Abstract
Seedlings grown in darkness, i.e., etiolated seedlings, lack chlorophyll and most other components of the photosynthetic apparatus. On illumination, the plastids become photosynthetically competent through the production of chlorophylls and proteins encoded by certain chloroplast and nuclear genes. There are two types of photosynthetic cells in leaves of the C4 plant maize: bundle sheath cells (BSC) and adjacent mesophyll cells (MC). Some proteins of the maize photosynthetic machinery are solely or preferentially localized in MC and others in BSC. A particular gene may be photoregulated up in one cell type and down in the other. Transcripts of the nuclear gene rpl29, encoding the chloroplast ribosomal protein L29, increase in abundance about 17-fold during light-induced maturation of plastids. There is about 1.5 times more L29 protein in ribosomes of greening leaves than in ribosomes of unilluminated leaves; the L29 contents of MC and BSC are about the same. However, L21 is present about equally in plastid ribosomes of unilluminated and illuminated seedlings. In contrast to both L29 and L21, the fraction of the ribosome population containing L2 is about the same in MC and BSC of etiolated leaves but, on illumination, the proportion of the ribosome population with L2 increases in BSC but not in MC. The existence of different subpopulations of plastid ribosomes—e.g., those with and without L21 and/or L29 during development—evokes interesting, but as yet unanswered, questions about the roles of different types of ribosomes in differentiation.
When germinated and grown in darkness, angiosperm seedlings are etiolated. They lack chlorophyll and many protein components of the photosynthetic apparatus. On illumination, the required components of the photosynthetic apparatus form, and the seedlings become photosynthetically competent.
Maize is a C4 plant. Certain steps in photosynthetic carbon fixation are limited to the mesophyll cells (MC) of the leaf, and other photosynthetic processes, including carbon dioxide fixation by ribulose bisphosphate carboxylase, are limited to the adjacent bundle sheath cells (BSC). MC have both of the two photosynthetic energy transducing photosystems (PSI and PSII), but BSC are poor in the oxygen evolving PSII. MC and BSC are morphologically distinct in etiolated leaves. In the course of light-induced development, genes for some components of the photosynthetic apparatus are expressed in both MC and BSC, but others are expressed differently in the two types of cells.
Plastid genes whose transcript levels change on illumination of dark-grown seedlings have been identified by hybridizing mRNAs prepared from unilluminated and illuminated leaf tissues against restriction fragments of plastid DNA (e.g., 1–3). Genes for the large subunit of ribulose bisphosphate carboxylase and genes for components of the energy-transducing elements of the photosynthetic apparatus are prominent among those whose expression has been shown to be promoted on illumination of dark-grown maize seedlings.
To broaden the search for light-induced genes that are expressed differently in MC and BSC beyond plastid genes and certain known nuclear genes, we undertook a differential display analysis (DDA) of cDNA fragments generated from populations of polyadenylated mRNAs of etiolated and greening maize leaves. Although some chloroplast mRNAs are polyadenylated (4), the vast majority of plant polyadenylated mRNAs are of nuclear origin. We report here that the subpopulations of ribosomes in MC and BSC of etioplasts and of chloroplasts of greening leaves differ in regard to the presence or absence of L29 as well as ribosomal protein L2. The fraction of the population containing L21 does not appear to change.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth.
Maize seed (Zea mays: FR 9 cms × Fr 37; Illinois Foundation seeds, Champaign, IL) were germinated and grown in moist vermiculite at 28°C in a dark room for 9 or 10 days. Seedlings then were either kept in the dark (etiolated plants) or illuminated with incandescent lamps (designated “W” or “white”) of approximately 1,500 lux or red light (“R”) [fluorescent tubes (GE F4OR Red) filtered through Roscolux (Rosco, Port Chester, NY) Fire no. 19 plastic] for 8, 24, or 36 hr (plants illuminated for these periods are designated W8, W24, W36, R8, R24, and R36).
To investigate the diurnal and circadian rhythms of abundance of L29 mRNA, plants were grown at 28°C in a growth chamber under 16-h light/8-h night cycles for 10 days. Illumination was provided by white fluorescent lamps (1,500 lux) from 6 a.m. to 10 p.m. In continuous light experiments, plants were maintained in constant light after 10 p.m. In continuous darkness experiments, plants were kept in darkness after 6 a.m.
mRNA Differential Display Analyses.
Three hundred ng of total RNA (after DNase I treatment) from the second leaves of 10-day-old etiolated seedlings or dark-grown seedlings illuminated with incandescent lamps or with red light for 8, 24, or 36 hr before harvest were reverse transcribed in 20-μl reaction mixtures with Superscript II reverse transcriptase (Life Technologies, Gaithersburg, MD). A degenerate oligo(dT) T12MG, T12MC, T12MA, or T12MT (where M represents a mixture of dG, dA, or dC) was used as a primer (Integrated DNA Technologies, Coralville, IA). Reactions were carried out in a thermocycler (Ericomp, San Diego): 65°C for 5 min, 37°C for 1 hr, and then 95°C for 5 min. Reverse transcriptase was added after 10 min at 37°C. Reverse transcriptase was omitted from control reactions.
