Abstract
Among mucus-secreting cells, the gastric gland mucous cells, Brunner’s glands, accessory glands of pancreaticobiliary tract, and pancreatic ducts exhibiting gastric metaplasia are unique in that they express class III mucin identified by paradoxical Con A staining composed of periodate oxidation, sodium borohydride reduction, Con A, and horseradish peroxidase reaction. Recently it was shown that these mucous cells secrete glycoproteins having GlcNAcα1→4Galβ→R at nonreducing terminals of the carbohydrate moieties. Herein we describe the expression cloning of a cDNA encoding a human α1,4-N-acetylglucosaminyltransferase (α4GnT), a key enzyme for the formation of GlcNAcα1→4Galβ1→R. COS-1 cells were thus cotransfected with a stomach cDNA library and a leukosialin cDNA. Transfected COS-1 cells were screened by using monoclonal antibodies specific for GlcNAcα1→4Galβ→R and enriched by fluorescence-activated cell sorting. Sibling selection of recovered plasmids resulted in a cDNA clone that directs the expression of GlcNAcα1→4Galβ→R. The deduced amino acid sequence predicts a type II membrane protein with 340 amino acids, showing no significant similarity with any other proteins. The α4GnT gene is located at chromosome 3p14.3, and its transcripts are expressed in the stomach and pancreas. An in vitro GlcNAc transferase assay by using a soluble α4GnT revealed that α1,4-linked GlcNAc residues are transferred most efficiently to core 2 branched O-glycans (Galβ1→4GlcNAcβ1→6(Galβ1→3)GalNAc), forming GlcNAcα1→4Galβ1→4GlcNAcβ1→6(GlcNAcα1→4Galβ1→3)GalNAc. Transfection of α4GnT cDNA into gastric adenocarcinoma AGS cells produced class III mucin, indicating that α4GnT is responsible for the formation of class III Con A reactivity. These results indicate that the α4GnT is a glycosyltransferase that forms α1,4-linked GlcNAc residues, preferentially in O-glycans.
Gastrointestinal mucins, heavily glycosylated glycoproteins, play protective roles against extracellular environments by forming a continuous mucous gel layer (1–3). To understand the physiological roles of the gastrointestinal mucins more precisely, various histochemical techniques have been developed to detect alimentary mucosubstances (4, 5). Among them, paradoxical Con A staining, a sequential staining method composed of periodate oxidation, sodium borohydride reduction, Con A binding, and horseradish peroxidase reaction, is particularly unique (6). In the normal human tissues, this method specifically identifies mucous glycoproteins, termed class III mucin, secreted from gland mucous cells (cardiac and pyloric gland mucous cells and mucous neck cells) of the stomach, Brunner’s glands of the duodenum, and accessory glands of the pancreaticobiliary tract (7, 8). In addition, class III mucin is also detected in pancreatic ducts that show gastric metaplasia, resembling pyloric mucosa of the stomach (8, 9), and thus these mucous glycoproteins as a whole have been regarded as the gastric gland mucous cell-type mucin. Interestingly, class III mucin also can be frequently detected in gastric and pancreatic carcinoma cells (8–10). Consequently, the paradoxical Con A staining has been used widely during studies on pathogenesis as well as carcinogenesis of these cancers (11–13). However, the carbohydrate epitope responsible for the class III Con A reactivity has not been fully elucidated because of complexity of O-glycans in this mucin (14).
Recently, a series of mAb, HIK1083, PGM36, and PGM37 against rat or pig gastric mucins, has been developed (15, 16). The analysis of the rat gastric mucins by using NMR spectroscopy revealed that HIK1083 antibody recognizes peripheral α1,4-GlcNAc attached to Gal residues of core 2 branched O-glycan, GlcNAcα1→4Galβ1→4GlcNAcβ1→6(GlcNAcα1→4Galβ1→3)GalNAc (15). Similarly, it was also demonstrated that PGM36 and PGM37 antibodies require a peripheral α-linked GlcNAc for binding (16). In parallel, immunohistochemical studies in human digestive tract indicate that these mAbs specifically react with gland mucous cells of the stomach, Brunner’s glands of the duodenum, accessory glands of the pancreaticobiliary tract, and pancreatic duct cells that exhibit gastric metaplasia (7, 8). These results all together are consistent with the previous reports that O-glycans having α1,4-linked GlcNAc in nonreducing terminal are present only in gastric and duodenal mucins of rat and pig (17, 18). In addition, the histochemical comparison between paradoxical Con A staining and immunostaining by using these mAbs in human alimentary tract indicated that the staining pattern of HIK1083, PGM36, or PGM37 antibody is quite similar to that of class III mucin, suggesting that peripheral α1,4-linked GlcNAc residues are involved in class III Con A reactivity (7, 8).
As the first step to understanding the roles of mucous glycoproteins having peripheral α1,4-linked GlcNAc residues, we describe here the expression cloning and functional analysis of a cDNA encoding human α1,4-N-acetylglucosaminyltransferase (α4GnT). By using the cDNA isolated, we have established that α4GnT forms GlcNAcα1→4Galβ→R preferentially in core 2 branched oligosaccharides and that the transcripts of α4GnT are predominantly expressed in the stomach and pancreas. We have also shown that GlcNAcα1→4Galβ→R is responsible for class III Con A reactivity by expressing α4GnT.
EXPERIMENTAL PROCEDURES
Expression Cloning of a cDNA Encoding Human α4GnT.
