Abstract
The antitumor antibiotic sparsomycin is a universal and potent inhibitor of peptide bond formation and selectively acts on several human tumors. It binds to the ribosome strongly, at an unknown site, in the presence of an N-blocked donor tRNA substrate, which it stabilizes on the ribosome. Its site of action was investigated by inducing a crosslink between sparsomycin and bacterial, archaeal, and eukaryotic ribosomes complexed with P-site-bound tRNA, on irradiating with low energy ultraviolet light (at 365 nm). The crosslink was localized exclusively to the universally conserved nucleotide A2602 within the peptidyl transferase loop region of 23S-like rRNA by using a combination of a primer extension approach, RNase H fragment analysis, and crosslinking with radioactive [125I]phenol-alanine-sparsomycin. Crosslinking of several sparsomycin derivatives, modified near the sulfoxy group, implicated the modified uracil residue in the rRNA crosslink. The yield of the antibiotic crosslink was weak in the presence of deacylated tRNA and strong in the presence of an N-blocked P-site-bound tRNA, which, as was shown earlier, increases the accessibility of A2602 on the ribosome. We infer that both A2602 and its induced conformational switch are critically important both for the peptidyl transfer reaction and for antibiotic inhibition. This supposition is reinforced by the observation that other antibiotics that can prevent peptide bond formation in vitro inhibit, to different degrees, formation of the crosslink.
Keywords: sparsomycin-tRNA crosslink, [125I]phenol-alanine-sparsomycin, RNase H analysis
Sparsomycin (Fig. 1) is one of very few antibiotics that can inhibit protein synthesis in bacteria, archaea, and eucarya. Moreover, the antibiotic and some of its derivatives selectively act on several different human tumors (1). Although some side effects can occur, including eye toxicity, active testing programs with sparsomycin and its derivatives continue, and it has been used successfully as a potentiate of cis-platinum treatment of tumors (1). Sparsomycin is one of several antibiotics that interfere with peptide bond formation, a process that is central to protein biosynthesis and takes place on the large ribosomal subunit (reviewed in refs. 2 and 3). For many of these antibiotics, mutational evidence and/or rRNA footprinting studies on free ribosomes using nucleotide-specific chemicals implicate the peptidyl transferase loop region of 23S-like rRNA, from one or more domains of life, in their binding site (reviewed in ref. 2). Here, they perturb or block peptide bond formation in subtly different ways that are only partially understood.
Figure 1.
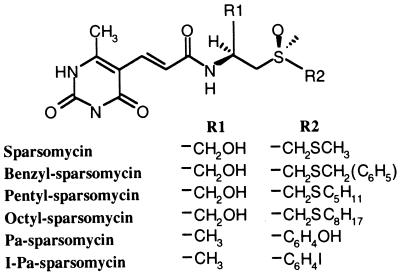
The chemical structures of sparsomycin and the derivatives benzyl-sparsomycin, pentyl-sparsomycin, octyl-sparsomycin, phenol-alanine-sparsomycin (Pa-sparsomycin), and iodo-phenol-alanine-sparsomycin (I-Pa-sparsomycin).
Sparsomycin differs from the other peptidyl transferase drugs in that it does not produce a chemical footprint on 23S-like rRNA of either free ribosomes or N-Ac-Phe-tRNA-ribosome complexes (4, 5). It is also special in that it binds very weakly, if at all, to ribosomes lacking an N-blocked aminoacyl-tRNA (6). This effect is reciprocated in that sparsomycin stimulates P′-site binding of N-blocked tRNA fragments containing the 3′-terminal-CCA-end (4, 7). These observations, combined with the capacity of sparsomycin to inhibit protein biosynthesis in the three domains of life, implies that it recognizes a universally conserved structural motif in the peptidyl transferase center that may involve components from ribosomal protein and/or rRNA. Because this motif is likely to be involved in binding of the 3′-terminal adenosine of the P/P′-site-bound tRNA (8), we can infer that one mechanism by which the drug acts is by stabilizing this interaction and, thereby, inhibiting movement of the 3′-end of the tRNA during elongation.
