Abstract
Aim: To characterise the phenotype and identify the underlying genetic defect in a family with deafness segregating with a North Carolina-like macular dystrophy (NCMD).
Methods: Details of the family were obtained from the Moorfields Eye Hospital genetic clinic database and comprised eight affected, four unaffected members, and two spouses. Pedigree data were collated and leucocyte DNA extracted from venous blood. Positional candidate gene and genetic linkage strategies utilising polymerase chain reaction (PCR) based microsatellite marker genotyping were performed to identify the disease locus.
Results: The non-progressive ocular phenotype shared similarities with North Carolina macular dystrophy. Electro-oculography and full field electroretinography were normal. Progressive sensorineural deafness was also present in all affected individuals over the age of 20 years. Hearing was normal in all unaffected relatives. Haplotype analysis indicated that this family is unrelated to previously reported families with NCMD. Genotyping excluded linkage to the MCDR1 locus and suggested a potential novel disease locus on chromosome 14q (Z=2.92 at θ=0 for marker D14S261).
Conclusion: The combination of anomalies segregating in this family represents a novel phenotype. This molecular analysis indicates the disease is genetically distinct from NCMD.
Keywords: North Carolina macular dystrophy, linkage, sensorineural deafness
The inherited macular degenerations constitute a significant, usually progressive, as yet untreatable cause of visual loss in both children and adults. A wide clinical spectrum is recognised, though in recent years evidence from genetic linkage studies has made classification more robust. Most progress has been made with autosomal dominant macular dystrophies where a number of disease loci and causative genes have been identified.1–2
Lefler3 and Frank4 were the first to describe a “progressive” foveal dystrophy now termed North Carolina macular dystrophy (NCMD).5 Extensive genealogical studies have revealed that several other diseases, thought originally to be distinct entities, central areolar pigment epithelial dystrophy,6 central pigment epithelial and choroidal degeneration,7 and central retinal pigment epithelial dystrophy,8 arise in descendants of the original NCMD founders and are in fact manifestations of the variable expressivity of the condition.9 Subsequently, families with NCMD unrelated to the original pedigree have been reported in the United Kingdom,10 Texas, United States,11 Central America,12 and France.13
The gene for NCMD has been shown to reside within a 3.1 cM interval termed the MCDR1 locus on 6q14–q16.214–15 ). Interestingly, NCMD co-localises with bifocal chorioretinal atrophy with which it shares some clinical similarities2 and lies adjacent to the loci for Stargardt-like dominant progressive macular dystrophy,16 autosomal dominant macular atrophy,17 cone-rod dystrophy 7,18 autosomal dominant drusen and macular degeneration,19 and the recessive retinitis pigmentosa 25 locus.20
NCMD is a fully penetrant dominant disease with variable expressivity and onset in infancy. It is now agreed that the condition is not progressive.21–22 Visual acuity varies from normal to significantly compromised and the posterior segment findings can be classified into one of three grades depending on severity: fine “drusen” within 3° of fixation (grade I); confluent elevated subretinal deposits sometimes associated with retinal pigment epithelial (RPE) changes (grade II); central macular chorioretinal atrophy with hypertrophic fibrous tissue (grade III).10
Deafness is a common congenital anomaly with an incidence of 1 in 1000 live births.23 About 50% of cases can be attributed to genetic factors, with hearing impairment which is typically progressive and sensorineural, inherited either as an isolated anomaly or as part of over 400 clinical syndromes.24 Associations between sensorineural hearing loss and retinal degenerations are well described, presumably arising because of shared tissue gene expression. By far the most frequent association is with retinitis pigmentosa (Usher syndrome),25,26 but deafness has been described together with several other rare hereditary peripheral retinal degenerations including Alport syndrome,27 a rod-cone dystrophy,28 a cone dystrophy,29 a form of X linked retinitis pigmentosa,30 the Mohr-Tranebjaerg syndrome,31 and a mitochondrially inherited pigmentary retinopathy with maternally inherited adult onset diabetes (MIDD).32,33 In only one family34 and in a single case35 has deafness been reported in association with a macular dystrophy. In both instances, the genetic defect was found to reside in the mitochondrial genome.
