Abstract
In addition to their essential catalytic role in protein biosynthesis, aminoacyl-tRNA synthetases participate in numerous other functions, including regulation of gene expression and amino acid biosynthesis via transamidation pathways. Herein, we describe a class of aminoacyl-tRNA synthetase-like (HisZ) proteins based on the catalytic core of the contemporary class II histidyl-tRNA synthetase whose members lack aminoacylation activity but are instead essential components of the first enzyme in histidine biosynthesis ATP phosphoribosyltransferase (HisG). Prediction of the function of HisZ in Lactococcus lactis was assisted by comparative genomics, a technique that revealed a link between the presence or the absence of HisZ and a systematic variation in the length of the HisG polypeptide. HisZ is required for histidine prototrophy, and three other lines of evidence support the direct involvement of HisZ in the transferase function. (i) Genetic experiments demonstrate that complementation of an in-frame deletion of HisG from Escherichia coli (which does not possess HisZ) requires both HisG and HisZ from L. lactis. (ii) Coelution of HisG and HisZ during affinity chromatography provides evidence of direct physical interaction. (iii) Both HisG and HisZ are required for catalysis of the ATP phosphoribosyltransferase reaction. This observation of a common protein domain linking amino acid biosynthesis and protein synthesis implies an early connection between the biosynthesis of amino acids and proteins.
Protein synthesis requires the association of amino acids with the nucleotide triplets of the genetic code, a reaction mediated by tRNA adapters and their specific aminoacyl-tRNA synthetases (aaRSs). As reflected in the absence of some of the canonical 20 aaRSs in contemporary organisms and the duplication and truncation of aaRS in others, some variation in components involved in proteins synthesis has persisted over evolution (1–3). For example, contemporary archaebacterial and bacterial species possess transamidation pathways that use glutamyl-tRNAGln and aspartyl-tRNAAsn (produced by GluRS and AspRS, respectively) as substrates (4–6). These transamidation pathways account for the absence of GlnRS and AsnRS in these species. The aaRSs also regulate the biosynthetic operons responsible for tryptophan, branched-chain amino acids, and histidine (7–9) by attenuation mechanisms that couple transcription of the operon to translation of leader peptides rich in codons specific for the amino acids in question. Notably, both the transamidation pathways and the regulation by attenuation are dependent on the same aminoacylation reactions that generate aminoacylated tRNA for protein synthesis.
The further involvement of aaRS or aaRS-like proteins in amino acid biosynthesis is also suggested by the existence of proteins that are based on the catalytic domains of an aaRS yet do not catalyze the aminoacylation reaction. A striking illustration is the asparagine synthetase A (AsnA), whose recently solved structure contains a class II aaRS catalytic domain (closely related to AspRS and AsnRS). The role of AsnA is to convert aspartate to asparagine via an amidation reaction involving a transient aspartyl-adenylate (10). The high degree of structural homology between AsnA and AspRS and the observation that truncated aaRS catalytic domains retain residual catalytic activity [e.g., MetRS (11), AlaRS (12), and HisRS (13)] lend support to proposals that synthetases arose by fusion of specialized tRNA interaction and editing domains to primordial catalytic domains capable of amino acid activation (14). Obtaining corroborating evidence for this theory is hindered by the difficulty in distinguishing between homologous proteins that might have been antecedents to the aaRS and proteins that might have started as functional aaRS but lost aminoacylation capacity over evolution.
We therefore sought to address this model by identifying and characterizing proteins in contemporary organisms that contain isolated aaRS functional domains. The studies reported herein were undertaken to determine the relationship between naturally occurring fragments of HisRSs and fragments produced biochemically (13). Herein, we describe a protein family (the HisZ family) related to the catalytic core of the contemporary class II HisRS whose members lack aminoacylation activity, as predicted by sequence comparisons with functional synthetases. The first member of the HisZ family was originally identified in Lactococcus lactis as an ORF of unknown function with significant homology to class II aaRSs, as indicated by sequences resembling the three diagnostic sequence motifs (15). We present herein genetic and biochemical evidence that support its direct involvement as an essential subunit of ATP phosphoribosyltransferase (HisG), the first enzyme in histidine biosynthesis. Other members of the HisZ family are identified, and the implications of these observations for aaRS evolution are discussed.
MATERIALS AND METHODS
Materials.
Escherichia coli 16S/23S RNA is from Boehringer Mannheim. The hairpin ribozyme RNA was a gift from J. Burke and J. Heckman (University of Vermont, Burlington). HT-Tuffryn (0.45-μm porosity) membranes were obtained from Gelman Sciences, nitrocellulose membranes (BA-85, 0.45-μm porosity) were purchased from Schleicher & Schuell, and positively charged nylon membranes (Hybond-N+ )were obtained from Amersham Pharmacia. Ni-nitrilotriacetic acid agarose was provided by Qiagen (Chatsworth, CA). [3H]Histidine and [γ-32P]ATP were supplied by Amersham Pharmacia Biotech. T4 polynucleotide kinase and bacterial alkaline phosphatase were provided by New England BioLabs and Amersham Lab Science, respectively.
Gene Manipulation and Protein Purification.