We first found that COS-1 cells were negative for HIK1083, PGM36, or PGM37 staining (15, 16) even after transfection with pRcCMV-leu harboring human leukosialin cDNA (19), indicating that COS-1 cells do not express peripheral α-linked GlcNAc residues. For the expression cloning, COS-1 cells (1.2 × 107 cells) were thus transfected with 30 μg of a human stomach cDNA library (provided by CLONTECH) constructed in a mammalian expression vector, pcDNAI together with 30 μg of pRcCMV-leu by using LipofectAmine (Life Technologies, Grand Island, NY), as described previously (20). After 60 hr, COS-1 cells expressing peripheral α-linked GlcNAc residues on their cell surface were enriched by fluorescence-activated cell sorting by using FACStar (Becton Dickinson) and an antibody mixture composed of HIK1083, PGM36, and PGM37. Subsequently, plasmid DNAs were isolated by the Hirt procedure (21) from the sorted cells and then transformed into the host bacteria, MC1061/P3 cells by using Cell-Porator (Life Technologies). Because the pcDNAI vector encodes a sup F gene that corrects the defect of both ampicillin- and tetracycline-resistant genes in the P3 episome, MC1061/P3 cells transformed by pcDNAI become resistant to both antibiotics. In contrast, MC1061/P3 cells transformed by pRcCMV-leu alone are resistant to ampicillin but not to tetracycline. Because of this difference for the antibiotics, only plasmids derived from the human stomach cDNA library were selectively rescued and amplified in the presence of ampicillin and tetracycline. These transformed MC1061/P3 cells were then divided into 10 small pools for the subsequent sibling selection.
The plasmids derived from each pool were independently transfected into COS-1 cells together with pRcCMV-leu, and the transfectants were screened by immunofluorescence microscopy by using the antibody mixture to identify a single plasmid encoding a human α4GnT, pcDNAI-α4GnT. The nucleotide sequence was determined in both directions by the dye dideoxy nucleotides chain-termination method (22) by using a 373A DNA sequencer (Applied Biosystems).
Northern Blot Analysis of Various Human Tissues.
Human multiple tissue Northern blots for normal adult tissues (CLONTECH) were hybridized with gel-purified cDNA insert of pcDNAI-α4GnT after labeling with [α-32P]dCTP by random-oligonucleotide priming by using Prime-It II labeling kit (Stratagene), as described before (23).
Detection of GlcNAcα1→4Galβ→R or Class III Mucin.
To detect GlcNAcα1→4Galβ→R in various human tissues, immunohistochemistry by using HIK1083 antibody was carried out as described (7, 8). Briefly, formalin-fixed and paraffin-embedded normal human tissue specimens selected from the pathology files of Central Clinical Laboratories, Shinshu University Hospital, Matsumoto, Japan, were immersed in absolute methanol containing 0.3% H2O2 for 30 min. Immunohistochemical staining with HIK1083 antibody was performed by using the indirect immunostaining method (24). For the secondary antibody, goat anti-mouse Ig conjugated with horseradish peroxidase (DAKO) was used, and peroxidase activity was visualized with a diaminobenzidine-hydrogen peroxide solution. A control experiment was done by omitting the primary antibody from the staining procedure, and no specific staining was found.
For detection of class III mucin, paradoxical Con A staining was performed as described (6). Briefly, the formalin-fixed specimens were oxidized with 1% periodate for 60 min, and then reduced with 0.2% sodium borohydride for 2 min. After washing, the specimens were incubated with 0.1% Con A (Sigma) for 60 min at room temperature and then immersed in a 0.001% horseradish peroxidase solution for 30 min. The peroxidase activity was developed in a diaminobenzidine-hydrogen peroxide solution.
In Vitro GlcNAc Transferase Assay and Product Characterization.
To construct a vector encoding a soluble form of chimeric α4GnT, the COOH-terminal segment including catalytic domain of this enzyme (nucleotides 82 to 1,035) was amplified by using PCR with pcDNAI-α4GnT as a template. Upstream and downstream primers used for this PCR were 5′-CGGGATCCGAGCTGCCTCTTCTGTTTGC-3′ (BamHI site shown in italics) and 5′-CCGCTCGAGCGAGTGTTAGCTTTATTTGTTAC-3′ (XhoI site shown in italics), respectively. The amplified product was digested by BamHI and XhoI and then cloned into the BamHI and XhoI sites of pcDNAI-A (23), resulting in pcDNAI-A-α4GnT.
CHO cells were transfected with pcDNAI-A-α4GnT, and a soluble chimeric α4GnT fused with protein A was recovered by using IgG-Sepharose (Amersham Pharmacia Biotech), as described previously (23). The enzyme was assayed by using various acceptors in a reaction mixture essentially as described previously (23), except that 5 mM MnCl2 instead of EDTA was added. For this assay, Galβ1→4GlcNAcβ1→6(Galβ1→3)GalNAcα→p-nitrophenol (pNP) was enzymatically synthesized from GlcNAcβ1→6(Galβ1→3)GalNAcα→pNP (Toronto Research Chemicals, Downsview, ON, Canada), as described before (25). In addition, GlcNAcβ1→3GalNAcα→pNP was purchased from Toronto Research Chemicals, whereas the rest of the oligosaccharides were from Sigma.
To produce a large amount of the enzymatic product for NMR analysis, the enzymatic reaction was scaled up to a total volume of 300 μl under the same conditions as described above. After incubation at 37°C overnight, the sample was purified by a Sep-Pak C18-cartridge column as described before (25). The product was more than 99% pure judging from HPLC analysis and was then subjected to NMR analysis. The NMR spectra were acquired on a 500-MHz Varian Unity-Plus spectrometer. All spectra of the acceptor molecule and its precursors starting from α-d-GalNAc→pNP were used to assign the chemical shifts. The spectra were taken in D2O after repeated exchange with D2O. The 1H NMR spectrum was assigned through a combination of two-dimensional (2D) double-quantum-filtered correlation spectroscopy and 2D total correlation spectroscopy (50 ms) experiments. To get a better resolution, spectra were taken by varying the temperature (5°C, 22°C, and 30°C). The 13C assignments and glycosidic linkages were confirmed with the help of heteronuclear multiple quantum coherence spectroscopy and 2D rotating frame nuclear Overhauser effect spectroscopy (150 ms and 300 ms), respectively.