In this paper, we describe the phenotype and genetic linkage of a four generation English kindred with a dominantly inherited macular disease, clinically similar to North Carolina macular dystrophy in whom genetic linkage to the MCDR1 locus has been excluded. Segregating with the ocular phenotype is progressive adult onset sensorineural deafness.
MATERIALS AND METHODS
Phenotyping
Ethical approval was granted by the ethics committee of Moorfields Eye Hospital before the commencement of this study. The genetic eye clinic database of Moorfields Eye Hospital, London, provided details of the proband. All family members gave written consent, provided a history and underwent clinical evaluation which included acuity, dilated fundal examination with photography, audiology and, in selected cases, ocular electrophysiology (electroretinograms and electro-oculograms were recorded in accordance with the ISCEV protocols).
Genotyping
EDTA sequestered venous blood samples were obtained from both affected and unaffected individuals and DNA extracted using the Nucleon II extraction kit (Scotlab Bioscience). Pedigree data were collated and polymerase chain reaction (PCR) based microsatellite marker genotyping was undertaken utilising version 2.0 of the fluorescent labelled ABI MD-10 (10 cM resolution) and HD-5 (5 cM resolution) linkage mapping sets (Applied Biosystems) derived from the Genethon human linkage map.36,37
PCR reactions were carried out for each marker individually in a 5 μl reaction volume, containing 25 ng DNA, 15 mM TRIS-HCl (pH 8.0), 50 mM KCl, 2.5 mM MgCl2, 250 μM each dNTP, 1.25 pmol of each primer, and 0.25U AmpliTaq Gold (Applied Biosystems). Reactions were performed on a Perkin Elmer 9600 with a standard thermocycling profile for all markers. This consisted of an initial denaturation of 12 minutes, immediately followed by 10 cycles of 95°C for 15 seconds, 55°C for 15 seconds and 72°C for 30 seconds, and then by 20 cycles of 89°C for 15 seconds, 55°C for 15 seconds, and 72°C for 30 seconds, with a single final extension step of 72°C for 10 minutes.
PCR products for selected sets of markers were pooled, diluted, and denatured in formamide and size fractionated using an ABI 3100 Genetic Analyser. PCR products were sized by the 3100 Data Collection Software version 1.0.1 program and scored using the GeneMapper version 2.0 program.38–39 Data were checked for genotyping errors using the pedcheck program.40
Linkage analysis
Subjects were classified as affected, unaffected, or of unknown status according to their clinical status. Linkage analysis was carried out using standard lod score methods. For lod score analysis the fastlink program was used via the HGMP Genetic Linkage User Environment (GLUE, http://www.hgmp.co.uk).
A dominant model with a disease allele frequency of 0.0001 and a penetrance of 1 was set. Marker allele frequencies were arbitrarily calculated in GLUE by assuming equal frequencies.
RESULTS
The phenotype
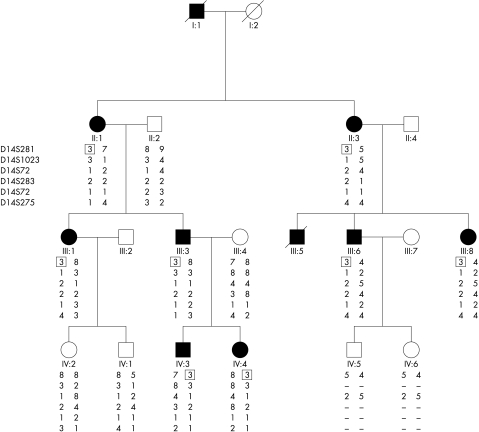
Eight affected, four unaffected, and two spouses agreed to participate in the study and some clinical details of two dead relatives were also available. Autosomal dominant inheritance was supported by the presence of affected members in each generation, equal numbers of affected males and females, and male to male transmission (Fig 1). The ocular phenotype was fully penetrant and exhibited variable expressivity not only among different family members but also between eyes of the same patient. Affected individuals had fine drusen-like subretinal deposits and pigmentary disturbance at the level of the retinal pigment epithelium (RPE) centred on the macula. Others had, in addition, a well demarcated subfoveal area of chorioretinal atrophy with pigment hypertrophy and fibrosis at the edge (Fig 2). There were no other fundal abnormalities.