Expression vectors (derived from pQE30; Qiagen) for preparation of HisG and HisZ were constructed by inserting the corresponding genes downstream from the sequence of a (His)6 affinity tag, as described (16). Purification of HisZ and HisG as a protein complex was accomplished by use of an expression vector in which both genes were inserted in tandem, with a (His)6 affinity tag appended to HisZ. In the HisZ-HisG tandem expression construct, the (His)6 affinity tag was appended to HisZ but not to HisG. All proteins were overexpressed in E. coli JM105, [thi rpsL (Strr)endA sbcB15 hsdR4 supE D(lac-proAB) F′:traD36 pro AB laclqZDM15] and were purified to homogeneity by affinity chromatography (16, 17). After elution from the Ni-nitrilotriacetic acid resin with a gradient of 0–0.5 M imidazole, the peak fractions were identified by SDS/PAGE, pooled, dialyzed against wash buffer (50 mM potassium phosphate, pH 6.0/300 mM NaCl/10 mM 2-mercaptoethanol/10% glycerol), concentrated, and stored at −20°C in 50% glycerol. The wild-type HisRS protein from E. coli lacking an affinity tag was purified as described (18).
Synthesis of in Vitro tRNAHis Transcript and Aminoacylation Reaction.
A synthetic gene encoding L. lactis tRNAHis downstream from the T7 RNA polymerase promoter was constructed in pGFIB and transcribed in vitro as described (16). Transcripts were purified from 12% polyacrylamide/urea gels to single-nucleotide resolution and ethanol-precipitated. The concentrations of tRNA stock solutions were determined by absorbance measurements at 260 nm by using a conversion factor of 40 μg/μl per OD unit. Aminoacylation assays of L. lactis tRNAHis were conducted as described (16).
Filter Binding Experiments.
Filter binding assays were performed as described (19). Typical assays contained 10–30 nM of in vitro-transcribed RNA molecules 5′-end-labeled with 32P that was incubated at 25°C with HisZ protein over a wide concentration range (5 nM–1 mM) in a standard reaction buffer [50 mM potassium phosphate, pH 5.5/100 mM KCl/10 mM MgCl2/0.5 μg of yeast (nonspecific) RNA/2 mM 2-mercaptoethanol]. Labeling of tRNA transcripts at their 5′ ends was performed with [γ-32P]ATP and T4 polynucleotide kinase on molecules previously dephosphorylated with alkaline phosphatase (20). Reactions were filtered immediately after binding through a triple-membrane sandwich (Tuffryn, nitrocellulose, and nylon) to allow quantification of both bound and free RNAs (21). After washing, the membranes were separated and dried, and the radioactivity was measured by using a PhosphorImager (Bio-Rad GS-525).
Bacterial Strains and Complementation Experiments.
L. lactis strains were grown at 30°C on chemically defined medium [CDM contains (per liter) 0.5 g of NH4Cl, 9.0 g of KH2PO4, 7.5 g of K2HPO4, 0.2 g of MgCl2, 5 mg of FeCl2, 50 mg of CaCl2, 5 mg of ZnSO4, 2.5 mg of CoCl2, 0.05 g of tyrosine, 0.1 g of asparagine, 0.1 g of cysteine, 0.1 g of glutamine, 0.1 g of isoleucine, 0.1 g of leucine, 0.1 g of methionine, 0.1 g of tryptophan, 0.1 g of valine, 0.1 g of histidine, 0.2 g of arginine, 0.2 g of glycine, 0.2 g of lysine, 0.2 g of phenylalanine, 0.2 g of threonine, 0.3 g of alanine, 0.3 g of proline, 0.3 g of serine, 10 mg of para-aminobenzoic acid, 10 mg of biotin, 1 mg of folic acid, 1 mg of nicotinic acid, 1 mg of panthotenic acid, 1 mg of riboflavin, 1 mg of thiamine, 2 mg of pyridoxine, 1 mg of cyanocobalamin, 5 mg of orotic acid, 5 mg of 2-deoxythymidine, 5 mg of inosine, 2.5 mg of dl-6,8-thioctic acid, 5 mg of pyridoxamine, 10 mg of adenine, 10 mg of guanine, 10 mg of uracil, 10 mg of xanthine, and 5 g of glucose]. Growth was measured kinetically with a Microbiology Reader Bioscreen C (Labsystems, Chicago). The wild-type L. lactis strain (WT) was NCDO2118 (22). The HisZ strain is a mutant of NCDO2118 containing a 600-bp in-frame deletion of the 983-bp HisZ gene. For complementation studies, E. coli strains were grown in minimal medium supplemented with the necessary components as required (23). The prototroph E. coli strain is TG1 [supE Dthi (lac-proAB) hsdD5 (F′+ traD36 proAB lacIqZΔM15)] and the E. coli HisG− mutant is strain JC411 from the Genetic Stock Center [leuB6, fhuA2, lacY1, supE44, gal-6, λ−, hisG1, rfbD1, galP63, argG6, rpsL104, malT1(λR), xyl-7, mtl-2, metB1].
ATP Phosphoribosyltransferase Assays.