Establishment of AGS Cell Lines Stably Expressing GlcNAcα1→4Galβ→R.
A preliminary study revealed that human gastric adenocarcinoma AGS cells express the transcripts for two different core 2 β1,6-N-acetylglycosaminyltransferases, C2GnT-M (23) and C2GnT-L (26), but not peripheral α1,4-linked GlcNAc residues on their cell surface. Thus, AGS cells were cotransfected with pcDNAI-α4GnT and pSV2neo in a 10:1 molar ratio by using LipofectAmine. For a control experiment, mock transfection was performed by using pcDNAI in place of pcDNAI-α4GnT. After selection with G418, clonal cell lines stably expressing GlcNAcα1→4Galβ→R on their cell surface were isolated by immunofluorescent staining with HIK1083 antibody. These cells were cultured as monolayers in Lab-Tek chamber slides (Nunc) and then subjected to paradoxical Con A staining (6).
Chromosomal Mapping of α4GnT Gene.
The chromosomal localization of α4GnT gene was assigned by use of a Stanford G3 Radiation Hybrid Panel (27), by using PCR as described previously (23). The upstream and downstream primers specific for human α4GnT were 5′-TGTGGGAATGCATGGAAAAC-3′ and 5′-AGTGTGTTGCTTCCTCTAATC-3′, respectively. The PCR mixtures were denatured for 10 min at 95°C, before 30 cycles at 94°C for 30 sec, 60°C for 30 sec, and 72°C for 23 sec, followed by 72°C for 3 min 30 sec. The amplified product was electrophoresed through 3.0% agarose gels. The results of the 83 radiation hybrid DNA clones (0000001000 0000001000 0000000000 0000000000 1011001000 0000000000 0000000100 0000001100 010) were sent through the World Wide Web to the RH server at the Stanford Human Genome Center (http://www-shgc.stanford.edu/RH/rhserverformnew.html).
RESULTS
Isolation of a cDNA Clone That Determines the Expression of GlcNAcα1→4Galβ→R Structure in the Cell Surface of COS-1 Cells.
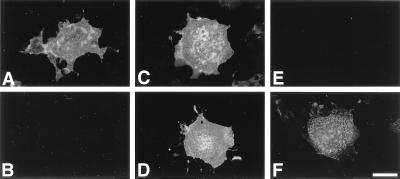
COS-1 cells transfected with pRcCMV-leu encoding human leukosialin were negative for either HIK1083, PGM36, or PGM37 staining, indicating the absence of peripheral α-linked GlcNAc residues (Fig. 1 A and B). Therefore, we transiently cotransfected 1.2 × 107 COS-1 cells with the human stomach cDNA library together with pRcCMV-leu. Sixty hours after the transfection, COS-1 cells expressing peripheral α-linked GlcNAc residues were enriched by fluorescence-activated cell sorting by using the antibody mixture composed of HIK1083, PGM36, and PGM37 under conditions where only highly positive cells were selected. Plasmid DNAs were rescued from these sorted cells and amplified in Escherichia coli MC1061/P3 cells in the presence of ampicillin and tetracycline. Thus, 325 colonies of the transformed MC1061/P3 cells were obtained from 258 sorted cells and then divided into 10 small pools for the subsequent sibling selection. One of the small pools was found to produce strong cell-surface staining detected by the antibody mixture when cotransfected with pRcCMV-leu. Thus, we selected this particular pool for the subsequent rounds of sibling selection with sequentially smaller active pools and eventually identified a single plasmid, pcDNAI-α4GnT.
Figure 1.
Expression of GlcNAcα1→4Galβ→R by pcDNAI-α4GnT. COS-1 cells were cotransfected with pcDNAI-α4GnT (C and D) or pcDNAI (A and B) together with pRcCMV-leu, or transfected with pcDNA-α4GnT alone (E and F). Sixty hours after transfection, the cells were fixed and examined by incubation with HIK1083 antibody (B, D, and F) or anti-leukosialin antibody, 1G10 (PharMingen) (A, C, and E), followed by FITC-conjugated anti-mouse IgM (for HIK1083) or anti-mouse IgG (for 1G10). (Bar = 50 μm).
COS-1 cells transiently cotransfected with the pcDNAI-α4GnT together with pRcCMV-leu were positive for immunostaining with HIK1083 antibody (Fig. 1 C and D), establishing that the cloned α4GnT forms GlcNAcα1→4Galβ→R. Interestingly, weaker staining was observed when only pcDNAI-α4GnT was transfected (Fig. 1 E and F), suggesting that the HIK1083 antibody preferentially reacts with α1,4-GlcNAc attached to clustered O-glycans. Other mAbs, PGM36 and PGM37, also gave positive cell-surface staining on the transfected COS-1 cells, as did HIK1083 (data not shown).
Predicted Amino Acid Sequence of Human α4GnT.