Figure 1.
Abridged pedigree and haplotype of six adjacent microsatellite markers located on 14p ordered sequentially from the telomere. Solid and open symbols indicate affected and unaffected individuals respectively. The disease allele is boxed.
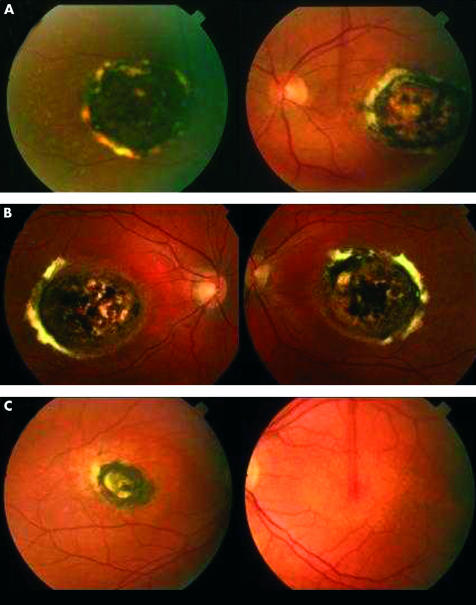
Figure 2.
The ocular phenotype. Colour fundus photographs: (A) individual II:1 a 67 year old female) showing centred on both foveae a well demarcated subfoveal area of chorioretinal atrophy with pigment hypertrophy and fibrosis at the edge and (B) her son (III:3, age 41 years), similar appearances both eyes; (C) her daughter (III:1, age 42 years) showing right eye, similar appearances; left eye, drusen-like deposits and retinal epithelial atrophy centred on the macula.
The clinical appearances were non-progressive and in all but one individual (III:8) had been noted soon after birth. Acuity was well preserved in those with the milder phenotype. Only one individual (II:3) described progressive visual loss in the latter years of life, at least in part because of the development of cataract. In those with the milder phenotype, acuity was well preserved. One child (IV:4) had developed strabismic amblyopia. There was an over-representation of low myopia among affected individuals (typically −1.50 dioptres).
Electrophysiology on all affected individuals was not possible as I:1 and III:5 were dead, and II:1, III:1, and III:8 declined to be tested. The clinical findings are summarised in Table 1. Electroretinography was essentially normal in those tested whereas the EOG was mildly subnormal.
Table 1.
Clinical details for affected individuals
| Pedigree number | Age (years) | Retinal appearances | Visual acuity right, left | Electrophysiological findings | Sensorineural hearing loss (age detected, years) | Other ocular findings |
| I:1 | died aged 72 | Bilateral “macula chorioretinitis” | Poor vision | Yes (40s) | ||
| II1 | 67 | Both eyes: well demarcated area of chorioretinal atrophy with pigment hypertrophy centred on the macula | 6/18, 6/24 | Declined testing | Yes (50) | Left exotropia, iris heterochromia |
| II3 | 77 | Both eyes: well demarcated area of chorioretinal atrophy with pigment hypertrophy centred on the macula | HM, 6/18 | PERG: normal | Yes (60) | Low myopia |
| EOG: mildly subnormal | Right squint surgery as child | |||||
| ERG: normal | ||||||
| III1 | 42 | Right eye: well demarcated area of chorioretinal atrophy with pigment hypertrophy | Declined testing | Yes (36) | ||
| Left eye: multiple yellow drusen-like deposits at the macula | ||||||
| III3 | 41 | Both eyes: well demarcated area of chorioretinal atrophy with pigment hypertrophy centred on the macula | 6/9, 6/9 | Yes (20) | Low myopia | |
| III5 | died aged 41 | Bilateral macular dystrophy | Not known | Hearing loss | ||
| III:6 | 51 | Multiple yellow drusen-like deposits centred on both maculas | 6/9, 6/9 | PERG: mild reduction | Yes (40s) | Low myopia |
| EOG: normal | Left squint surgery aged 4 | |||||
| ERG: normal | ||||||
| III8 | 44 | Multiple yellow drusen-like deposits centred on both maculas | 6/6, 6/6 | Declined testing | No | |
| IV3 | 15 | Multiple yellow drusen-like deposits centred on both maculas | 6/6, 6/6 | ERG: normal | Not detected | Low myopia |
| EOG: mildly subnormal | Left strabismic amblyopia | |||||
| IV4 | 13 | Multiple yellow drusen-like deposits centred on both maculas | 6/9, 6/24 | ERG: normal | Not detected | |
| EOG: mildly subnormal |
ERG = electroretinogram; PERG = pattern ERG; EOG = electro-oculography.