ATP phosphoribosyltransferase activity was monitored at A290 as an increase in the formation of N-1-(5′-phosphoribosyl)-ATP (PR-ATP), as described (24). The measurements were performed at room temperature in 100 mM Tris⋅HCl (pH 8.5) containing 10 mM MgCl2, 150 mM KCl, 5 mM ATP, 5 mM 2-mercaptoethanol, and inorganic pyrophosphatase at 2 μg/ml. The purified proteins were added to a final concentration of 100 nM. 5-Phosphoribosyl 1-pyrophosphate (PRPP; 0.5 mM) was added last to start the reaction. Assays were conducted in a U2000 UV/Vis spectrophotometer (Hitachi, Tokyo) in a 1-cm quartz cuvette. The absorbance was detected every 9 s for 10 min after initiating the reaction. The baseline absorbance was established in a similar titration solution without PRPP.
RESULTS
HisZ Constitutes a Family of Proteins in Diverse Bacterial Species.
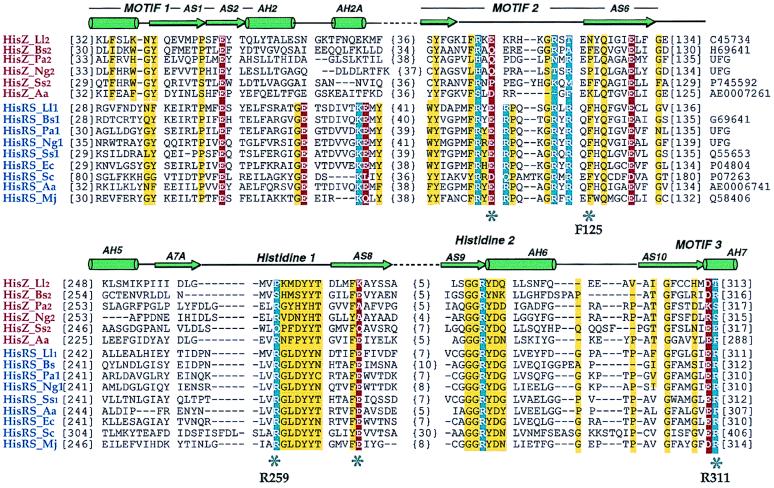
The first member of the HisRS-like family (designated herein as HisZ) was identified as the second ORF (Orf3) in the histidine biosynthesis operon of L. lactis (15). This HisZ protein possesses all three sequence motifs diagnostic for the class II aaRSs and, because of its truncation immediately after motif 3, is missing the mixed α/β anticodon binding domain (25) found in the class IIa subgroup. Notably, HisZ also lacks several essential catalytic residues that have been shown by mutagenesis to be important for the aminoacylation function by class II aaRS in general and by the E. coli HisRS in particular (25–28) (Fig. 1). The genome of L. lactis also includes a full-length version of HisRS that presumably fulfills the typical role of a functional aaRS in protein synthesis (29). Subsequent to the description of the L. lactis HisZ, we identified additional HisRS/HisZ pairs in Bacilllus subtilis, Pseudomonas aeruginosa, Neisseria gonorrhoeae, Synechocystis sp. PC6803, and Aquifex aeolicus (Fig. 1). The absence of one or both of the catalytic arginines in the histidine 1 peptide and the GLER peptide of motif 3 (27) suggests that none of these HisZ proteins is a functional aaRS in vivo. This is in marked contrast to the paralogous LysRS (LysU) and ThrRS (thrZ) in E. coli and B. subtilis, respectively, which are functional aaRSs (30, 31).
Figure 1.
Structure-based sequence alignment of HisRS-like proteins (HisZ) and representative functional histidyl-tRNA synthetases (HisRSs). Species names are abbreviated as follows: Ll, L. lactis; Bs, B. subtilis; Pa, P. aeriginosa; Ng, N. gonorrhoeae; Ss, Synechocystis sp.; Aa, A. aeolicus; Ec, E. coli; Sc, Saccharomyces cerevisae; Mj, Methanococcus jannaschii. Numbers at the left of the sequences indicate positions of first aligned residues in the block, whereas the bracketed numbers indicate spacing between the elements of the blocks. Swiss-Prot, Protein Identification Resource, and National Center for Biotechnology Information (NCBI)/GenBank accession numbers are indicated to the right. UFG indicates sequences retrieved from the unfinished genomes of the NCBI server. The three diagnostic class II motifs and HisRS-specific peptide motifs are indicated and are overlaid by a schematic representation of HisRS secondary structure (25, 28). Highly conserved neutral, acidic, and basic residues are outlined in yellow, red, and blue, respectively; and key residues (E. coli HisRS numbering) that denote significant differences between the HisRS functional and HisRS-like proteins are labeled.
HisZ Lacks Aminoacylation and Pyrophosphate Exchange Activities.
The functional analysis of HisZ from L. lactis was initiated by expression and purification of the protein to homogeneity by nickel affinity chromatography (17). HisZ migrates on denaturating polyacrylamide as a single chain with a molecular mass of 36 kDa. Preliminary sedimentation velocity experiments indicate that HisZ is multimeric, compared with the monomeric truncated version of HisRS produced biochemically (13) (data not shown). Sequence changes between HisZ and HisRS may, therefore, be related to different multimeric states. In vitro experiments were subsequently performed to investigate the functional relationship between HisZ and the E. coli HisRS. HisZ was unable to aminoacylate an in vitro transcript that was based on the L. lactis tRNAHis sequence and did not catalyze the ATP-pyrophosphate (PPi) exchange reaction (data not shown). The E. coli HisRS is able to efficiently charge the L. lactis tRNAHis, allowing competition experiments to be performed. These results showed that up to a 100-fold excess of HisZ had no effect on the aminoacylation plateau characteristic of HisRS (data not shown). The absence of canonical aaRS function demonstrated by these experiments confirmed the predictions from sequence analysis and raised the possibility that HisZ is involved in biological functions other than translation.