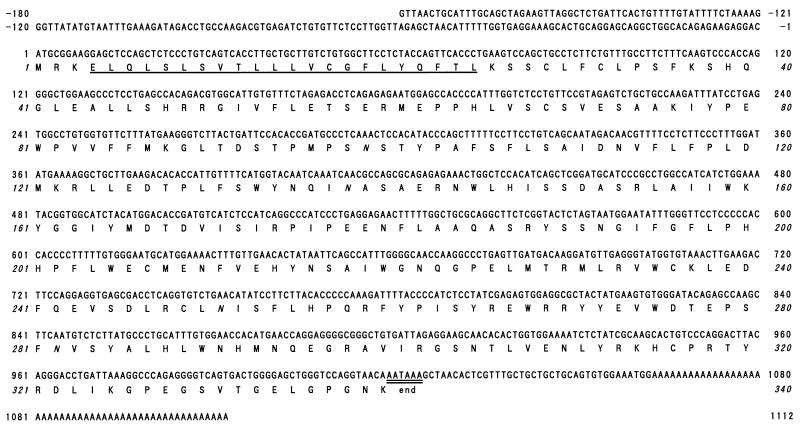
The cDNA insert of pcDNAI-α4GnT, which consists of 1,292 base pairs, contains an ORF predicting a protein of 340 amino acid residues with a molecular mass of 39,497 Da (Fig. 2). Four potential N-glycosylation sites are present in this protein. A hydropathy plot analysis predicts that this protein has a type II membrane topology, as is found in almost all mammalian glycosyltransferases cloned to date (28). Namely, a very short cytoplasmic NH2-terminal segment of three amino acid residues is followed by a transmembrane/signal anchoring domain (amino acid residues 4–25), which is flanked by basic amino acid residues. A large COOH-terminal segment consisting of stem and catalytic domains presumably resides in the Golgi lumen. A consensus sequence for the polyadenylation signal is present at nucleotides 1,019–1,024, and interestingly, TAA of the AATAAA also served as a terminal codon of the translation. No significant similarity was found between this protein sequence and other sequences including glycosyltransferases reported in GenBank.
Figure 2.
The nucleotide and deduced amino acid sequence of human α4GnT. The signal/membrane-anchoring domain is singly underlined, and a consensus polyadenylation signal is doubly underlined. Potential N-glycosylation sites are shown in italics. The sequences are numbered relative to the translation initiation site.
Transcripts of α4GnT Are Expressed in the Stomach and Pancreas.
The expression of α4GnT mRNA in various normal human tissues was examined by Northern blot analysis (Fig. 3). A transcript of α4GnT approximately 1.7 kb in size was detected in the stomach and pancreas. In addition, 2.1-kb and 0.7-kb transcripts were detected in stomach and pancreas, respectively.
Figure 3.
Northern blot analysis of α4GnT in various human tissues. Each lane contained 2 μg of poly(A)+ RNA. The blot was probed by 32P-labeled α4GnT cDNA.
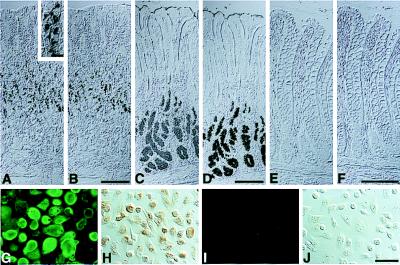
To test whether the expression of cloned α4GnT is correlated with the expression of GlcNAcα1→4Galβ→R, the normal human tissue specimens examined for the Northern blot analysis in this study were subjected to immunohistochemistry by using HIK1083 antibody. As shown previously (7, 8), and as further confirmed here, glycoproteins having GlcNAcα1→4Galβ→R are strongly expressed in mucous neck cells and pyloric gland cells of the gastric mucosa (Fig. 4 A and C) as well as accessory glands and gastric metaplasia of the pancreatic ducts. In other tissues, for example in colon (Fig. 4E), the peripheral α1,4-linked GlcNAc residues are not detectable. These results, taken together, strongly suggest that the cloned α4GnT directs the expression of GlcNAcα1→4Galβ→R specifically found in gland mucous cells of the stomach and accessory glands as well as metaplastic ducts of the pancreas.
Figure 4.
Expression of GlcNAcα1→4Galβ→R structures and class III mucin in the human gastrointestinal mucosa and the gastric adenocarcinoma ASG cells stably transfected by α4GnT cDNA. Gastric fundic mucosa (A and B), gastric pyloric mucosa (C and D), and colonic mucosa (E and F) were stained with HIK1083 antibody (A, C, and E) or subjected to paradoxical Con A staining (B, D, and F). Similarly, AGS cells stably transfected with pcDNAI-α4GnT (G and H) or pcDNAI (I and J) were stained with HIK1083 antibody followed by FITC-conjugated anti-mouse IgM (G and I) or subjected to paradoxical Con A staining (H and J). (Bars in B, D, and F = 200 μm; bar in the inset of A = 20 μm; bar in J = 50 μm).
α4GnT Forms GlcNAcα1→4Galβ→R Most Efficiently in Core 2 Branched O-Glycans.
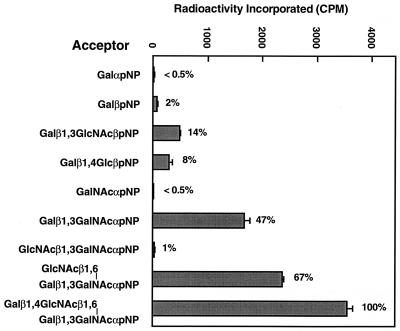
To determine the acceptor specificity of α4GnT, a soluble α4GnT fused with protein A was expressed, and its activity was assayed by using various synthetic acceptors. The results shown in Fig. 5 demonstrate that α4GnT incorporates GlcNAc most efficiently to core 2 branched oligosaccharides, Galβ1→4GlcNAcβ1→6(Galβ1→3)GalNAcα→pNP. Interestingly, α4GnT acts on GlcNAcβ1→6(Galβ1→3)GalNAcα→pNP better than a core 1 acceptor, Galβ1→3GalNAcα→pNP. Product analysis by exoglycosidase treatment demonstrated that only GlcNAcα1→4Galβ1→3(GlcNAcβ1→6)GalNAcα→pNP was formed from Galβ1→3(GlcNAcβ1→6)GalNAcα→pNP (data not shown). These results suggest that the addition of β1,6-linked GlcNAc may change the conformation of the acceptor, resulting in a better acceptor for α4GnT. Similar findings were observed for the action of iGnT on I-branched oligosaccharides (29).