Typically, the pattern of sensorineural hearing loss segregating with the ocular phenotype was progressive (loss up to 100 dB) bilateral, symmetrical and high frequency. Tympanometry revealed normal middle ear pressures and compliance indicating an inner ear defect. Typically, hearing loss became significant from the fourth decade. In one individual (III:3), hearing loss was noted aged 20. No audiological abnormality was detected when the two youngest members of the family (IV:3, aged 15 and IV:4, aged 13) were tested.
Linkage analysis
Two point linkage analysis for microsatellite markers spanning the MCDR1 locus on 6q (Table 2) excluded genetic linkage of our family to the North Carolina macular dystrophy gene. Indeed, haplotype analysis of these markers (data not shown) confirms that this family is unrelated to any previously reported NCMD kindred.
Table 2.
Two point lod scores between disease phenotype and markers linked to the North Carolina macular dystrophy locus on chromosome 6q
| Recombination fraction | ||||||||||
| Microsatellite marker | 0.00 | 0.05 | 0.10 | 0.20 | 0.30 | 0.40 | Z max | θ max | Excln θ | Excln cM |
| D6S251 | −∞ | −2.02 | −1.39 | −0.76 | −0.41 | −0.17 | 0.00 | 0.50 | 0.05 | 5 |
| D6S252 | −∞ | −1.15 | −0.65 | −0.25 | −0.10 | −0.03 | 0.00 | 0.50 | 0.01 | 1 |
| D6S1717 | −∞ | −0.33 | −0.12 | 0.01 | 0.02 | 0.01 | 0.02 | 0.30 | <0.001 | <0.1 |
| D6S1543 | −∞ | −0.92 | −0.45 | −0.11 | −0.02 | −0.00 | 0.00 | 0.50 | 0.01 | 1 |
| D6S468 | −∞ | −1.17 | −0.65 | −0.23 | −0.06 | −0.01 | 0.00 | 0.50 | 0.02 | 2 |
| D6S283 | −∞ | −0.02 | 0.23 | 0.36 | 0.30 | 0.17 | 0.36 | 0.20 | <0.001 | <0.1 |
Excln θ is the recombination fraction at a lod score of −2; Excln cM is the genetic distance (in centimorgans) around the marker which is excluded from linkage.
Linkage analysis initially targeted candidate regions identified by the presence of a cloned and/or mapped gene causing retinal disease (Table 3). These included the regions containing ABCA4 on 1p, CORD8 on 1q, STGD4 on 4p, GCAP, CORD7, and ELOVL4 on 6p, CORD5 and CACD on 17p, CORD4 and UNC119 on 17q, CORD1 on 18q, CORD2 and CRX on 19q and TIMP3 on 22q. Since none of these regions produced significant lod scores, the analysis was extended to a genome wide search. In total ∼50% of the genome was screened involving genotyping of 195 markers. A significantly positive lod score was obtained with marker D14S261 (Z=2.92 at θ=0) which lies in the centromeric region of chromosome 14. This marker is placed by GeneMap (http://corba.ebi.ac.uk/RHdb/) in the telomeric region of the short p arm (pter) but at q11.2 adjacent to the centromere by Ensembl (http://www.ensembl.org) and UCSC (http://genome.ucsc.edu/). Since the telomeric region of this essentially acrocentric chromosome appear to be devoid of genes, we will adhere to the latter location.