HisZ Is an RNA Binding Protein But Does Not Regulate the His Operon Transcription.
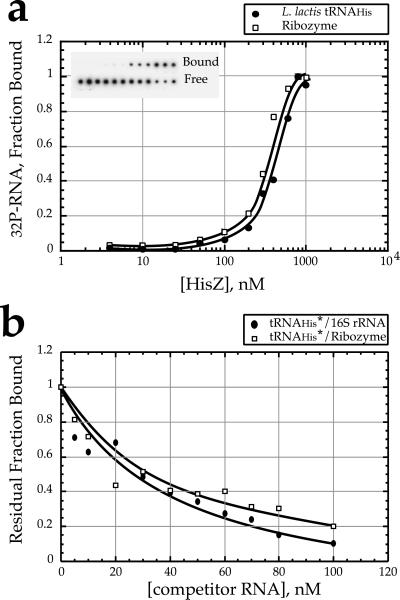
As a next hypothesis, we considered the possibility that HisZ serves a role associated with the regulation of transcription of the his operon. Notably, the genes for both the L. lactis and the B. subtilis HisZ proteins (Fig. 1, sequence HisZ_Bs2) are located within the promoter-proximal portions of the histidine operons, and the role of HisRS in attenuation control of the E. coli and Salmonella typhimurium his operons is well known (32). The his operon of L. lactis contains a “T-box” consensus sequence downstream of the promoter that is involved in a transcriptional antitermination mechanism. According to the model originally proposed for the regulation of aaRS and amino acid biosynthetic operon in B. subtilis and other Gram-positive bacteria, uncharged tRNA directly interacts with the specifier sequence of the mRNA and the T-box sequence to stabilize the antiterminator stem and promote transcriptional antitermination (22, 33). In a variant of this model suggested by the retention of basic and polar side chains associated with RNA binding (Fig. 1), HisZ could inhibit antitermination by altering the balance between charged and uncharged tRNAs or could increase antitermination by stabilizing tRNA–mRNA complex. Filter-binding experiments subsequently confirmed the ability of HisZ to bind tRNA with a dissociation constant of (2.4 ± 0.3) × 10−7 M (Fig. 2a). However, a similar Kd was determined for a RNA that was based on the hairpin ribozyme, suggesting that the interaction with RNA is nonspecific. Both the ribozyme RNA and 16S/23S rRNA from E. coli also competed efficiently for tRNAHis binding (Fig. 2b). In other experiments, no binding discrimination between charged and uncharged tRNAs could be detected (data not shown). The relatively nonspecific nature of the interaction makes it unlikely that HisZ would be able to effectively regulate the his operon by altering the T-box antitermination mechanism. This lack of binding discrimination is consistent with the absence of the domain present in functional HisRS homologs that is responsible for anticodon recognition.
Figure 2.
RNA binding by L. lactis HisZ determined by filter binding experiments. Assays were performed as described (19). (a) 32P-labeled L. lactis tRNAHis (●) and hairpin ribozyme (□) were incubated at 25°C with HisZ protein over a concentration range of 5 nM to 1 mM. The binding constant for tRNA reported in the text represents the global fit of five experiments. (b) Competition experiments between labeled L. lactis tRNAHis transcripts (<5 nM) and unlabeled nonspecific RNA competitors over the range of 0 to 100 nM (●, E. coli 16S/23S RNA; □, hairpin ribozyme) and in the presence of 500 nM HisZ.
Additional genetic experiments conducted to rule out regulatory models also suggested a direct role for HisZ in histidine biosynthesis, as opposed to a role in transcriptional regulation. A his-Promoter/T-box/luciferase fusion was used to determine the effect of HisZ on his operon expression, because previous work had suggested an effect of HisZ overexpression on the regulation of the his operon (29). This fusion was integrated on the chromosome in the his promoter locus in a wild-type strain and in a hisZ− strain. In the wild-type strain, the luciferase activities were 1.3 × 103 lux/OD unit ±1 × 102 lux/OD unit and 92 × 103 lux/OD unit ±7 × 103 lux/OD unit, in the presence and absence of histidine (but supplemented with histidinol), respectively. In the hisZ− strain, the luciferase activities were 2.5 × 103 lux/OD unit ±1 × 102 lux/OD unit and 190 × 103 lux/OD unit ±4 × 103 lux/OD unit under the same conditions, respectively. Thus, rates of antitermination (expressed as the ratio of luciferase in absence and presence of histidine) were similar for the wild-type strain and for the hisZ− strain (71 versus for 76). Similar results were obtained with plasmid constructs (data not shown).