Figure 5.
Incorporation of 3H-GlcNAc from UDP-[3H] GlcNAc to various acceptors by a soluble chimeric α4GnT. The acceptor concentration was 1.0 mM, except for 0.7 mM for Galβ1→4GlcNAcβ1→6(Galβ1→3)GalNAcα→pNP. The incorporated radioactivity was shown relative to that obtained when Galβ1→4GlcNAcβ1→6(Galβ1→3)GalNAcα→pNP was used as an acceptor. SEs are shown by bars.
The α4GnT acted on Galβ1→3GlcNAcβ→pNP slightly better than Galβ1→4Glcβ1→pNP, but acted much better on Galβ1→4GlcNAcβ1→6(Galβ1→3)GalNAcα→pNP or Galβ1→3GalNAcα→pNP (Fig. 5). These results indicate that this enzyme prefers β1,4-linked or β1,3-linked Gal residues on O-glycans. On the other hand, α4GnT did not use UDP-GalNAc when these acceptors were tested (data not shown).
To determine the structure of the oligosaccharide product, the enzymatic reaction was performed by using 300 nmol of Galβ1→4GlcNAcβ1→6(Galβ1→3)GalNAcα→pNP as an acceptor and then subjected to the NMR analysis. The 1H NMR spectrum showed signature signals of doublets for six anomeric protons, as shown in Table 1. These results are consistent with those reported previously (15). Assignments of the non-anomeric proton signals were made on the basis of crosspeaks observed in the 2D-double-quantum-filtered correlation spectroscopy and 2D-total correlation spectroscopy spectra as far as H-4. The 13C NMR spectrum of the product was analyzed by 2D-H-detected heteronuclear 1H-13C correlation by using the inverse mode. The heteronuclear multiple quantum coherence spectroscopy spectrum indicated that C-4 positions of β-d-Gal residues are substituted. In the 2D-rotating frame nuclear Overhauser effect spectroscopy spectrum, two crosspeaks appeared at around δ 3.96 ppm for α-d-GlcNAc H-1/β-d-Gal H-4, indicating the (1→4) linkage between them. Other crosspeaks in the spectrum were also consistent with the proposed structure (details on NMR analysis will be published elsewhere).
Table 1.
Chemical shifts (δ) in ppm of 1H and 13C spectra
| Sugars | H-1 | C-1 | H-2 | C-2 | H-3 | C-3 | H-4 | C-4 | H-6 | C-6 |
|---|---|---|---|---|---|---|---|---|---|---|
| α-d-GalNAc | 5.8 (3.5 Hz) | 97.5 | 4.6 | 49.7 | 4.35 | 78.0 | 4.25 | 70.5 | 3.57 | — |
| 2× α-d-GlcNAc | 4.87 (3.5 Hz) | 99.8 | 3.89 | 54.7 | 3.78 | 71.8 | 3.54 | 70.2 | — | — |
| 4.88 (3.5 Hz) | 99.9 | |||||||||
| 2× β-d-Gal | 4.59 (7 Hz) | 105.5 | 3.56 | 71.2 | 3.72 | 72.1 | 3.96 | 78.3 | — | — |
| 4.42 (8 Hz) | 104.9 | 3.58 | 3.70 | 3.95 | ||||||
| β-d-GlcNAc | 4.44 (8 Hz) | 102.5 | 3.56 | 56.0 | 3.62 | 71.8 | 3.74 | 76.2 | — | — |
—, not determined.
From the above-mentioned spectroscopic data, it can be concluded that the two d-GlcNAc residues were added by the α4GnT to the β-d-Gal moieties through α-(1→4) linkage.
GlcNAcα1→4Galβ→R Formed by α4GnT Is Responsible for Class III Con A Reactivity.
Class III mucin demonstrated by paradoxical Con A staining is expressed in the gastric gland mucous cells where GlcNAcα1→4Galβ→R structures are also detected (in Fig. 4, compare A with B and C with D), suggesting that the peripheral α1,4-linked GlcNAc residues are involved in class III Con A reactivity. To test this hypothesis, AGS cells stably expressing GlcNAcα1→4Galβ→R, termed AGS-α4GnT cells, were established by cotransfection with pcDNAI-α4GnT and pSV2neo, followed by G418 selection. Presence of GlcNAcα1→4Galβ→R structure on the cell surface of AGS-α4GnT cells was confirmed by immunostaining with HIK1083 antibody (Fig. 4G). By using paradoxical Con A staining, class III mucin was detected on AGS-α4GnT cells but not on mock-transfected AGS cells (Fig. 4 H and J). These results establish that GlcNAcα1→4Galβ→R formed by α4GnT is responsible for class III Con A reactivity.
α4GnT Gene Is Mapped to Chromosome 3.
Results by using the Stanford G3 Radiation Hybrid Panel showed that α4GnT gene locus is mapped to chromosome 3, seven cRs from SHGC-53479 (lod score = 10.68). Currently this marker has not been aligned on the radiation hybrid G3 map, but the closest marker of SHGC-53479 is found to be SHGC-33458 (lod score = 4), which is mapped between D3S1569 and D3S1550. These results indicate that the human α4GnT gene locus residues at chromosome 3p14.3.