One-tenth of the cDNA was then amplified by PCR in a solution containing 4 nM dNTP, 1 nM of the corresponding T12MN, and 0.8 nM arbitrary 10 mer (OP-differential display reverse transcription 1–26, Operon Technologies, Alameda, CA) in a total of 20 μl of PCR reaction (Perkin–Elmer/Cetus). Parameters for the 40-cycle PCR were: denaturations at 94°C for 20 sec, annealing at 40°C for 75 sec, and extension at 72°C for 20 sec. The last cycle was followed by a 5-min extension at 72°C. Radio-labeled PCR amplification products were analyzed by electrophoresis in denaturing 6% polyacrylamide gels. To confirm the reproducibility of amplification for selected bands, we repeated the reactions at least three times with different preparations of cDNA.
Recovery and Reamplification.
The cDNA band (No. 40), later found to contain L29 mRNA, was excised from the gel, rehydrated in 100 μl distilled H2O for 10 min, and boiled for 15 min. Debris was removed by centrifugation, and the cDNA in the supernatant was precipitated by the addition of 250 μl 100% ethanol. The pellet was redissolved in 10 μl H2O. Then, 5 μl of the cDNA solution was used to reamplify the DNA for 35 cycles in a total volume of 40 μl with the same primer set and PCR conditions that were used in the initial RT-PCR, except that no radioactive dNTP was included, and 20 nM dNTPs was used. The PCR products in 30 μl of the above were run on a 1.5% agarose gel and stained with ethidium bromide. A DNA fragment of about 400 bp was excised, recovered, and used as a probe for Northern blot analyses.
5′ Rapid Amplification of cDNA Ends (RACE) Assays and cDNA Cloning.
A cDNA fragment predicted to encode the amino-terminal region of maize L29 was isolated by using the Marathon cDNA Amplification Kit (CLONTECH). One μg poly(A+) RNA was extracted from the second leaves of 10-day-old illuminated with incandescent lamps for 8 hr before harvest. The antisense primer 5′-GTCTGACCACAATGCTCTGCTTCC-3′ was used in the amplification of L29 cDNA. PCR amplification was performed according to the manufacturer’s instructions. DNA products were subjected to electrophoresis in a 1% agarose gel, recovered, and cloned into the PCR II vector by using the TA cloning kit (Invitrogen).
Isolation of Bundle Sheath Strands (BSS) and Mesophyll Protoplasts (MP).
BSS and MP were prepared essentially as described previously (7).
RNA Isolation and Northern Blot Hybridization.
To prepare RNA, second leaves of etiolated or greening plants or frozen BSS were ground to a fine powder in liquid N2 and extracted with acid guanidinium thiocyanate solution. The extract was treated with phenol and chloroform as described previously (8). Frozen MP were put directly into the guanidium thiocyanate solution. RNA samples (20–30 μg) were denatured, resolved by electrophoresis in 1% agarose gel containing 2.2 M formaldehyde, and transferred to Gene Screen membrane (Dupont/NEN, Boston, MA) by standard capillary blotting techniques. Specific probes were generated by labeling reamplified or cloned cDNA fragments with [α-32P]dCTP (Dupont/NEN) with a random primer DNA labeling kit (Life Technologies). After hybridization at 42°C (for probe from mRNA differential display experiments) and high-stringency washes at 63°C in 0.2× SSC/0.1% SDS solution, the blots were exposed to Kodak X-omat SR films with intensifying screens. Hybridization with a partial cDNA fragment of the maize 23S rRNA was used as a loading control.
DNA Sequencing and Computer Analysis.
Cloned reamplified PCR fragments or 5′-RACE products were completely sequenced from two directions automatically by using an Applied Biosystems model 373A DNA sequencer at the Boston University Medical Center. The nucleic acid and deduced amino acid sequences were compared with sequences in the database server at the National Center for Biotechnology Information by using the blast network service (9).
Antigen, Antibodies, and Immunoblotting.
By using keyhole limpet hemocyanin as the carrier protein, an antibody was raised in rabbits against a synthetic peptide corresponding to amino acids 128 to 146 (EQGINKRLSRKLDRLWKQS) of L29. The peptide was synthesized and the antibody was generated at Research Genetics (Huntsville, AL).
Antibodies against maize chloroplast ribosomal proteins L2 (10, 11) and L21 (12) were kindly provided by Alap Subramanian.