Previously, crosslinks were shown to form between sparsomycin derivatives carrying affinity labels near the sulfoxy group (Fig. 1) and unidentified ribosomal proteins (9). Weak evidence also exists for a direct antibiotic-rRNA interaction because haloarchaeal mutants carrying single-site mutations at positions 2438, 2451, 2499, 2500, or 2584 (lack of modification) within the peptidyl transferase loop region of 23S rRNA (Escherichia coli numbering) exhibit enhanced sparsomycin resistance (10–12). However, given the small size of the latter effects, the mutated nucleotides are likely to produce general perturbations in the peptidyl transferase center rather than constitute the sparsomycin binding site. If sparsomycin interacts with 23S-like rRNA, then mutation of its rRNA binding site may produce a dominant lethal phenotype that would escape selection.
In the present work, we investigate whether sparsomycin can interact with rRNA or ribosomal protein. On irradiating with low energy ultraviolet light, a direct crosslink was induced at the universally conserved A2602 of 23S-like rRNA within bacterial, archaeal, and eukaryotic ribosomes whereas no crosslink was observed to ribosomal proteins. Parallel studies with sparsomycin derivatives implicated the modified uracil moiety of the drug in the rRNA interaction. A rationale is provided for the requirement of the N-blocked P-site-bound tRNA for strong drug binding.
MATERIALS AND METHODS
Preparation of Ribosomes and 23S rRNA.
Ribosomes and ribosomal subunits were prepared from E. coli as described (13) except that the final ethanol precipitation step was replaced by centrifuging in a fixed-angle rotor at 100,000 × g. Ribosomes were prepared from Bacillus megaterium, Halobacterium halobium, and Saccharomyces cerevisiae as described (5, 14, 15).
Crosslinking of Sparsomycin to 70S Ribosomes from E. coli.
Sparsomycin (200 nM to 150 μM) was incubated with 70S ribosomes (30 nM to 150 nM) in 20–100 μl of 20 mM Tris⋅HCl (pH 7.5), 50 mM NH4Cl, and 10 mM MgCl2 for 20 min at 37°C. In most experiments, deacylated tRNA or N-Ac-Phe-tRNA (1.3 mol/mol ribosomes) was added in the presence of poly(U) (1 μg/pmol ribosome) (16). In the antibiotic interference experiments, other peptidyl transferase drugs were added either to sparsomycin complexed with ribosomes.poly(U).N-Ac-Phe-tRNA or directly to the ribosomal complex before adding sparsomycin. After complex formation, samples were placed in a microtiter tray, on an ice-water bath, and were irradiated at 254, 312, or 365 nm in a Stratalinker 1800 (5 × 8 W bulbs, Stratagene) for 1–30 min. A Petri dish (Sarstedt) was sometimes used as a filter that efficiently eliminates shorter wavelengths (<300 nm). Samples were extracted with phenol, phenol:chloroform, and chloroform and were precipitated with ethanol to remove protein and noncrosslinked antibiotics. The rRNA then was subjected to primer extension by using avian myeloblastosis virus reverse transcriptase (Life Sciences, St. Petersburg, FL) and different 5′-labeled deoxyoligonucleotides complementary to either 5S rRNA, 16S rRNA, or 23S rRNA (17). The extension products were separated in denaturing polyacrylamide gels and were autoradiographed.
Isolation of the ≈83 Fragment from E. coli 23S rRNA.