DISCUSSION
In the present study, we have isolated a cDNA encoding human α4GnT, a key enzyme for the biosynthesis of a unique glycan, GlcNAcα1→4Galβ→R, by using expression cloning. This carbohydrate is characteristic for the gastric gland mucous cell-type mucin and is attached to core 2 branched O-glycans (7, 8, 15). To identify α4GnT cDNA by expression cloning, we used COS-1 cells as recipient cells because COS-1 cells express core 2 β1,6-N-acetylglusoaminyltransferase responsible for core 2 branch (26) but not terminal α-linked GlcNAc residues. To efficiently display carbohydrates having the peripheral α-linked GlcNAc residues on the cell surface of transfected COS-1 cells, we cotransfected a cDNA encoding human leukosialin together with a cDNA library of the human stomach. Leukosialin is a major membrane-bound sialoglycoprotein of leukocytes and contains 80 O-glycans in its extracellular domain (19). Although a protein carrying GlcNAcα1→4Galβ→R structure has not yet been identified, we envisaged that leukosialin could be a preferred substrate for α4GnT, because GlcNAcα1→4Galβ→R structures were found to be attached to core 2 branched O-glycans (15, 17, 18). In fact, when COS-1 cells were transfected with pcDNAI-α4GnT alone, the staining of peripheral α1,4-linked GlcNAc residues by the HIK1083 antibody was weak (see Fig. 1F), indicating that the cotransfection with a leukosialin cDNA was critical for this cloning. This finding is similar to that found during the cloning of C2GnT-L, and coexpression of leukosialin was also critical in that cloning (26). It is also noteworthy that COS-1 cells have an additional advantage of high-level replication of the transferred pcDNAI vectors. The binding of SV40 T antigen expressed in COS-1 cells to the ori sequence for SV40 replication in pcDNAI results in a high number of replicated pcDNAI.
To date, nine different kinds of GlcNAc transferase cDNAs involved in the biosynthesis of glycoproteins have been isolated from mammals, including human. Among them, C2GnT-M and C2GnT-L catalyze the biosynthesis of O-glycans (23, 26), whereas GnT-I to GnT-V are exclusively involved in the N-glycan biosynthesis (30–35). IGnT and iGnT, on the other hand, act on both O-glycans and N-glycans (36, 37). However all the GlcNAc transferases cloned so far transfer β-GlcNAc residues to their specific acceptor carbohydrates, and there has been no report on GlcNAc transferase that adds α-GlcNAc residues to acceptor carbohydrates. The α4GnT demonstrated here is the first cloned GlcNAc transferase that forms GlcNAcα1→4Galβ→R in O-glycans. Mucous glycoproteins having peripheral α1,4-linked GlcNAc residues are shown to be specifically expressed in the gastric gland mucous cell-type mucin found in the stomach, duodenum, and pancreas (7, 8), but much less attention has been paid to this particular carbohydrate because of the complexity of this mucin. Thus, α4GnT cDNA will allow us to elucidate the biosynthetic pathway leading into complex carbohydrates present in the gastric gland mucous cell-type mucin.
Among the mucin core proteins cloned from the gastric mucosa (38, 39), MUC6 can be a candidate for a carrier molecule of the carbohydrates having GlcNAcα1→4Galβ→R, because MUC6 is exclusively expressed in the mucous neck cells and pyloric glands of the gastric mucosa (40). However, Brunner’s glands of the duodenum, which are also positive for HIK1083, lack MUC6 (40). In this study, we did not examine the expression of α4GnT mRNA in the duodenum. Future studies are required to determine whether GlcNAcα1→4Galβ→R is actually attached to MUC6, and whether α4GnT is expressed in Brunner’s glands of the duodenum.
The α4GnT was found to act more efficiently on core 2 branched O-glycans and less efficiently on core 1 oligosaccharide. However, this enzyme hardly transferred GlcNAc to core 3 oligosaccharide, GlcNAcβ1→3GalNAcα→pNP (Fig. 5). The fact that α4GnT weakly acts on Galβ1→3GlcNAcβ→pNP suggests that N-glycans may also serve as substrates for α4GnT, because Galβ1→3GlcNAcβ→R structures can be found not only in O-glycans but also in N-glycans (19, 41). Further study will be of significance to determine whether α4GnT act on N-glycans.
Most Con A reactivity of carbohydrate moieties is lost when they are oxidized with periodate. However, the Con A affinity to particular mucin, termed class III mucin, is paradoxically enhanced after oxidization followed by reduction (6). Previously, Hotta et al. (14) suggested that GlcNAc residues at nonreducing terminal of the gastric glycoproteins are responsible for class III Con A reactivity by analyzing Con A affinity to the isolated human gastric mucins after oxidation, followed by reduction in vitro. In the present study, we have demonstrated that the AGS cells stably transfected with α4GnT cDNA actually express class III mucin (see Fig. 4 G and H). This result established that GlcNAcα1→4Galβ→R synthesized by α4GnT is responsible for class III Con A reactivity. This conclusion is corroborated by the fact that the transcripts for α4GnT are present in the stomach and pancreas, where class III mucin can be detected (7, 8).