Table 3.
Two point linkage analysis across the critical region of chromosome 14
| Recombination fraction | ||||||
| Microsatellite marker | 0.00 | 0.05 | 0.10 | 0.20 | 0.30 | 0.40 |
| D14S261 | 2.92 | 2.66 | 2.39 | 1.81 | 1.19 | 0.56 |
| D14S1023 | −∞ | −0.04 | 0.14 | 0.20 | 0.15 | 0.08 |
| D14S72 | −∞ | 0.51 | 0.65 | 0.61 | 0.44 | 0.23 |
| D14S283 | 0.54 | 0.46 | 0.37 | 0.22 | 0.09 | 0.02 |
DISCUSSION
We describe the clinical evaluation and molecular genetic analysis of a four generation English pedigree with dominantly inherited non-progressive macular dystrophy and progressive sensorineural hearing loss. The differential diagnosis of macular dystrophies includes dominantly inherited drusen, Sorsby’s pseudoinflammatory macular dystrophy, Stargardt disease, progressive foveal dystrophy, Best disease, and adult vitelliform macular, pattern, and central areolar dystrophies. It is unlikely that our family has a variant of any of these based on the observed inheritance pattern, age of onset, asymmetry, lack of progression, and electrophysiological findings.
The behaviour and clinical features of the ocular phenotype are strikingly similar to North Carolina macular dystrophy (NCMD).22 The drusen-like subretinal deposits with RPE disturbance and the areas of well demarcated chorioretinal atrophy and pigment epithelial hypertrophy would be consistent with grade I and III NCMD lesions, respectively.10 Furthermore, the presence of normal electrophysiology parallels the findings in NCMD4 and suggests that the deficit is restricted to the central retina. Also, similar to NCMD reasonable acuity was generally retained even in those with subfoveal chorioretinal lesions. It has been hypothesised this might arise if foveal disruption occurs sufficiently early in retinal development to allow neural specialisation at an eccentric point.10 Holz et al41 have described an Indian family with autosomal dominant macular dystrophy with appearances that overlapped between NCMD, pattern dystrophy, fundus flavimaculatus, and drusen but there was no associated hearing impairment.
There are more than 400 syndromes of which hearing loss is a part and more than 70 loci for non-syndromic deafness.42 Severe to profound hearing loss with onset in the first postnatal year is characteristic of recessive disease. Those with dominantly inherited disease generally present after speech development with sensorineural deficit. Characteristically, the auditory frequencies affected are similar among members of the same family. The hearing impairment in our family is therefore consistent with this latter inheritance pattern.
Genes implicated in heritable deafness are known to encode a diverse group of proteins involved in structure, mitochondrial function, adhesion, and transcriptional regulation.43 Up to 50% of autosomal recessive non-syndromic deafness can be attributed to mutations in the gene that encodes the gap junctional protein, connexin 26.44 Associations between hearing loss and retinal disease are well described.45 The structural similarities between the hair cells of the cochlear and retinal photoreceptors suggest a common evolutionary origin. Indeed, advances in understanding of the genetic defects that underlie Usher syndrome (sensorineural hearing loss and retinitis pigmentosa) and most recently X linked retinitis pigmentosa (RPGR, retinitis pigmentosa GTPase regulator mutations46) have revealed examples of shared gene expression.47,48 Usually, however, the pattern of retinal involvement in these syndromes is widespread (involving both rod and cone photoreceptor systems) and progressive. It is very rare for a primarily macular dystrophy to be associated with deafness. There have been only two reports both implicating a mitochondrial gene defect; the first in a family with diabetes, deafness, and pattern dystrophy with A3243G heteroplasmy34 and the second, a 17 year old male with diabetes, deafness, cataract, and maculopathy who was found to have a 7 kb mitochondrial gene deletion.35 Thus, this is the first report of autosomal inheritance for a syndrome comprising macular dystrophy and deafness.