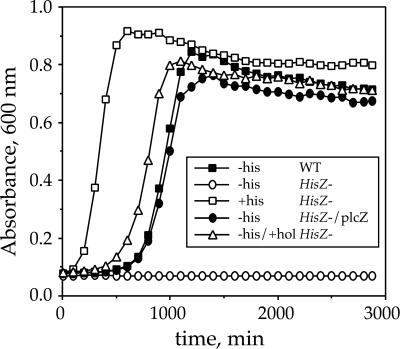
Additional experiments using a strain carrying an in-frame deletion of HisZ demonstrated that the deletion conferred a failure to grow on minimal medium in the absence of exogenously supplied histidine (Fig. 3). This deficiency could be relieved by trans complementation with HisZ or by inclusion of histidinol, the substrate for histidinol dehydrogenase (HisD) and the last intermediate in the histidine biosynthetic pathway. These results confirm that HisZ is unlikely to be involved in regulation of the his operon. However, failure to grow in the absence of added histidine or histidinol indicated a role in histidine biosynthesis at a step before the step catalyzed by HisD.
Figure 3.
Genetic evidence for the role of HisZ in histidine biosynthesis. L. lactis strains differing by the presence (WT) or the absence (HisZ−) of HisZ were grown at 30°C on chemically defined medium (CDM) without histidine (−his) or supplemented with 0.01% histidine (+his) or histidinol (+hol). plcZ stands for a high-copy-number plasmid derivative of pJIM2279 (22) Emr (erythromycin resistance) and containing HisZ gene under control of a constitutive promoter.
HisZ Is a Functional Subunit of the ATP Phosphoribosyltransferase.
Having eliminated the regulatory model, we begun to focus on a direct role of HisZ in histidine biosynthesis. Early literature in the his operon field showed that the ATP phosphoribosyltransferase (HisG, first enzyme involved in the histidine biosynthesis pathway) from enteric bacteria binds tRNA with an affinity of about 100 nM (34). The observed affinity for tRNA and the apparent involvement of HisZ in histidine biosynthesis prompted consideration of a model in which HisZ serves as a functional subunit of the HisG enzyme. In support of this model, revertants of a L. lactis HisZ− deletion strain were selected, and these mutations were mapped within HisG (data not shown). Furthermore, we noticed a striking covariation between the presence of HisZ in bacterial genomes and the length of the HisG polypeptides, as deduced from the translational sequence (Table 1). In all cases, the lengths of the HisG ORFs in genomes from taxa containing HisZ were shorter by some 80–100 amino acids, whereas other his biosynthetic enzyme polypeptide lengths remained unchanged. Sequence alignments of HisG genes reveal that the reduced length is accounted for by the deletion of a contiguous region of the C terminus (data not shown).
Table 1.
Covariation between the presence of HisZ and the length of HisG polypeptides in various bacterial species
| Organisms | Polypeptide length, no. residues
|
HisZ homolog | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HisA | HisB | HisC | HisD | HisF | HisG | HisH | HisIE | ||
| E. coli | 245 | 356* | 356 | 434 | 258 | 299 | 196 | 204 | − |
| H. influenzae | 249 | 362* | 367 | 427 | 258 | 303 | 199 | 221 | − |
| S. typhimurium | 246 | 355* | 359 | 433 | 258 | 299 | 194 | 203 | − |
| S. solfataricus | 232 | 193 | 376 | 398 | 251 | 290 | 199 | 85 | − |
| M. janischii | 237 | 197 | 373 | 429 | 272 | 288 | 198 | 222 | − |
| L. lactis | 240 | 200 | 360 | 430 | 244 | 208 | 202 | 212 | + |
| Synechocystis sp. | 256 | 161 | 352 | 409 | 261 | 210 | 210 | 230 | + |
| B. subtilis | 238 | 192 | 354 | 426 | 253 | 214 | 212 | 212 | + |
| A. aeolicus | 238 | 192 | 354 | 426 | 253 | 205 | 207 | 205 | + |
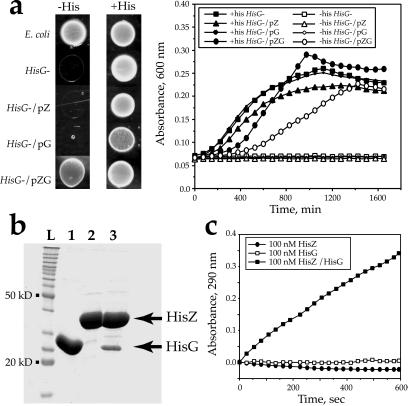
The HisG enzyme catalyzes the transfer of ATP to PRPP producing PR-ATP and PPi. PR-ATP is then converted by a series of nine enzymatic steps to histidine in a process elucidated in E. coli (37) and S. typhimurium (38). To prove the involvement of HisZ in PRPP-ATP transferase function, we did both genetic and biochemical experiments. An E. coli strain containing a deletion in the chromosomal copy of HisG was transformed with plasmids encoding L. lactis HisG alone, L. lactis HisZ alone, or both L.lactis HisG and HisZ. Only the construct containing both genes in tandem was able to confer histidine prototrophy (Fig. 4a). This genetic complementation experiment provides strong evidence that HisZ serves as a functional subunit of HisG in vivo. This evidence raised the possibility that a physical association between the two proteins is a requirement for the transferase reaction. To test this model, an expression vector was constructed by inserting the genes for L. lactis HisZ and L. lactis HisG in tandem, with a (His)6-affinity tag appended to HisZ. As shown in Fig. 4b (lane 3), an additional polypeptide of 27 kDa copurified with HisZ, whereas no additional protein copurified with HisZ in the absence of the tandem HisG (Fig. 4b, lane 2). Subsequently, the identity of both proteins was confirmed by N-terminal sequencing. Only HisZ carries an affinity tag in this experiment, so that HisG must have copurified as a result of a direct physical interaction between HisZ and HisG. To confirm that the interaction between HisG and HisZ was specific and not a reflection of an intrinsic affinity of HisG for the nitrilotriacetic acid-agarose support, additional immunoprecipitation experiments with a monoclonal antibody specific for the (His)6 affinity tag were performed, and these showed that HisG was coprecipitated with HisZ, despite the absence of an affinity tag on the former (data not shown).