Clinicopathological studies demonstrated that class III mucin is frequently detected not only in gastric adenocarcinoma but also in pancreatic ductal carcinoma (8–10). In addition, this mucin is found in the mucinous bronchioloalveolar cell carcinoma of the lung (42) and the so-called adenoma malignum of the uterine cervix (43). Despite the fact that the latter three tumors do not originate from the gastric mucosa, they characteristically show gastric phenotypes such as the presence of pepsinogen II (42–44). It is of significance to determine whether class III mucin plays a role in the pathogenesis of gastric cancer and other tumors including pancreatic cancer exhibiting the gastric phenotypes. It was also demonstrated that in the normal pancreas, class III mucin is expressed in accessory glands, but not in the duct epithelial cells themselves (8, 9). However, the expression of class III mucin in the pancreatic duct is strikingly increased in association with gastric metaplasia, histologically simulating pyloric mucosa of the stomach. This alteration frequently occurs with increased age and often coexists with pancreatic cancer (45). However, the pathogenesis of gastric metaplasia of the pancreatic ducts has not been well understood. The α4GnT cDNA obtained in the present study will be a powerful tool to determine the biological roles of class III mucin carrying GlcNAcα1→4Galβ1→R residues found in the stomach and pancreas under normal and pathological conditions.
Acknowledgments
We thank Drs. K. Hotta and K. Ishihara for the kind gifts of HIK1083, PGM36, and PGM37 antibodies, Dr. H. Ota for useful discussion, Drs. I. Ueno and M. Momose for their skillful technical assistance, and Dr. E. Ong for critical reading of the manuscript. This work is supported by a Grant-in-Aid for Scientific Research on Priority Area (10178104) from the Ministry of Education, Science, Sports and Culture of Japan, Public Trust Haraguchi Memorial Cancer Research Fund, and in part by the Ribosome Engineering Project (Taka-Aki Sato, Ph. D.) in Organized Research Combination System (to J.N.), a grant for Gastric Mucin Research Forum from the Otsuka Pharmaceutical Company, Ltd. (to T.K.), and National Institutes of Health Grants CA33000, CA48737, and CA71932 (to M.F.).
ABBREVIATIONS
- α4GnT
α1,4-N-acetylglucosaminyltransferase
- pNP
p-nitrophenol
- 2D
two-dimensional
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF141315).
References
- 1.Allen A. In: Handbook of Physiology, Vol. III. Rauner B B, editor. New York: Oxford Univ. Press; 1989. pp. 359–382. [Google Scholar]
- 2.Ota H, Katsuyama T. Histochem J. 1992;24:86–92. doi: 10.1007/BF01082444. [DOI] [PubMed] [Google Scholar]
- 3.Matsuo K, Ota H, Akamatsu T, Sugiyama A, Katsuyama T. Gut. 1997;40:782–789. doi: 10.1136/gut.40.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katsuyama T, Ono K, Nakayama J, Kanai M. In: Gastric Mucus and Mucus Secreting Cells. Kawai K, editor. Tokyo: Excerpta Medica; 1985. pp. 3–18. [Google Scholar]
- 5.Spicer S S, Schulte B A. J Histochem Cytochem. 1991;40:1–38. doi: 10.1177/40.1.1370305. [DOI] [PubMed] [Google Scholar]
- 6.Katsuyama T, Spicer S S. J Histochem Cytochem. 1978;26:233–250. doi: 10.1177/26.4.351046. [DOI] [PubMed] [Google Scholar]
- 7.Ota H, Nakayama J, Momose M, Kurihara M, Ishihara K, Hotta K, Katsuyama T. Histochem Cell Biol. 1998;110:113–119. doi: 10.1007/s004180050272. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura N, Ota H, Katsuyama T, Akamatsu T, Ishihara K, Kurihara M, Hotta K. J Histochem Cytochem. 1998;46:793–801. doi: 10.1177/002215549804600702. [DOI] [PubMed] [Google Scholar]
- 9.Matsuzawa K, Akamatsu T, Katsuyama T. Hum Pathol. 1992;23:925–933. doi: 10.1016/0046-8177(92)90407-t. [DOI] [PubMed] [Google Scholar]
- 10.Fujimori Y, Akamatsu T, Ota H, Katsuyama T. Hum Pathol. 1995;26:725–734. doi: 10.1016/0046-8177(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 11.Ota H, Katsuyama T, Akamatsu T, Fujimori Y, Matsuzawa K, Ishii K, Honda T, Nakayama J, Furihata K, Ono K, Maeda K. Acta Histochem Cytochem. 1995;28:43–58. [Google Scholar]
- 12.Furihata C, Tatematsu M, Miki K, Katsuyama T, Sudo K, Miyagi N, Kubota T, Jin S -S, Kodama K, Ito N, et al. Cancer Res. 1984;44:727–733. [PubMed] [Google Scholar]
- 13.Tatematsu M, Furihata C, Katsuyama T, Mera Y, Inoue T, Matsushima T, Ito N. J Natl Cancer Inst. 1987;78:771–777. [PubMed] [Google Scholar]
- 14.Hotta K, Gosa K, Kato Y. Histochemistry. 1982;76:107–112. doi: 10.1007/BF00493289. [DOI] [PubMed] [Google Scholar]