Western blotting was performed according to the procedures described in Santa Cruz Biochemicals Research Applications (Santa Cruz Biotechnology Santa Cruz, CA) with the following exceptions. For L29 ribosomal proteins alone, the blots were blocked for 1 hr at room temperature with 4% milk in TBST (tris-buffered saline plus 0.05% Tween). Primary antibody incubation was for 1 hr [1:400 in 2% milk plus 2% BSA (Sigma)], followed by three 5-min washes in TBST. Secondary antibodies were incubated for 1 hr against rabbit IgG (purchased from Santa Cruz Biochemicals) (1:2,000 in 2 + 2% solution) followed by three 5-min washes in TBST and one 5-min wash in TBS. Blots were developed in Luminol for 1 min, per the Santa Cruz Biochemicals protocol. For L2, L21, and L29 Western blots, blots were blocked for 1 hr at room temperature in 4% milk in TBST. Primary antibodies were incubated for 1 hr at room temperature (L29 at 1:200; L2 at 1:2,000; L21 at 1:1,000) in 2% milk plus 2% BSA in TBS + t, followed by three 5-min washes in TBS + t. Secondary antibodies were incubated as above.
RESULTS
mRNA Differential Display.
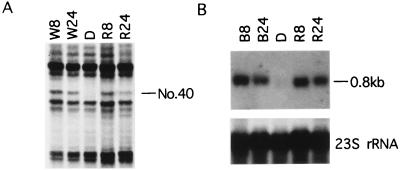
DDA was used to identify fragments of transcribed sequences of genes whose expression is stimulated by light during the development of etiolated Z. mays leaves. With anchor primer T12MG and arbitrary primer AP12 (GATCTAACCG), a unique band (No. 40, Fig. 1A) was detected only in the samples from greening leaves illuminated with white or red light for 8 or 24 hr. By using a cDNA fragment (399 bp, indicated by the two arrows in Fig. 2A), amplified from the DNA eluted from this band as a probe, and RNA samples prepared from etiolated leaves or greening leaves, an intense message about 0.8 kbp in length was detected by Northern blot analysis of the sample from greening leaves (Fig. 1B). Both blue and red light induced the expression of gene No. 40 to the same extent, indicating that phytochrome may be involved in inducing expression of this gene.
Figure 1.
DDA of cDNAs derived from mRNAs from etiolated and illuminated maize leaves and Northern blot analyses of these mRNAs. (A) Part of a gel from a DDA done with primers T12MG and AP12 (GATCTAACCG) is shown. Total RNA preparations from 10-day-old etiolated maize leaves (D) and greening leaves illuminated with white light (W) or red light (R) for 8 hr or 24 hr were subjected to differential display reverse transcription-PCR. Band No. 40 represents the light-regulated cDNA segment of L29. (B) Confirmation of the differential expression pattern of L29. Twenty-five μg of total RNA from etiolated leaves (D) or greening leaves illuminated with blue light (B) or red light (R) for 8 hr or 24 hr was fractionated electrophoretically in a 1% formaldehyde agarose gel, transferred, and probed with [α-32P]dCTP-labeled cDNA of reamplified fragment No. 40. A single band of about 0.8 kbp was detected in both blue light- and red light-treated samples but not in the etiolated samples. Equal loading was confirmed by 23S rRNA hybridization.
Figure 2.
L29 is a maize homologue of B. stearothermophilus ribosomal protein L29. (A) The nucleotide sequence and the deduced amino acid sequences are numbered at the left. The two primers used in the DDA are labeled with arrows. Four bases in the AP12 primer did not match the cDNA sequence. The gene-specific primer used in a 5′-RACE reaction is underlined. Three N-myristylation sites are boxed, and four potential protein kinase C phosphorylation sites are marked with asterisks. The boldfaced letters (amino acids 133–136) designate the potential cAMP/cGMP-dependent protein kinase phosphorylation site (accession no. AF 147725). (B) The amino acid alignment of the homologous regions of maize L29, B. stearothermophilus (BACST) and Bacillus subtilis (BACSLI) is shown. Double and single dots indicate amino acid identity and similarity, respectively. Gaps represented by dashes were inserted to maximize the sequence identity. Consensus amino acids are residues that are identical in the three proteins. Numbers in parentheses indicate the position of the start of an amino acid sequence with respect to the initiator methionine of the corresponding protein. B. stearothermophilus L29 exhibited the highest identity with maize L29 of all sequences in the NCBI protein databases at the time of submission. The region of L29 that can be crosslinked to 23S rRNA in ribosomes of B. stearothermophilus (16) are underlined, and the crosslinked lysyl residues are in boldface type (K). Accession numbers for the bacterial L29 proteins shown: B. stearothermophilus (SPIP04457) and B. subtilis (SPIP12873).
Isolation and Sequence Analysis of L29.