Figure 4.
HisZ and HisG are both required for PRPP-ATP transferase activity. (a) Complementation of an E. coli HisG mutant strain by L. lactis HisG and HisZ. E. coli strains differing by the presence (E. coli) or the absence (HisG−) of HisG were grown at 37°C on minimal medium supplemented with 0.0025% amino acids (arginine, leucine, and methionine) without (−his) or with (+his) histidine. Growth on plates was assayed by spotting 5-μl drops of washed rich-medium overnight culture on plates containing minimal medium, and growth kinetics were measured. The HisG− strain was complemented with a high copy number plasmid vector (pBS, Stratagene) containing various L. lactis his DNA fragments. Plasmid pZG [formally pIL704 (15)] has a 2.2-kb fragment containing the HisZ and HisG genes in tandem. Plasmids pZ (expressing the HisZ gene) and pG (expressing the HisG gene) are derivatives of pZG with a 538-bp deletion in the HisG gene or a 600-bp deletion in the HisZ gene, respectively. (b) Physical association of HisZ and HisG. Typical SDS/polyacrylamide gel showing L. lactis HisG (lane 1), HisZ (lane 2), and HisZ/HisG (lane3) purified by affinity chromatography (17) from overexpression E. coli strains. For homogenous HisG and HisZ (lanes 1 and 2), expression vectors were as described above. L stands for a protein ladder (BenchMark, GIBCO/BRL). (c) Effect of L. lactis HisZ and/or L. lactis HisG proteins on the PRPP conversion, detected by A290. The purified proteins were provided at a final concentration of 100 nM, and the reaction was initiated by the addition of PRPP (0.5 mM). The HisZ-HisG trace refers to a preparation from the tandem expression construct. The accumulation of PR-ATP product was monitored by A290 (24).
The activity of PRPP-ATP transferase can be assayed by monitoring an absorbance increase at 290 nm over time, which reflects the accumulation of PR-ATP (24). Assays were performed with purified HisZ, HisG, and the HisZ/HisG preparation obtained from the tandem construct. As shown in Fig. 4c, neither HisZ nor HisG alone (in the presence of PRPP, ATP, and inorganic pyrophosphatase) were able to convert PRPP into PR-ATP. However, incubation with HisZ and HisG led to a significant increase in A290 compared with either protein alone. This activity was completely inhibited by the presence of 1 mM histidine, which is characteristic of the E. coli or S. typhimurium HisG enzymes (39) (data not shown).
DISCUSSION
The HisZ subfamily described herein plays an important role in histidine biosynthesis that was not anticipated previously on the basis of sequence analysis. However, both the functional HisRS and a class of HisRS-like proteins specific to eukaryotes (GCN2) are involved in regulation of histidine biosynthesis (32, 40). The GCN2 class possesses a Ser-Thr kinase domain and a HisRS-like domain (40, 41). Binding of uncharged tRNA by the HisRS-like domain activates the kinase, which through phosphorylation of eIF2α increases the translation of the general amino acid control factor GCN4. These three families (functional HisRS, HisZ, and GCN2) share limited but significant sequence identity and are likely to share a common protein fold, as suggested by the class II aaRS catalytic domain. A phylogenetic analysis of the HisRS family suggests that the HisZ subfamily is the result of an early gene duplication in the bacteria, whereas the GCN2 subfamily represents a separate duplication of a primordial HisRS gene in the Eukarya (J.B. and C.F., unpublished results). The independent nature of these early duplications is supported by the fact that the functional roles of HisZ and GCN2 are distinct. Notably, we have no evidence at present that the apparent nonspecific RNA binding properties of HisZ have any functional significance.
The evolution of histidine biosynthetic pathways was likely to be important in the development of cellular physiology, as a result of the essential catalytic role of histidine in the active sites of enzymes and because of the direct connections between nitrogen metabolism in general and purine, pyrimidine, and tryptophan biosynthesis in particular. The high energetic costs associated with the biosynthesis of each molecule of histidine likely exerted selective pressure on primitive organisms to develop multilevel regulatory circuits. Although the transcriptional controls observed in enteric bacteria require several cell generations for the new steady-state level to be reached, feedback inhibition of the first enzyme of the pathway serves as a major control that provides rapid regulation of biosynthetic activity as a function of the availability of exogenous amino acid (42). The precise role of the HisZ subunit(s) in the transferase remains to be elucidated, but a model suggested by conservation of histidine binding site residues in HisZ (Fig. 1) is that it allows allosteric control of the transferase by histidine and AMP. Notably, mutations in the E. coli HisG that confer unresponsiveness to feedback inhibitors map to the C-terminal part of HisG (43), possibly in the segment absent in the short versions of HisG found in Gram-positive bacteria. This C-terminal extension of HisG shares no obvious sequence homology with aaRS or with any other known RNA binding domain (data not shown).