Results of our molecular genetic analysis provide strong evidence that the family is unrelated to any previously published NCMD pedigree and excludes genetic linkage to the MCDR1 locus. The linkage data also identify a novel putative disease locus for both retinal and otological disease within a 1.1 cM region on 14q (Z=2.92 at θ=0 for D14S261). The lod value falls just below the threshold score of 3 for full statistical significance but with the present family size, even with full microsatellite marker informativity, such a score is not attainable. Confirmation of this locus will need to await study of further families with this rare disorder. A search of online databases reveals a number of genes and expressed sequence tags within the critical interval. However, none is known to be expressed in both retina and cochlear and cannot therefore be considered good candidates for mutation analysis.
Identification of the disease causing mutation in this family will advance understanding of the biology of both hearing and vision, particularly given the progressive nature of the hearing loss and the congenital stationary retinal dystrophy. Furthermore, the presence of drusen-like deposits and subfoveal lesions (clinically reminiscent of the sequelae of neovascular membrane formation) suggest that dissection of the molecular mechanisms underlying the pathology seen in this family might provide insights into the biology of drusen, a frequent finding in age related macular degeneration.
3rd Asia Pacific Forum on Quality Improvement in Health Care 3–5 September 2003, Auckland, New Zealand.
We are delighted to announce this forthcoming conference in Auckland, New Zealand.
The themes of the 3rd Asia Pacific Forum on Quality Improvement in Health Care are:
Agenda for quality: Improving equity in health care delivery
Improving safety
Leadership for improvement
Measuring quality and benchmarking for change
Evidence based knowledge and education for quality improvement
Improving health systems
Patient/consumer centred quality improvement
Presented to you by the BMJ Publishing Group (London, UK) and Institute for Healthcare Improvement (Boston, USA), supported by the New Zealand Ministry of Health, ACC, and Standards New Zealand.
For more information about the Forum or to register contact: quality@bma.org.uk or go to: www.quality.bmjpg.com Tel: +44 (0)20 7383 6409 Fax: +44 (0)20 7383 6869
Acknowledgments
We thank the family for their participation, Mr Andrew Dearlove, UK Human Genome Mapping Project Resource Centre, Babraham, Cambridge, Mr David Calver for providing clinical details of some of the family members and Mr Wilfred Rwapunga, senior audiologist for carrying out the audiometry. This work was supported by grants from the Wellcome Trust and the British Retinitis Pigmentosa Society.
REFERENCES
- 1.Nichols B, Sheffield V, Vandenburgh K, et al. Butterfly shaped pigment dystrophy of the fovea caused by a point mutation in the RDS gene. Nat Genet 1993;3:202–7. [DOI] [PubMed] [Google Scholar]
- 2.Kelsell R, Godfrey B, Evans K, et al. Localisation if the gene for progressive bifocal chorioretinal atrophy (PBCRA) to chromosome 6q. Hum Mol Genet 1995;4:1653–6. [DOI] [PubMed] [Google Scholar]
- 3.Lefler W, Wadsworth J, Sidbury J. Hereditary macular degeneration and aminoaciduria. Am J Ophthalmol 1971;71:224–30. [DOI] [PubMed] [Google Scholar]
- 4.Frank H, Landers M, Williams R, et al. A new dominant progressive foveal dystrophy. Am J Ophthalmol 1974;78:903–16. [DOI] [PubMed] [Google Scholar]
- 5.Gass J. Stereoscopic atlas of macular diseases. St Louis: Mosby, 1987.
