) (blue in Fig. 3A).
In the trimer (Fig. 3C), the N-terminal helices form a triple-stranded coiled coil as observed earlier in TBHA2 (Fig. 1). The short, antiparallel α-helices pack in the grooves between two of the long N-terminal α-helices. The monomer chain then crosses over the long α-helix, near the short perpendicular α-helix (residues 146–153) and proceeds antiparallel in the adjacent groove between the long α-helices (Fig. 3C). The most N-terminal and C-terminal residues that are ordered then converge at one end of the rod-shaped molecule to form a helical N cap.
N-Terminal Coiled Coil Is Terminated by a Four-Residue, Threefold Annular N Cap.
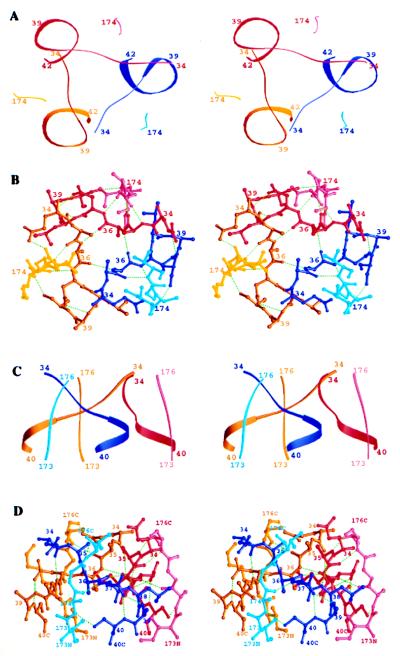
Unlike the other amide groups in α-helices, three amides at the N terminus of an α-helix do not form hydrogen bonds with carbonyl residues of preceding residues in the helix. As a consequence, α-helices are often stabilized and terminated by adjacent residues, sometimes negatively charged, termed the N cap, that form hydrogen bonds with these unpaired amide groups (38, 39). The four residues (34–37) of HA2 appear to have evolved both to cap the N terminus of the core α-helix and to terminate the triple-stranded coiled coil. Proceeding N-terminally, the last helical residue is residue 38. The aspartic acid side chain of residue 37 hydrogen bonds to the two terminal amide groups at residues 39 and 40, serving as an N cap. The segment 34–37 then crosses (clockwise, Fig. 4 A and B) to cap the neighboring α-helix of the three-stranded coiled coil by forming a hydrogen bond from the carbonyl group of residue 34 to the third free helical amide group, that of residue 38 (Fig. 4B).
Figure 4.
N cap domain of EHA2(23–185). (A) Stereo ribbon diagram of the N-terminal residues (dark blue, dark red, dark yellow) and the C-terminal residues (light colors) of EHA2 (23–185), viewed down the molecular threefold-symmetry axis. (B) Stereoatomic diagram of the N-terminal residues (colors and view as in A) and the C-terminal residues of EHA2 (23–185). Potential hydrogen bonds are green dashed lines. (C) Stereoribbon diagram as in A but viewed perpendicular to the molecular threefold-symmetry axis. (D) Stereoatomic diagram as in B but viewed as in C. Figure prepared with ribbons (43).
The three N capping segments (residues 34–37) as they extend laterally from one helix to the adjacent one also interact with each other around the threefold molecular symmetry axis to form a small annulus (Fig. 4 A and B). The annulus is stabilized by the threefold contacts among the methyl groups of Ala-36 from each monomer, forming a nonpolar layer around the trimer axis and another similar layer from Ala-35 stacked on top of it. Three symmetric hydrogen bonds cycle around the trimer axis from the carbonyl group of residue 36 in one monomer to the amide group of the same residue (residue 36) on the next monomer counterclockwise around the threefold axis (Fig. 4B).
Besides these interactions with each other, the four capping residues also make extensive contacts with the C-terminal-most ordered residues of HA2, residues 174–178 (Fig. 4 C and D). The C-terminal residues of HA2 run antiparallel to the N-terminal α-helix, packing in the groove between α-helices in the coiled coil (Fig. 3C). Residues 35 and 37, which flank the annular set of hydrogen bonds formed by residue 36, make three hydrogen bonds with the ascending C-terminal residues 174, 175, and 176 (amide 37 to carbonyl 174, carbonyl 35 to amide 176, and amide 35 to carbonyl 176). The side chains of Ile-173 and Val-176 make nonpolar contacts to the N-terminal N capping segment below and above the central alanines (residues 36 and 35) (Fig. 4 B and D).
Thus, the N- and C-terminal residues of HA2 form an extensive set of interactions that both act as an N cap to terminate the central three-stranded coiled coil and fix the N- and C-terminal regions of the molecule together at one extreme end of the rod-like structure.
Comparison with the Neutral pH Structure of Viral HA.