According to current models, the aaRS evolved from simpler enzymes based on the canonical class I and class II catalytic domains that possessed amino acid activation function but were limited in their ability to catalyze sequence specific aminoacylation of tRNA (44). This follows from the modular organization of the aaRS, which is evident in their three-dimensional structures and in the retention of limited catalytic activity by the isolated class conserved core domains produced biochemically (11–13, 44). Two noteworthy contemporary proteins that are based on isolated aaRS catalytic or tRNA binding domains are AsnA and the biotin repressor (BirA), which show homology to the isolated catalytic domains of AspRS and SerRS, respectively (10, 45). Both AsnA and BirA catalyze reactions that include adenylated intermediates, which is in marked contrast to the HisZG enzyme. The absence of adenylation function in HisZ and its putative role as the functional binding site for regulation by histidine lends credence to the model that the first aaRS progenitors may in fact have been simple amino acid binding proteins. Later evolutionary improvements might have included the ability to activate amino acids by condensation with ATP and the ability to bind tRNA in a sequence specific fashion. This latter step required the accrual of additional RNA binding domains capable of folding independently. Along these lines, the recent report that ribosomal protein L25, which plays a critical role in 5S RNA binding, is structurally related to the anticodon binding domains of GlnRS is highly significant (46).
The occurrence of HisZ and other aaRS fragments in contemporary genomes is also consistent with proposals that the aaRS are ancient proteins that may have played crucial roles in the transition from an RNA-based metabolism. Because of the participation of ATP as a direct biosynthetic constituent of histidine, some authors have speculated that histidine itself may be a molecular remnant of a catalytic moiety of an oligonucleotide (47). The examples provided by HisZ, AsnA, and BirA raise the possibility of an early close connection between the biosynthesis of amino acids, cofactors, and the aminoacylation reaction required in all living organisms.
Acknowledgments
We thank J. Burke and J. Heckman for the gift of hairpin ribozyme RNA, the Protein Chemistry Laboratory (University of Texas Medical Branch, Galveston) for polypeptide sequencing, and D. Johnson for helpful comments on the manuscript. This work is supported by National Institutes of Health Grant GM54899 and Contract B104-CT96-0498 from the Commission of the European Communities.
ABBREVIATIONS
- aaRS
aminoacyl-tRNA synthetase
- PR-ATP
N-1-(5′-phosphoribosyl)-ATP
- PRPP
5-phosphoribosyl 1-pyrophosphate
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Bult C J, White D, Olsen G, Zhou L, Fleischmann R, Sutton G, Blake J, Fitzgerald L, Clayton R, Gocayne J, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 2.Kunst F, Ogasawara N, Moszer I, Albertini A, Alloni G, Azeveda V, Bertero M, Bessieres P, Bolotin A, Borchert S, et al. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 3.Deckert G, Warren P, Gaasterland T, Young W, Lenox A, Graham D, Overbeek R, Snead M, Keller M J, Aujay M, et al. Nature (London) 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 4.Curnow A W, Ibba M, Söll D. Nature (London) 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 5.Curnow A W, Tumbula D L, Pelaschier J T, Min B, Söll D. Proc Natl Acad Sci USA. 1998;95:12838–12843. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker H D, Kern D. Proc Natl Acad Sci USA. 1998;95:12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landick R, Turnbourgh J C L, Yanofsky C. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1263–1286. [Google Scholar]
- 8.Umbarger H E. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 352–367. [Google Scholar]
- 9.Ames B N, Tsang T H, Buck M, Christman M F. Proc Natl Acad Sci USA. 1983;80:5240–5242. doi: 10.1073/pnas.80.17.5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatsu T, Kato H, Oda J. Nat Struct Biol. 1998;5:15–19. doi: 10.1038/nsb0198-15. [DOI] [PubMed] [Google Scholar]
- 11.Kohda D, Yokoyama S, Miyazawa T. J Biol Chem. 1987;262:558–563. [PubMed] [Google Scholar]
- 12.Lechler A, Martin A, Zuleeg T, Limmer S, Kreutzer R. Nucleic Acids Res. 1997;25:2737–2744. doi: 10.1093/nar/25.14.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Augustine J G, Francklyn C S. Biochemistry. 1997;36:3473–3482. doi: 10.1021/bi962395y. [DOI] [PubMed] [Google Scholar]