- 6.Hermsen V, Judsch J. Central areolar pigment epithelial dystrophy. Ophthalmologica 1984;189:69–72. [DOI] [PubMed] [Google Scholar]
- 7.Leveille A, Morse P, Kiernan J. Autosomal dominant central pigment epithelial and choroidal degeneration. Ophthalmology 1982;89:1407–13. [DOI] [PubMed] [Google Scholar]
- 8.Klein R, Bresnick G. An inherited central retinal pigment epithelial dystrophy. Birth Defects 1982;19:281–96. [PubMed] [Google Scholar]
- 9.Small K, Hermsen V, Gurney N, et al. North Carolina macular dystrophy and central areolar pigment epithelial dystrophy. Arch Ophthalmol 1992;110:515–18. [DOI] [PubMed] [Google Scholar]
- 10.Reichel M, Kelsell R, Fan J, et al. Phenotype of a British North Carolina macular dystropy family linked to chromosome 6q. Br J Ophthalmol 1998;82:1162–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia C, Callardo G, Mullen L, et al. A new North Carolina macular dystrophy (MCDR1) family in Texas maps to chromosome 6q16. Invest Ophthalmol Vis Sci 1995;36(suppl):892. [Google Scholar]
- 12.Rabb M, Mullen L, Yechits S, et al. Belize macular dystrophy maps to chromosome 6q16 (the north carolina macular dystrophy locus, MCDR1. Invest Ophthalmol Vis Sci 1995;36(suppl):892. [Google Scholar]
- 13.Yechits S, Puech B, Mullen L, et al. A new French family with the North Carolina macular dystrophy phenotype maps to the MCDR1 locus. Am J Human Genet 1996;59:A391. [Google Scholar]
- 14.Small K, Weber J, Roses A, et al. North Carolina macular dystrophy is assigned to chromosome 6. Genomics 1992;13:681–5. [DOI] [PubMed] [Google Scholar]
- 15.Small K, Weber J, Roses A, et al. North Carolina macular dystrophy (MCDR1). A review and fine mapping to 6q14-q16.2. Ophthal Paed Genet 1993;14:143–50. [DOI] [PubMed] [Google Scholar]
- 16.Stone E, Nichols B, Kimura A, et al. Clinical features of a Stargardt-like dominant progressive macular dystrophy with genetic linkage to chromosome 6q. Arch Ophthalmol 1994;112:765–72. [DOI] [PubMed] [Google Scholar]
- 17.Griesinger I, Sieving P, Ayyagari R. Autosomal dominant macular atrophy at 6q14 excludes CORD7 and MCDR1 loci. Invest Ophthalmol Vis Sci 2000;41:248–55. [PubMed] [Google Scholar]
- 18.Kelsell R, Gregory-Evans K, CY G, et al. Localisation of a gene (CORD7) for a dominant cone-rod dystrophy to chromosome 6q. Am J Human Genet 1998;63:274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stefko S, Zhang K, Gorin M, et al. Clinical spectrum of chromosome 6-linked autosomal dominant drusen and macular degeneration. Am J Ophthalmol 2000;130:203–8. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz A, Borrego S, Marcos I, et al. A major locus for autosomal recessive retinitis pigmentosa on 6q, determined by homozygosity mapping of chromosomal regions that contain gamma-aminobutyric acid-receptor clusters. Am J Human Genet 1998;62:1452–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Small K. North Carolina macular dystrophy. Ophthalmology 1989;96:1747–54. [DOI] [PubMed] [Google Scholar]
- 22.Small K, Killian J, WCMcLean. North Carolina’s dominant progressive foveal dystrophy: how progressive is it? Br J Ophthalmol 1990;75:401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marazita M, Ploughman L, Rawlings B, et al. Genetic epidemiological studies of early-onset deafness in the US school-age population. Am J Med Genet 1993;46:486–91. [DOI] [PubMed] [Google Scholar]
- 24.Tekin M, Arnos K, Pandya A. Advances in hereditary deafness. Lancet 2001;358:1082–90. [DOI] [PubMed] [Google Scholar]
- 25.Kimberling W, Moller C. Clinical and molecular genetics of Usher syndrome. J Am Acad Audiol 1995;6:63–72. [PubMed] [Google Scholar]
- 26.Eudy J, Sumegi J. Molecular genetics of Usher syndrome. Cell Mol Life Sci 1999;56:258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colville D, Savige J. Alport syndrome. A review of the ocular manifestations. Ophthalmic Genet 1997;18:161–73. [DOI] [PubMed] [Google Scholar]