A homologue of the cDNA sequence in clone No. 40 (from our DDA reverse transcription PCR experiments) could not be identified among listed sequences with known functions in any of the DNA databases we examined. Consequently, we used 5′-RACE to clone the full-length cDNA. With a gene-specific primer (underlined sequence in Fig. 2A) along with a RACE adapter primer, we amplified a 580-bp cDNA fragment from a total RNA preparation from greening leaves illuminated with white light for 8 hr. As shown in Fig. 2A, ligation of these two fragments resulted in a full-length cDNA clone (782 bp, as expected from Northern blot analysis) of L29 (light-inducible ribosomal protein L29). The sequence identified by DDA was the same as that of the cDNA clone that had been isolated (from base 384 to the 3′-end), except that there were three mismatched bases located in the upstream primer AP12, which confirmed degenerate binding of the upstream primers that had been noted previously (5, 6). Two methionine codons were found at the beginning of the ORF (Fig. 2A). The nucleotide sequence surrounding the first methionine codon corresponded well to the Kozak consensus translation initiation sequence, with purines at the −3 and at the +4 positions (13). Therefore, the first of these methionines is likely to be the start codon for this protein. Translation of the ORF in the L29 clone predicts a protein of 161 amino acids, of which 51 are charged residues (32 basic and 19 acidic amino acids). This protein would be expected to have a mass of 18,093 Da. Within the predicted protein sequence, there are: one potential cAMP/cGMP-dependent protein kinase phosphorylation site, four potential protein kinase C phosphorylation sites, and three N-myristylation sites (Fig. 2A). The protein would be basic, with a calculated isoelectric point of 11.3 (pI 11.3). A hydropathy plot showed no highly hydrophobic domains typical of proteins that span the membrane in an α-helix conformation (data not shown).
A search of the National Center for Biotechnology Information (NCBI) DNA databases failed to reveal the L29 nucleotide sequence. A search of the NCBI protein database against the predicted protein sequence of L29 revealed that L29 might encode a maize homologue of eubacterial ribosomal protein L29. As shown in Fig. 2B, the middle part of L29 (from amino acid 61 to 133) had more than 45% identity to the Bacillus stearothermophilus ribosomal protein L29 (65% similarity). We also detected sequence similarity to ribosomal protein L29 of other species, such as Halobacterium halobium (SPIP28538, 34% identity, 63% similarity), E. coli (SPIP02429, 26% identity, 63% similarity), and to the yeast 60S ribosomal protein L35 (SPIP39741, 32% identity, 59% similarity). Most of the prokaryotic L29 ribosomal proteins are about 70 aa in length, whereas the eukaryotic homologues of L29 [such as that of yeast (14) and rat L35 (15)] have C-terminal extensions of 56 aa. Similarly, the maize L29 has a 29-aa C-terminal extension, but L29 also has a 69-aa-long N-terminal extension; database searches failed to identify its homologue. The N-extension with moderate hydrophobicity was rich in Ala residue (21 of 69 amino acids; 30%). All three potential N-myristylation sites (amino acids 34–39, 35–40, and 43–48) were located in this region (Fig. 2A). It is possible that the plant N-extension functions as a trafficking signal and/or subcellular plastid localization signal.
The two regions of the sequence underlined in Fig. 2B have been found to crosslink to 23S rRNA in B. stearothermophilus ribosomes by using UV and α-iminothiolane (16). These two sequences are also found in the conserved region of maize L29; the second sequence (amino acids 120–133) is particularly well conserved. It is likely that L29 binds to the ribosome through interaction between this conserved site and 23S rRNA.
Light-Dependent Accumulation of L29 mRNA.
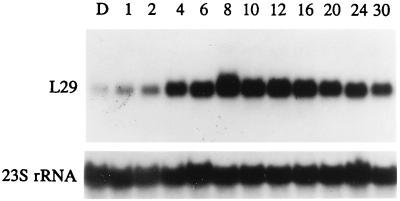
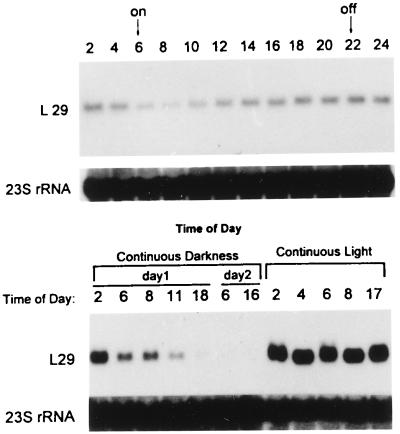
We studied the time course of L29 mRNA expression induced in dark-grown seedlings by white light using full-length cDNA as a probe in Northern blot experiments. Leaves from 10-day-old dark-grown seedlings and leaves from seedlings illuminated with white light for the indicated times were collected separately from each sample, and total RNA was extracted for electrophoresis and Northern analyses. Low but detectable hybridization signals were observed in samples from etiolated leaves (lane D in Fig. 3). L29 mRNA increased about 3-fold after 2-hr illumination with white light; the maximum increase of about 17-fold occurred after 8-hr illumination. After 30 hr of illumination, there was only about five times more L29 mRNA than in etiolated leaves (Fig. 3). Within the next 6 hr of illumination, the amount of L29 mRNA increased a little initially then declined to the baseline level after illumination for 36 hr. But with further illumination, the amount of L29 mRNA increased again, peaked at 40 hr at a somewhat lower level than the first peak, then declined to a level about 5-fold above that in etiolated leaves by 48 hr of illumination (data not shown). Inasmuch as similar fluctuations had been observed in light-harvesting chlorophyll a/b binding protein mRNA (17) during continuous illumination of etiolated seedlings, it seemed possible that L29 mRNA fluctuations, like those of LHCP, might be circadian. Fig. 4A shows that the amount of L29 mRNA in leaves of 10-day-old maize seedlings growing under a 16-hr day/8-hr night cycle differs in light and in darkness. In darkness, the mRNA level declined, but it reached a minimum after only 2 hr in light. Then it rose and peaked at the end of the light period (2200). The maximum and minimum levels differed by more than 3-fold. To investigate whether a circadian rhythm is involved in the control of L29 mRNA accumulation, a set of 10-day-old maize seedlings grown under a 16-hr day (6 a.m.–10 p.m.)/8-hr night cycle was maintained in continuous darkness after 10 p.m., and another set was placed in continuous light at 8:00 a.m. As shown in Fig. 4B, transfer of plants to continuous darkness led to a decrease in L29 mRNA levels over the indicated time to a barely detectable level at 1600 at day 2 (i.e., 42 hr in darkness). In contrast, transfer of plants to continuous light resulted in sustained high levels of L29 mRNA. These results suggest that L29 expression is light dependent but not circadian.