- 14.Schimmel P, Giegé R, Moras D, Yokoyama S. Proc Natl Acad Sci USA. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delorme C, Ehrlich S D, Renault P. J Bacteriol. 1992;174:6571–6579. doi: 10.1128/jb.174.20.6571-6579.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan W, Augustine J, Francklyn C. Biochemistry. 1996;35:6559–6568. doi: 10.1021/bi952889f. [DOI] [PubMed] [Google Scholar]
- 17.Crowe J, Dobeli H, Gentz R, Hochuli E, Stuber D, Henco K. Methods Mol Biol. 1994;31:371–387. doi: 10.1385/0-89603-258-2:371. [DOI] [PubMed] [Google Scholar]
- 18.Francklyn C, Harris D, Moras D. J Mol Biol. 1994;241:275–277. doi: 10.1006/jmbi.1994.1498. [DOI] [PubMed] [Google Scholar]
- 19.Yarus M, Berg P. Anal Biochem. 1970;35:450–465. doi: 10.1016/0003-2697(70)90207-1. [DOI] [PubMed] [Google Scholar]
- 20.Silberklang M, Gillum A M, RajBhandary U L. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- 21.Wong I, Lohman T M. Proc Natl Acad Sci USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delorme C, Ehrlich S D, Renault P. J Bacteriol. 1999;181:2026–2037. doi: 10.1128/jb.181.7.2026-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 24.Voll M J, Appella E, Martin R G. J Biol Chem. 1967;242:1760–1767. [PubMed] [Google Scholar]
- 25.Arnez J G, Harris D C, Mitschler A, Rees B, Francklyn C S, Moras D. EMBO J. 1995;14:4143–4155. doi: 10.1002/j.1460-2075.1995.tb00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavarelli J, Eriani G, Rees B, Ruff M, Boeglin M, Mitschler A, Martin F, Gungloff J, Thierry J, Moras D, et al. EMBO J. 1994;13:327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnez J G, Augustine J G, Moras D, Francklyn C S. Proc Natl Acad Sci USA. 1997;94:7144–7149. doi: 10.1073/pnas.94.14.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aberg A, Yaremchuk A, Tukalo M, Rasmussen B, Cusack S. Biochemistry. 1997;36:3084–3094. doi: 10.1021/bi9618373. [DOI] [PubMed] [Google Scholar]
- 29.Renault P, Godon J-J, Goupil N, Delorme C, Corthier G, Ehrlich S D. Dev Biol Stand. 1995;85:431–441. [PubMed] [Google Scholar]
- 30.Brevet A, Chen J, Lévèque F, Blanquet S, Plateau P. J Biol Chem. 1995;270:14439–14444. doi: 10.1074/jbc.270.24.14439. [DOI] [PubMed] [Google Scholar]
- 31.Putzer H, Laalami S, Brakhage A A, Condon C, Grunberg-Manago M. J Mol Microbiol. 1995;16:709–718. doi: 10.1111/j.1365-2958.1995.tb02432.x. [DOI] [PubMed] [Google Scholar]
- 32.Yanofsky C. Nature (London) 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- 33.Grundy F J, Henkin T M. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 34.Kovach J S, Phang J M, Blasi F, Barton R W, Ballesteros-Olmo A, Goldberger R F. J Bacteriol. 1970;104:787–792. doi: 10.1128/jb.104.2.787-792.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alifano P, Fani R, Liò P, Lazcano A, Bazzicalupo M, Carlomagno M S, Bruni C B. Microbiol Rev. 1996;60:44–69. doi: 10.1128/mr.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fani R, Mori E, Tamburini E, Lazcano A. Origins Life Evol Biosphere. 1998;28:555–570. doi: 10.1023/a:1006531526299. [DOI] [PubMed] [Google Scholar]
- 37.Bruni C B, Musti A M, Frunzio R, Blasi F. J Bacteriol. 1980;142:32–42. doi: 10.1128/jb.142.1.32-42.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldberger R F, Kovach J S. Curr Top Cell Regul. 1972;5:285–308. doi: 10.1016/b978-0-12-152805-8.50014-9. [DOI] [PubMed] [Google Scholar]
- 39.Bell R M, Parsons S M, Dubravac S A, Redfield A G, Koshland D E J. J Biol Chem. 1974;249:4110–4118. [PubMed] [Google Scholar]
- 40.Hinnebusch A G. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 41.Zhu S, Sobolev A Y, Wek R C. J Biol Chem. 1996;271:24989–24994. doi: 10.1074/jbc.271.40.24989. [DOI] [PubMed] [Google Scholar]
- 42.Brenner M, Ames B N. In: Metabolic Pathways. Vogel H J, editor. Vol. 5. New York: Academic; 1971. pp. 349–387. [Google Scholar]
- 43.Sterboul C C, Kleeman J E, Parsons S M. Arch Biochem Biophys. 1977;181:632–642. doi: 10.1016/0003-9861(77)90269-7. [DOI] [PubMed] [Google Scholar]
- 44.Buechter D D, Schimmel P. Crit Rev Biochem Mol Biol. 1993;28:309–322. doi: 10.3109/10409239309078438. [DOI] [PubMed] [Google Scholar]
- 45.Artymiuk P J, Rice D W, Poirrette A R, Willet P. Nat Struct Biol. 1994;1:758–760. doi: 10.1038/nsb1194-758. [DOI] [PubMed] [Google Scholar]
- 46.Stoldt M, Wohnert J, Gorlach M, Brown L R. EMBO J. 1998;17:6377–6384. doi: 10.1093/emboj/17.21.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fani R, Liò P, Lazcano A. J Mol Evol. 1995;41:760–774. doi: 10.1007/BF00173156. [DOI] [PubMed] [Google Scholar]