- 15.Ishihara K, Kurihara M, Goso Y, Urata T, Ota H, Katsuyama T, Hotta K. Biochem J. 1996;318:409–416. doi: 10.1042/bj3180409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurihara M, Ishihara K, Ota H, Katsuyama T, Nakano T, Naito M, Hotta K. Comp Biochem Physiol. 1998;121B:315–321. doi: 10.1016/s0305-0491(98)10113-x. [DOI] [PubMed] [Google Scholar]
- 17.Kochetkov N K, Derevitskaya V A, Arbatsky N P. Eur J Biochem. 1976;67:129–136. doi: 10.1111/j.1432-1033.1976.tb10641.x. [DOI] [PubMed] [Google Scholar]
- 18.Van Halbeek H, Gerwig G J, Vliegenthart J F G, Smits H L, Van Kerkhof P J M, Kramer M F. Biochim Biophys Acta. 1983;747:107–116. doi: 10.1016/0167-4838(83)90128-0. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda M. In: Cell Surface Carbohydrates and Cell Development. Fukuda M, editor. Boca Raton: CRC; 1992. pp. 127–159. [Google Scholar]
- 20.Nakayama J, Fukuda M N, Fredette B, Ranscht B, Fukuda M. Proc Natl Acad Sci USA. 1995;92:7031–7035. doi: 10.1073/pnas.92.15.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirt B. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh J-C, Ong E, Fukuda M. J Biol Chem. 1999;274:3215–3221. doi: 10.1074/jbc.274.5.3215. [DOI] [PubMed] [Google Scholar]
- 24.Nakane P K. Ann NY Acad Sci. 1975;254:203–211. doi: 10.1111/j.1749-6632.1975.tb29170.x. [DOI] [PubMed] [Google Scholar]
- 25.Ujita M, McAuliffe J, Schwientek T, Almeida R, Hindsgaul O, Clausen H, Fukuda M. J Biol Chem. 1998;273:34843–34849. doi: 10.1074/jbc.273.52.34843. [DOI] [PubMed] [Google Scholar]
- 26.Bierhuizen M F A, Fukuda M. Proc Natl Acad Sci USA. 1992;89:9326–9330. doi: 10.1073/pnas.89.19.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter M A, Spillett D J, Thomas P, Weissenback J, Goodfellow P N. Nat Genet. 1994;7:22–28. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- 28.Schachter H. In: Molecular Glycobiology. Fukuda M, Hindsgaul O, editors. Oxford: IRL; 1994. pp. 88–162. [Google Scholar]
- 29.Ujita M, McAuliffe J, Suzuki M, Hindsgaul O, Clausen H, Fukuda M N, Fukuda M. J Biol Chem. 1999;274:9296–9304. doi: 10.1074/jbc.274.14.9296. [DOI] [PubMed] [Google Scholar]
- 30.Kumar R, Yang J, Larsen R D, Stanley P. Proc Natl Acad Sci USA. 1990;87:9948–9952. doi: 10.1073/pnas.87.24.9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarkar M, Hull E, Nishikawa Y, Simpson R J, Moritz R L, Dunn R, Schacher H. Proc Natl Acad Sci USA. 1991;88:234–238. doi: 10.1073/pnas.88.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Agostaro G A, Zingoni A, Moritz R L, Simpson R J, Schachter H, Bendiak B. J Biol Chem. 1995;270:15211–15221. doi: 10.1074/jbc.270.25.15211. [DOI] [PubMed] [Google Scholar]
- 33.Nishikawa A, Ihara Y, Hatakeyama M, Kangawa K, Taniguchi N. J Biol Chem. 1992;267:18199–18204. [PubMed] [Google Scholar]
- 34.Minowa M T, Oguri S, Yoshida A, Hara T, Iwamatsu A, Ikenaga H, Takeuchi M. J Biol Chem. 1998;273:11556–11562. doi: 10.1074/jbc.273.19.11556. [DOI] [PubMed] [Google Scholar]
- 35.Saito H, Nishikawa A, Gu J, Ihara Y, Soejima H, Wada Y, Sekiya C, Niikawa N, Taniguchi N. Biochem Biophys Res Commun. 1994;198:318–327. doi: 10.1006/bbrc.1994.1045. [DOI] [PubMed] [Google Scholar]
- 36.Bierhuizen M F A, Mattei M -G, Fukuda M. Genes Dev. 1993;7:468–478. doi: 10.1101/gad.7.3.468. [DOI] [PubMed] [Google Scholar]
- 37.Sasaki K, Kurata-Miura K, Ujita M, Angata K, Nakagawa S, Sekine S, Nishi T, Fukuda M. Proc Natl Acad Sci USA. 1997;94:14294–14299. doi: 10.1073/pnas.94.26.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toribara N W, Roberton A M, Ho S B, Kuo W -L, Gum E, Hicks J W, Gum J, R, Byrd J C, Siddiki B, Kim Y S. J Biol Chem. 1993;268:5879–5885. [PubMed] [Google Scholar]
- 39.Guyonnet Duperat V, Audie J P, Debailleul V, Laine A, Buisine M P, Zouitina-Galieque S, Pigny P, Degand P, Aubert J P, Porchet N. Biochem J. 1995;305:211–219. doi: 10.1042/bj3050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolos C D, Garrido M, Real F X. Gastroenterology. 1995;109:723–734. doi: 10.1016/0016-5085(95)90379-8. [DOI] [PubMed] [Google Scholar]
- 41.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 42.Honda T, Ota H, Ishii K, Nakamura N, Kubo K, Katsuyama T. Am J Clin Pathol. 1998;109:423–430. doi: 10.1093/ajcp/109.4.423. [DOI] [PubMed] [Google Scholar]
- 43.Ishii K, Hosaka N, Toki T, Momose M, Hidaka E, Tsuchiya S, Katsuyama T. Virchows Arch. 1998;432:315–322. doi: 10.1007/s004280050172. [DOI] [PubMed] [Google Scholar]
- 44.Sessa F, Bonato M, Frigerio B, Capella C, Solcia E, Prat M, Bara J, Samloff I M. Gastroenterology. 1990;98:1655–1665. doi: 10.1016/0016-5085(90)91104-e. [DOI] [PubMed] [Google Scholar]
- 45.Kozuka S, Sassa R, Taki T, Masamoto K, Nagasawa S, Saga S, Hasegawa K, Takeuchi M. Cancer. 1979;43:1418–1428. doi: 10.1002/1097-0142(197904)43:4<1418::aid-cncr2820430431>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]