Figure 3.
Time course of L29 mRNA accumulation in leaves on illumination of etiolated maize seedlings. Total RNA was extracted from leaves of 10-day-old etiolated (D) or greening plants illuminated with white light for the number of hours indicated and fractionated electrophoretically in a 1% formaldehyde agarose gel (25 μg/lane). After transfer to Gene Screen filter (DuPont/NEN), the filter was hybridized with [α-32P] dCTP-labeled maize L29 full-length cDNA, and 23S rRNA probe was used to assess loading differences.
Figure 4.
L29 mRNA levels during a day–night cycle. (Upper) Maize seedlings were grown under a 16 hr/8 hr day–night cycle for 10 days. Illumination was from 600 to 2200 daily. Leaf tissue was harvested every 2 hr. (Lower) Two sets of 10-day-old seedlings grown under the 16 hr/8 hr day–night cycle were transferred to continuous darkness at 2200 or continuous light at 600, respectively. Leaf tissue was harvested from these plants at the indicated times. The Northern blotting procedure is the same as in the experiment shown in Fig. 3.
L29 mRNA was about as abundant in leaves from 10-day-old seedlings growing under constant white light as in leaves of dark-grown seedlings illuminated for 6 hr (data not shown).
Association of L29 Protein with Ribosomes.
To determine whether the L29 protein is indeed a component of ribosomes and whether it is more abundant in greening than in etiolated leaves and in separated MC and BSC, a synthetic peptide corresponding to a portion of the L29 protein was synthesized and used as an antigen to raise antibodies in rabbits. The antibodies were used to detect L29 protein in Western blots of proteins in leaf ribosomes.
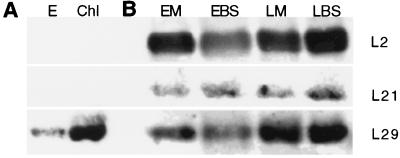
Fig. 5A shows that L29 protein is in plastids and that the amount of immunoreacting protein is much greater after illumination. The experiment was performed by isolating etioplasts and chloroplasts (from homogenates of leaves of 10-day-old dark-grown unilluminated seedlings and similar seedlings illuminated for 10 hr before harvest, respectively) by differential centrifugation as washed 1,200 × g pellets and preparing ribosomes pelleted through a 1 M sucrose cushion from the plastids (18).
Figure 5.
Association of L29 protein with ribosomes isolated from etioplasts and chloroplasts. L2, L21, and L29 in ribosomes from MC and BSC of etiolated and greening plants. (A) Immunoblot for ribosomal protein L29 in ribosomes isolated from purified etioplasts (E) and from chloroplasts (Chl) of dark-grown plants illuminated for 10 hr (18). (B) Representative immunoblots for ribosomal proteins L2, L21, and L29. Ribosomes were isolated from MC of etiolated leaves (EM), from BSC of etiolated leaves (EBS), from MC and BSC of dark-grown plants illuminated for 10 hr (LM and LBS, respectively) (7, 18). The lanes were loaded with equal amounts of rRNA.
There is about 1.5 times more L29 in MC and BSC ribosomes prepared from leaves of dark-grown seedlings after illumination for 10 hr [Fig. 5B, light MC (LM) and light BSC (LBS), respectively] than before illumination [Fig. 5B, etiolated MC (EM) and etiolated BSC (EBS): LM/EM = 1.64 ± 0.28; LBS/EBS = 1.56 ± 0.96. There was no significant difference between the amount of the protein in MC and BSC ribosomes (LM/LBS = 1.17 ± 0.61).