Despite the major refolding that occurs in the low pH-induced conformational change of HA2 (Fig. 3 B and C), where secondary structures refold and relocate, a number of secondary structural features are conserved. The N terminus of the central α-helix of EHA2 (23–185) (Fig. 3C) and the short outer α-helix of the neutral-pH HA2 conformation (Fig. 3B) both start at residue 38 and are N capped by the side chain of Asp-37. The β-sheet hairpin (residues ≈131–140) is packed against the same α-helical segment (residues 110–128) in both structures, although the direction of both has reversed. Beyond the β-sheet hairpin in the fusion-pH-induced conformation, a segment of structure that had been compact in neutral pH HA2 (including helices at residues 146–154 and 163–171) appears to be extruded to form a 23-residue (72-Å long) polyproline type II conformation that extends the length of the molecule (Fig. 3A).
Comparison with TBHA2, the Proteolytically Dissected Fragment of the Fusion-pH HA.
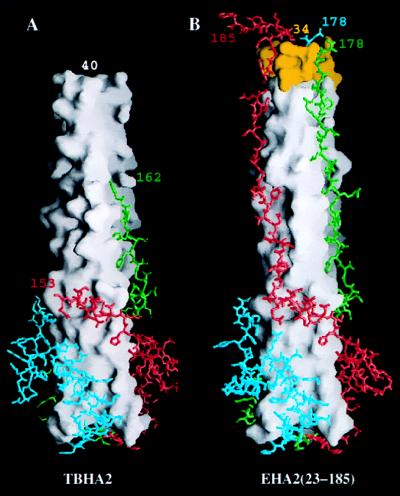
Native viral HA, when incubated at fusion pH (pH 5.0), aggregated or bound to nonionic detergents or lipid vesicles if present (7). Incubation with thermolysin removed the N-terminal fusion peptide of HA2 and generated a soluble, trimeric fragment, TBHA2 (Fig. 1), containing residues 1–27 of HA1 disulfide-linked to residues 38–175 of HA2 (10). The molecule studied here, EHA2 (23–185), lacks the HA1 residues of TBHA2 but is 15 residues longer at the N terminus and 10 residues longer at the C terminus, both extensions of regions that were disordered in the TBHA2 x-ray structure (disordered from 38–40 and 162–175) (compare Fig. 5 A and B).
Figure 5.
Comparison of TBHA2 from low pH-treated HA and EBHA2(23–185). (A) TBHA2 (residues 1–27 of HA1 and 38–175 of HA2) from thermolytic digestion of low pH-treated viral BHA. The white molecular surface is the central triple-stranded coiled coil. The atomic models are the C-terminal residues beyond residue 106 colored differently for each monomer. The last residues visible in the electron density at the termini are labeled. (B) EBHA2 (23–185) with the N-terminal residues from 34 to 40 colored yellow. The rendering is as in A with the visible terminal residues labeled. Figure prepared with grasp (44).
In EHA2(23–185), the N-terminal α-helix begins at residue 38, its terminus stabilized by an N cap formed by Asp-37. The TBHA2 molecule began at residue 38, lacked Asp-37, and the α-helix appeared to be slightly unraveled with the first residue clearly defined at residue 40. In the TBHA2 structure, the C termini are extended strands of 11 residues, 152–162, that were observed to adopt two conformations, ordered in two monomers and disordered in one monomer (Fig. 5A) (11). The observation that the final structured residues of HA2 in TBHA2 take different paths in the different monomers in crystals suggested that these residues can adopt alternate conformations of approximately the same energetic stability. By contrast, in EHA2 (23–185) the C-terminal residues are ordered as an extended chain from residue 152 to 178. They may be stabilized in the groove between the α-helices in the central coiled coil of EHA2 (23–185) as a result of the contacts that residues 173–176 make in the N cap (to yellow residues at the top of Fig. 5B) described in the section above.
Implications for Membrane Fusion and Viral Entry.
The three-dimensional structure of the recombinant protein EHA2 (23–185) extends our knowledge of the location and structure of the N- and C-terminal residues of the membrane fusion glycoprotein of influenza virus (HA) in its fusion-pH conformation. Residues disordered in a smaller thermolytic digestion fragment of fusion-pH viral HA, TBHA2 (38–175) (Fig. 1) are found ordered in a construct longer at both termini, EHA2 (23–185). N-terminal residues are ordered in an extended conformation from residue 37 to 33, about 20 residues from where the nonpolar fusion peptide is expected to insert into the target membrane. C-terminal residues are also ordered in an extended conformation to residue 178, about eight residues from the transmembrane anchor that would be embedded in the viral membrane. Furthermore, the most N- and C-terminal residues are in very close proximity in the trimer, which would permit close apposition of the two prefusion membranes. These conclusions are subject to the reservations that neither nonpolar membrane interacting segments nor lipids are present in these crystals and that the HA1 chain and the oligosaccharides of HA2 are absent.