- 28.Beighton P, Bartmann L, Bingham G, et al. Rod-cone dystrophy, sensorineurl deafness and renal dysfunction: an autosomal recessive syndrome? Am J Med Genet 1993;47:832–6. [DOI] [PubMed] [Google Scholar]
- 29.Meire F, Genderen MV, Lemmens K, et al. Thiamine-responsive megaloblastic anemia syndrome (TRMA) with cone dystrophy. Ophthalmic Genet 2000;21:243–50. [PubMed] [Google Scholar]
- 30.Rosenburg T, Parving A. A syndrome with retinitis pigmentosa, progressive hearing impairment, vestibular dysfunction and congenital cataract. Acta Ophthalmol Scand 1996;219 (suppl):50–3. [DOI] [PubMed] [Google Scholar]
- 31.Ponjavic V, Andreasson S, Tranebjaerg L, et al. Full-field electroretinograms in a family with Mohr-Tranebjaerg syndrome. Acta Ophthalmol Scand 1996;74:632–5. [DOI] [PubMed] [Google Scholar]
- 32.Smith P, Bain S, Good P, et al. Pigmentary retinal dystrophy an the syndrome of maternally inherited diabetes and deafness caused by the mitochondrial DNA 3243 tRNA(Leu) A to G mutation. Ophthalmology 1999;106:1101–8. [DOI] [PubMed] [Google Scholar]
- 33.Massin P, Virally-Monod M, Vialettes B, et al. Prevalence of macular pattern dystrophy in maternally inherited diabetes and deafness. ophthalmology 1999;106:1821–7. [DOI] [PubMed] [Google Scholar]
- 34.Harrison T, Boles R, Johnson D, et al. Macular pattern retinal dystrophy, adult-onset diabetes, and deafness: a family study of A3243G mitochondrial heteroplasmy. Am J Ophthalmol 1997;124:217–21. [DOI] [PubMed] [Google Scholar]
- 35.Souied E, Sales M, Soubrane G, et al. Macular dystrophy, diabetes, and deafness associated with a large mitochondrial DNA deletion. Am J Ophthalmol 1998;125:100–3. [DOI] [PubMed] [Google Scholar]
- 36.Gyapay G, Morisette J, Vignal A, et al. The 1993–1994 Genethon human linkage map. Nat Genet 1994;7:246–339. [DOI] [PubMed] [Google Scholar]
- 37.Dib C, Faure S, Fizames C, et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 1996;380:152–4. [DOI] [PubMed] [Google Scholar]
- 38.Schaffer A. Faster linkage analysis computations for pedigrees with loops or unused alleles. Hum Hered 1996;46:226–35. [DOI] [PubMed] [Google Scholar]
- 39.Cottingham R, Idury R, Schaffer A. Faster sequential genetic linkage computations. Am J Hum Genet 1993;53:252–63. [PMC free article] [PubMed] [Google Scholar]
- 40.O’Connell J, Weeks D. Program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998;63:259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holz F, Evans K, Gregory C, et al. Autosomal dominant macular dystrophy simulating North Carolina macular dystrophy. Arch Ophthalmol 1995;113:178–84. [DOI] [PubMed] [Google Scholar]
- 42.Resendes B, Williamson R, Morton C. At the speed of sound: gene discovery in the auditory system. Am J Hum Genet 2001;69:923–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keats B, Berlin C. Genomics and hearing impairment. Genome Res 1999;9:7–16. [PubMed] [Google Scholar]
- 44.Zelante L, Gasparini P, Estivill X, et al. Connexin26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum Mol Genet 1997;6:1605–9. [DOI] [PubMed] [Google Scholar]
- 45.Eudy J, Sumegi J. Molecular genetics of Usher syndrome. Cell Mol Life Sci 1999;56:258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vervoort R, Wright A. Mutations of RPGR in X-linked retinitis pigmentosa (RP3). Hum Mutat 2002;19:486–500. [DOI] [PubMed] [Google Scholar]
- 47.Joensuu T, Hamalainen R, Yuan B, et al. Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am J Hum Genet 2001;69:673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alagramam K, Yuan H, Kuehn M, et al. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet 2001;10:1709–18. [DOI] [PubMed] [Google Scholar]