These observations of the L29 protein drew our attention to the question of whether other chloroplast ribosomal proteins change when etiolated seedlings are illuminated. To address this issue, the same Western blots used in the L29 experiments described above were exposed to antibodies against chloroplast ribosomal proteins L2 (10, 11) and L21 (12). As is shown in the representative Western blots in Fig. 5B, unlike L29, the amounts of L21 protein in MC and BSC are about equal and are not affected by illumination: LM/EM = 1.06 ± 0.21; LBS/EBS = 0.93 ± 0.21. Furthermore, L2 protein increases in BSC but not MC on illumination of etiolated seedlings for 10 hr: LM/EM = 0.97 ± 0.17; LBS/EBS = 1.46 ± 0.36.
The relative amounts of a ribosomal protein present in a given cell type under different conditions were calculated from densitometer scans of four Western blots of proteins in ribosomal pellets. Extracts were prepared from MC and BSC isolated from dark-grown unilluminated or illuminated (10 hr) plants (7). Ribosomal pellets were prepared by centrifugation of ribosomes through 1 M sucrose cushions (18). Western blot gels were loaded with equal amounts of RNA. The entire experiment, from growing plants through Western blotting, was carried out independently twice, and the proteins in each pellet were analyzed twice in Western blots. Thus, MC and BSC were isolated from two separately grown sets of illuminated (LM and LBS) and unilluminated (EM and EBS) dark-grown seedlings and the proteins in each pellet were analyzed by Western blotting twice.
DISCUSSION
This research was undertaken initially to identify nuclear genes whose poly(A)-containing mRNAs change in abundance on illumination of dark-grown maize seedlings. One gene identified by DDA is rpl29, which encodes the plastid ribosomal protein L29. We have determined the sequence of the gene and have investigated several aspects of rpl29 expression, including alterations in the contents of mRNA in leaves of dark-grown unilluminated and illuminated seedlings. We have also assessed the L29 protein content of ribosomes in such seedlings. The later work led us to investigate the abundance of two other chloroplast ribosomal proteins, L2 and L21, in the ribosomes of BSC and MC of unilluminated and illuminated dark-grown seedlings. Most interestingly, these studies have revealed that the proportions of the populations of maize plastid ribosomes that contain or lack particular proteins vary substantially during photoregulated MC and BSC maturation.
Relatively small amounts of mRNA encoded by rpl29 are present in leaves of 10-day-old dark-grown maize seedlings. Transcripts of this gene are about 17-fold more abundant in leaves of such seedlings after they have been illuminated for 8 hr. In contrast, the pool of transcripts of the maize plastid gene rps4, encoding plastid ribosomal protein S4, increases only about 20% during 16 hr of illumination of dark-grown maize seedlings (19). Thus, transcripts of some, but not all, genes for maize plastid ribosomal proteins increase greatly during light-induced development.
The L29 protein is present in ribosomes of etiolated leaves, and it increases about 1.5-fold on illumination. There is no striking difference between the amounts of L29 protein in MC and BSC; this is the case in both etiolated or greening leaves.
Studies with E. coli ribosomes have shown L29 to be involved in peptidyl transferase activity (20). Yet, the viability of E. coli strain AM111.5 shows that the L29 protein is not required for growth and thus is not essential for protein synthesis by ribosomes. The E. coli strain AM is erythromycin dependent (21). AM111, an erythromycin-independent revertant of AM, lacks ribosomal proteins S17 and L29 (21, 22). AM111–5, selected as a faster-growing spontaneous derivative of AM111, lacks L29 alone (23). These observations, together with ours presented here, suggest that BSC and MC at least of etiolated maize leaves contain two different populations of functional chloroplast ribosomes with respect to L29: one class with and the other without L29. One way of looking at these data is that if all the ribosomes in leaves of dark-grown illuminated seedlings have L29, then only about 60% of the plastid ribosomes in leaves of unilluminated plants contain L29. Thus the data indicate that the proportions of the populations of plastid ribosomes with and without L29 are different in etiolated vs. greening leaf cells.
The chloroplast ribosomal protein L21, like L29, is encoded by a nuclear gene (12, 24). Unlike L29, the L21 content of the ribosomes is about the same in etiolated and greening leaves. The L2 protein, which in angiosperms is encoded in chloroplasts, behaves differently from either L21 or L29: L2 is present in about equal amounts in MC and BSC ribosomes of etiolated leaf tissue but on illumination of dark-grown seedlings, the amount of the protein in ribosomes increases in BSC but not MC. In other words, the fraction of chloroplast ribosomes containing L2 is greater in ribosomes of green than in etiolated BSC, but the L2-containing fraction of MC ribosomes is about constant. These observations on L29, L2, and L21 emphasize that there are substantial differences in expression patterns among chloroplast ribosomal protein genes and, consequently, in the protein compositions of plastid ribosome populations during cell differentiation.
A striking demonstration that a change in expression of a ribosomal protein gene can affect development is provided by the observation that a mutation in one of the three genes encoding cytoplasmic ribosomal protein S18 (S18A) in Arabidopsis results in plants with pointed first leaves, reduced fresh weight, and retarded growth (25). The three genes are all transcribed and encode completely identical proteins; however, no transcript is detected from the mutated gene, designated S18A. The three genes appear to be transcribed differently in various tissues of the organism. There are several other reports of individual copies of a plant ribosomal protein gene being transcribed in a tissue-specific manner as well as of single genes with multiple promoters that are active under different conditions (e.g., 26–31).