However, other evidence supports the conclusion that the N-terminal α-helix starts at residue 38 in intact HA, as observed here, including (i) 38 is the first residue of the protease-resistant core of fusion-pH HA whether produced by digestion of virus or purified HA (10); (ii) In neutral-pH viral HA, the same α-helix starts at residue 38 and is N capped by Asp-37 (40); (iii) Asp-37, which begins the N cap, and the two preceding alanines that end it, are conserved in the sequences of all influenza A and B viruses, where HA2 sequences have as little as 40% sequence identity.
The unexpected observation of a capping structure formed by the association of N- and C-terminal residues of the trimer (Fig. 4), suggests that the α-helical secondary structure is terminated so that a more flexible conformation of extended polypeptide will link the trimeric rod to the membrane embedded segments. This, and the observation that the last ordered C-terminal residues are also in an extended conformation, suggests that the linkage to the membrane embedded sequences do not need to be α-helical, as in the HIV-1 and SV5 fusion proteins (14, 16, 17, 21, 22), to cause fusion. This conclusion is consistent with the structures of the C-terminal-most ordered residues in the Ebola GP2 and HTLV-1 TM ectodomain constructs, which are also non-α-helical (18–20).
The EHA2 (23–185) structure, therefore, favors the type of nonsymmetric fusion intermediates with flexible linkage to at least the viral membrane, discussed previously (see Fig. 3 in ref. 14; refs. 11 and 14). In membrane-anchored HA, flexible linkage to the C-terminal membrane anchors would facilitate the HA conformational change (11) because it would allow the C termini in the viral membrane to migrate from an axial position in neutral pH structure to an outer layer position in the fusion-pH rod. In the neutral-pH conformation of HA, the three C-terminal membrane anchors could be near the molecular threefold-symmetry axis and may even be in contact with each other (5). After the conformational change, the three C-terminal anchors may again be near each other and the molecular threefold axis, but they are attached to a long extended strand on the outside layer of the two-layered fusion-pH conformation (Fig. 5).
It is possible that the rod-like conformation observed here for EHA2 (23–185) is the conformation of a prefusion intermediate (perhaps as a component of an oligomeric pore) or the postfusion state of the ectodomain. The distinction is that in the prefusion intermediate, the N and C termini are inserted in the cellular and viral membranes, respectively, whereas in the postfusion state, both sets of termini are inserted in the one remaining, fused membrane (19). An alternative possibility is that HA2 mediates membrane fusion when its ectodomain is in a transient conformation (possibly resembling TBHA2, Fig. 5A) preceding the conformation observed here. In fusion by influenza virus, therefore, the kinetic question may now be refined to determining when membrane fusion occurs relative to when the N cap domain is formed.
The observation of an apparently stabilizing N cap that also holds the C-terminal residues next to the N-terminal residues in EHA2 and its absence in the shorter TBHA2 (38–175) structure suggests that EHA2 (23–185) might be more thermodynamically stable than TBHA2. The midpoint of the thermal denaturation curve of EBHA2 (38–175) (27) that is missing the N cap residues, is 70°C, whereas F185 (Fig. 1) and EHA2 (23–185) that contain them denature at 80°C (30) and 78°C, consistent with this idea.
The free energy released in the fusion pH-induced conformational change in HA indicated by the change in thermal denaturation temperatures may be used in the process of membrane fusion. Finding the N cap in the more complete EHA2 (23–185) fragment suggests that the free energy of stabilization of the N cap is a driving force for the transition from structures like TBHA2 (38–175) (Fig. 5A) to those like EHA2 (23–185) (Fig. 5B); EHA2 (23–185) may be the minimum free-energy conformation toward which the fusion pH-induced structural changes are headed. Whether fusion occurs on completion of the conformational change or at a transient intermediate stage resembling for example, TBHA2, the fusion event could be coupled to the favorable free-energy change of adopting an N capped, EHA2 (23–185)-like conformation. The difference in stability between EHA2 (23–185) and EBHA2 (38–175), which lacks the N cap, suggests the availability of additional free energy for membrane fusion even at a step after the formation of the central triple-stranded coiled coil and insertion of the fusion peptide into the target membrane.
Acknowledgments
We thank Barbara Harris and Marlène Bouvier for helpful discussion, Dr. Eva Vareckova for the kind gift of antibody IIF4, Georg E. Schulz for an idea about the significance of the structure as a free energy target in membrane fusion, Martin Lawrence for help with data collection, and Xiaodong Zhang for help with initial ncs averaging and model building. This research was supported by the Medical Research Council (U.K.), the National Institutes of Health, and the Howard Hughes Institute of Medical Research (HHMI). D.C.W. is an investigator of the HHMI.