Other cases of differences in expression among plant ribosomal protein genes have been reported. Bisanz-Seyer et al. (32) found that proteins of the plastid 70S ribosomes did not accumulate synchronously during the germination of spinach seedlings: one group of ribosomal proteins could be detected in dry seeds or after 24 hr of imbibition, whereas a second group of ribosomal proteins accumulated only after 3–5 days of development. Koyama et al. (33) reported that during the greening of barley leaves, the amounts of two proteins present in cytoplasmic, 80S, ribosomes of etiolated leaves decreased during greening, but six other ribosomal proteins increased as greening proceeded. The number of copies of the other cytoplasmic ribosomal proteins did not change during greening.
It is not known whether plastid or E. coli ribosomes composed of different proteins, e.g., those with vs. those without L29 or with vs. without L21, etc., can all translate the same set of messages at all or with the same or different efficiencies. Koyama et al. (33) summarize reports of ribosome heterogeneity and ribosomal protein differences (particularly phosphorylation) that have been observed under various conditions in organisms ranging from E. coli to higher plants and human cells and the suggestions that have been made that such differences could affect translation. If there are such qualitative differences, are they important in differentiation?
We have dealt only with ribosomal proteins associated with ribosomes, but there are other ways, besides undemonstrated but possible qualitative effects on translation, in which changes in the populations of free ribosomal proteins could affect development. There are now a number of examples of ribosomal proteins that have extraribosomal functions. For example, the E. coli 30S ribosomal protein S16 is an endonuclease (34), the E. coli ribosomal protein L7/L12 has a helix-turn-helix motif similar to that found in DNA-binding regulatory proteins (35), and E. coli S1 is a component of the replicase of some RNA phages (36). Wool (37) has reviewed the literature on extraribosomal functions of ribosomal proteins and pointed out that a number of eubacterial, archeabacterial, and mammalian ribosomal proteins contain zinc finger motifs, that a number of rat ribosomal proteins have leucine zipper-like motifs, and that ribosomal proteins may also function in DNA replication, transcription, RNA processing, and DNA repair aside from their roles as part of the translational apparatus in the ribosome. Directly to the point here are two recent reports regarding bacterial L29. First, Dantec et al. (38) report that P46 of Spiroplasma citri has a L29 N-terminal domain and a C-terminal domain involved in DNA-inverted repeat sequence binding. They conclude that P46 is the L29 ribosomal protein of S. citri and that it is bifunctional. Second, Sharpe and Craig (39) have determined that in E. coli, L29 and the acyl carrier protein together stimulate the binding of the Tn7-encoded protein TnsD to the specific site attTn7 on the bacterial chromosome.
Coming to understand ribosome anatomy as well as the order of assembly of ribosomal proteins and how ribosomes function in protein synthesis stand as landmarks of biochemistry, molecular biology, and molecular genetics. Yet, so far we appear to have only a few glimpses of extraribosomal roles of a few of the proteins, and we are totally unaware of how alterations in ribosome composition come to influence development. Considering the large amount of work that has been done on ribosomes and ribosomal proteins over many years, we are surprised to realize that we know almost nothing about how organisms use them in cell differentiation and development.
Acknowledgments
We are indebted to Dr. Alap Subramanian for generous gifts of antibodies against spinach ribosomal proteins L2 and L21 as well as for helpful discussions. This research was supported in part by a grant from the National Institute of General Medical Sciences of the National Institutes of Health–United States Public Health Service.
ABBREVIATIONS
- MC
mesophyll cells
- BSC
bundle sheath cells
- DDA
differential display analysis
- RACE
rapid amplification of cDNA ends
- LM
light MC
- LBS
light BSC
- EM
etiolated MC
- EBS
etiolated BSC
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. AF147725).