ABBREVIATIONS
- HA
hemagglutinin
- BHA
soluble neutral-pH HA
- TBHA2
trimeric BHA2 fragment
- EBHA2
HA2 ectodomain
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1QU1).
References
- 1.Skehel J J, Waterfield M D. Proc Natl Acad Sci USA. 1975;72:93–97. doi: 10.1073/pnas.72.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klenk H-D, Rott R, Orlich M, Blodorn J. Virology. 1975;68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 3.Lazarowitz S G, Choppin P W. Virology. 1975;68:440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- 4.Brand C M, Skehel J J. Nat New Biol. 1972;238:145–147. doi: 10.1038/newbio238145a0. [DOI] [PubMed] [Google Scholar]
- 5.Wilson I A, Skehel J J, Wiley D C. Nature (London) 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 6.Wiley D C, Wilson I A, Skehel J J. Nature (London) 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 7.Skehel J J, Bayley P M, Brown E B, Martin S R, Waterfield M D, White J M, Wilson I A, Wiley D C. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doms R W, Helenius A, White J. J Biol Chem. 1985;260:2973–2981. [PubMed] [Google Scholar]
- 9.Daniels R S, Douglas A R, Skehel J J, Waterfield M D, Wilson I A, Wiley D C. In: The Origin of Pandemic Influenza Viruses. Laver W G, editor. New York: Elsevier; 1983. pp. 1–7. [Google Scholar]
- 10.Ruigrok R W, Aitken A, Calder L J, Martin S R, Skehel J J, Wharton S A, Weis W, Wiley D C. J Gen Virol. 1988;69:2785–2795. doi: 10.1099/0022-1317-69-11-2785. [DOI] [PubMed] [Google Scholar]
- 11.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 12.Carr C M, Kim P S. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 13.Skehel J J, Wiley D C. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 14.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 15.Fass D, Harrison S C, Kim P S. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 16.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 17.Caffrey M, Cai M, Kaufman J, Stahl S J, Wingfield P T, Covell D G, Gronenborn A M, Clore G M. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobe B, Center R J, Kemp B E, Poumbourios P. Proc Natl Acad Sci USA. 1999;96:4319–4324. doi: 10.1073/pnas.96.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weissenhorn W, Carfí A, Lee K H, Skehel J J, Wiley D C. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 20.Malashkevich V N, Schneider B J, McNally M L, Milhollen M A, Pang J X, Kim P S. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 22.Tan K, Liu J, Wang J, Shen S, Lu M. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson P I, Roth R, Morisaki H, Jahn R, Heuser J E. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 24.Poirier M A, Xiao W, Macosko J C, Chan C, Shin Y K, Bennett M K. Nat Struct Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- 25.Sutton R B, Fasshauer D, Jahn R, Brünger A T. Nature (London) 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 26.Blacklow S C, Lu M, Kim P S. Biochemistry. 1995;34:14956–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Wharton S A, Weissenhorn W, Calder L J, Hughson F M, Skehel J J, Wiley D C. Proc Natl Acad Sci USA. 1995;92:12205–12209. doi: 10.1073/pnas.92.26.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carr C M, Chaudry C, Kim P S. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Epand R F, Macosko J C, Russell C J, Shin Y K, Epand R M. J Mol Biol. 1999;286:489–503. doi: 10.1006/jmbi.1998.2500. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Skehel J J, Wiley D C. Biochemistry. 1998;37:13643–13649. doi: 10.1021/bi981098l. [DOI] [PubMed] [Google Scholar]
- 31.Vareckova E, Mucha V, Betakova T, Russ G. Arch Virol. 1993;130:45–56. doi: 10.1007/BF01318995. [DOI] [PubMed] [Google Scholar]
- 32.Navaza J, Saludjian P. Methods Enzymol. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 33.Brünger A T, Adams P D, Clore G M, Delano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project. Acta Cryst D. 1994;50:760–763. [Google Scholar]
- 35.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Cryst A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 36.Brünger A T. xplor, Version 3.843. New Haven, CT: Yale University; 1996. [Google Scholar]
- 37.Jones T A. In: Molecular Replacement. Dodson E J, Gover S, Wolf W, editors. Warrington, U.K.: SERC Daresbury Laboratory; 1992. pp. 91–105. [Google Scholar]
- 38.Presta L G, Rose G D. Science. 1988;240:1632–1641. doi: 10.1126/science.2837824. [DOI] [PubMed] [Google Scholar]
- 39.Richardson J S, Richardson D C. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 40.Weis W I, Brünger A T, Skehel J J, Wiley D C. J Mol Biol. 1990;212:737–761. doi: 10.1016/0022-2836(90)90234-D. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 42.Kraulis J P. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- 43.Carson M. J Mol Graphics. 1987;5:103–106. [Google Scholar]
- 44.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]