References
- 1.Bedbrook J R, Link G, Coen D M, Bogorad L, Rich A. Proc Natl Acad Sci USA. 1978;75:3060–3064. doi: 10.1073/pnas.75.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodermel S R, Bogorad L. J Cell Biol. 1985;100:463–476. doi: 10.1083/jcb.100.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haley J, Bogorad L. Plant Cell. 1990;2:323–333. doi: 10.1105/tpc.2.4.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haff L A, Bogorad L. Biochemistry. 1976;15:4110–4115. doi: 10.1021/bi00663a030. [DOI] [PubMed] [Google Scholar]
- 5.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 6.Liang P, Averbaoukh L, Pardee A B. Nucleic Acids Res. 1993;21:3269–3275. doi: 10.1093/nar/21.14.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheen J-Y, Bogorad L. Plant Physiol. 1985;79:1072–1076. doi: 10.1104/pp.79.4.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Anal Biochem. 1987;16:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Zurawski G, Bottomley W, Whitfeld P R. Nucleic Acids Res. 1984;12:6547–6558. doi: 10.1093/nar/12.16.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamp R M, Srinivasa B R, von Knoblauch K, Subramanian A R. Biochemistry. 1987;26:5866–5870. [Google Scholar]
- 12.Smooker P M, Kruft V, Subramanian A R. J Biol Chem. 1990;265:16699–16703. [PubMed] [Google Scholar]
- 13.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong T, Arndt K T. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki K, Olivera J, Wool I G. Biochem Biophys Res Commun. 1990;167:1377–1382. doi: 10.1016/0006-291x(90)90675-d. [DOI] [PubMed] [Google Scholar]
- 16.Urlaub H, Kraft V, Bischof O, Muller E C, Whittmann-Liebold B. EMBO J. 1995;14:4578–4588. doi: 10.1002/j.1460-2075.1995.tb00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulsen H, Bogorad L. Plant Physiol. 1988;88:1104–1109. doi: 10.1104/pp.88.4.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spedding G. Ribosomes and Protein Synthesis: A Practical Approach. Oxford: IRL Press; 1990. [Google Scholar]
- 19.Russell D, Bogorad L. Nucleic Acids Res. 1987;15:1853–1867. doi: 10.1093/nar/15.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bischof O, Kruft V, Wittmann-Liebold B. J Biol Chem. 1994;269:18315–18319. [PubMed] [Google Scholar]
- 21.Dabbs E R. J Bacteriol. 1979;140:734–737. doi: 10.1128/jb.140.2.734-737.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoffler-Meilicke M, Dabbs E R, Albrecht-Ehrlich R, Stoffler G. Eur J Biochem. 1985;150:485–490. doi: 10.1111/j.1432-1033.1985.tb09048.x. [DOI] [PubMed] [Google Scholar]
- 23.Dabbs E R, Hasenbank R, Kastner B, Rak K, Wartusch B, Stoffler G. Mol Gen Genet. 1983;192:301–308. doi: 10.1007/BF00392166. [DOI] [PubMed] [Google Scholar]
- 24.Martin W, Lagrange T, Li Y F, Bisanz-Seyer C, Mache R. Curr Genet. 1990;18:553–556. doi: 10.1007/BF00327027. [DOI] [PubMed] [Google Scholar]
- 25.Van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, Van Montagu M. EMBO J. 1994;13:3378–3388. doi: 10.1002/j.1460-2075.1994.tb06640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams M E, Sussex I M. Plant J. 1995;8:65–76. doi: 10.1046/j.1365-313x.1995.08010065.x. [DOI] [PubMed] [Google Scholar]
- 27.Lagrange T, Franzetti B, Axelos M, Mache R, Lerbs-Mache S. Mol Cell Biol. 1993;13:2614–2622. doi: 10.1128/mcb.13.4.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai Z, Gao J, An K, Lee J M, Edwards G E, An G. Plant Mol Biol. 1996;32:1055–1065. doi: 10.1007/BF00041389. [DOI] [PubMed] [Google Scholar]
- 29.Joanin P, Gigot C, Philipps G. Plant Mol Biol. 1993;21:701–704. doi: 10.1007/BF00014553. [DOI] [PubMed] [Google Scholar]
- 30.Lebrun M, Waksman G, Freyssinet G. Nucleic Acids Res. 1987;15:4360. doi: 10.1093/nar/15.10.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marty I, Meyer Y. Nucleic Acids Res. 1992;20:1517–1522. doi: 10.1093/nar/20.7.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bisanz-Seyer C, Li Y, Seyer P, Mache R. Plant Mol Biol. 1989;12:201–211. doi: 10.1007/BF00020505. [DOI] [PubMed] [Google Scholar]
- 33.Koyama K, Wada A, Maki Y, Tanaka A. Physiol Plant. 1996;96:85–90. [Google Scholar]
- 34.Oberto J, Bonnefoy E, Mouray E, Pellegini O, Wikstrom P M, Rouviere-Yaniv J. Mol Microbiol. 1996;19:1319–1330. doi: 10.1111/j.1365-2958.1996.tb02476.x. [DOI] [PubMed] [Google Scholar]
- 35.Rice P A, Steitz T A. Nucleic Acids Res. 1989;17:3757–3762. doi: 10.1093/nar/17.10.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamen R I. In: RNA Phages. Zinder N D, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1975. pp. 203–234. [Google Scholar]
- 37.Wool I G. In: Ribosomal RNA and Group I Introns. Green R, Schroeder R, editors. Austin, TX: Landes Bioscience; 1997. pp. 153–178. [Google Scholar]
- 38.Dantec L L, Castroviejo M, Bové J M, Saillard C. J Biol Chem. 1998;273:24379–24386. doi: 10.1074/jbc.273.38.24379. [DOI] [PubMed] [Google Scholar]
- 39.Sharpe P L, Craig N L. EMBO J. 1998;17:5822–5831. doi: 10.1093/emboj/17.19.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]