Sparsomycin (50 μM) was complexed to 70S ribosomes (50 pmol) in the presence of poly(U) (1 μg/pmol ribosome) and N-Ac-Phe-tRNA (1.3 mol/mol ribosomes) in 100 μl of 20 mM Tris⋅HCl (pH 7.5), 50 mM NH4Cl, and 10 mM MgCl2 for 20 min at 37°C and was irradiated at 365 nm as described above. Samples then were treated twice with phenol and were precipitated with ethanol. After resuspending the rRNA (50 pmol) in 20 μl of 20 mM Tris⋅HCl (pH 7.8) and 63.5 mM NH4Cl, 75 pmol of oligonucleotide EC2563 (5′-TCG CGT ACC ACT TTA; complementary to positions 2563–2577 of E. coli 23S rRNA) and oligonucleotide EC2654 (5′-CTC CGG TCC TCT CGT ACT; complementary to positions 2654–2671 of E. coli 23S rRNA) were added. After 5 min of incubation at 55°C, 3 μl of 10 mM magnesium acetate and 0.5 units of RNase H (Amersham Pharmacia) were added, and incubation was continued for 5 min. rRNA was precipitated with ethanol, and the ≈83-nt fragment released by RNase H (corresponding to ≈2,575–2,657) (18) was purified on a 5% polyacrylamide gel (containing 7 M urea) and was localized by UV-shadowing. Gel slices were transferred to 0.3 M sodium acetate (pH 6.0) and 2 mM EDTA and were extracted with phenol overnight at room temperature. RNA was recovered by ethanol precipitation, was suspended in double distilled H2O, and was subjected to primer extension analysis.
Sparsomycin Crosslinking to Ribosomes from Different Organisms.
Sparsomycin was crosslinked to ribosomes from B. megaterium, H. halobium, and S. cerevisiae as follows. Complexes of sparsomycin (50 mM), ribosomes (150 nM), poly(U) (1 μg/pmol ribosome), and N-Ac-Phe-tRNA (1.3 mol/mol ribosome) were formed in 20 μl of 20 mM Tris⋅HCl (pH 7.5), 50 mM NH4Cl, and 10 mM MgCl2 (B. megaterium and S. cerevisiae) or 70 mM Hepes⋅KOH (pH 7.8), 60 mM magnesium acetate, 3.0 M KCl, and 1 mM DTT (H. halobium) for 20 min at 37°C. Samples then were irradiated at 365 nm for 15 min and were treated as described for E. coli. Domain V of 23S-like rRNA was analyzed by reverse transcription by using regularly spaced primers (see refs. 5 and 14).
Crosslinking of [125I]Phenol-Alanine-Sparsomycin.
Phenol-alanine-sparsomycin (named according to ref. 9) was dissolved in 20 μl 50% DMSO at 7 mM and was iodinated with [125I]NaI (100 μCi, Amersham Pharmacia) by using a mild procedure (6, 19). [125I]Phenol-alanine-sparsomycin (50 μM) was subsequently complexed to 70S ribosomes (40 pmol) in the presence of poly(U) (1 μg/pmol ribosome) and N-Ac-[14C]Phe-tRNA (1.3 mol/mol ribosomes) in 80 μl of 20 mM Tris⋅HCl (pH 7.5), 50 mM NH4Cl, and 10 mM MgCl2 for 20 min at 37°C, and half of the sample was irradiated at 365 nm as described above. After phenol extraction and ethanol precipitation, 7.5 pmol of both the UV-irradiated and the control sample, each supplemented with 15 pmol of purified 23S rRNA, were digested with RNase H in the presence of oligonucleotides EC2563 and EC2654, as described above. The samples were subsequently coelectrophoresed with control samples (with no RNase H treatment) in an 8% denaturing polyacrylamide gel, were stained with toluidine blue, and were subjected to autoradiography. Additional RNase H analysis, using regularly spaced oligonucleotides on 23S rRNA (≈500 nucleotides apart) revealed no further radioactively labeled fragments outside of the A2602 region. Ribosomal proteins from the crosslinked samples (5 pmol) were subjected to one-dimensional SDS/PAGE.
RESULTS
Sparsomycin Crosslinks Directly to the Universally Conserved A2602.
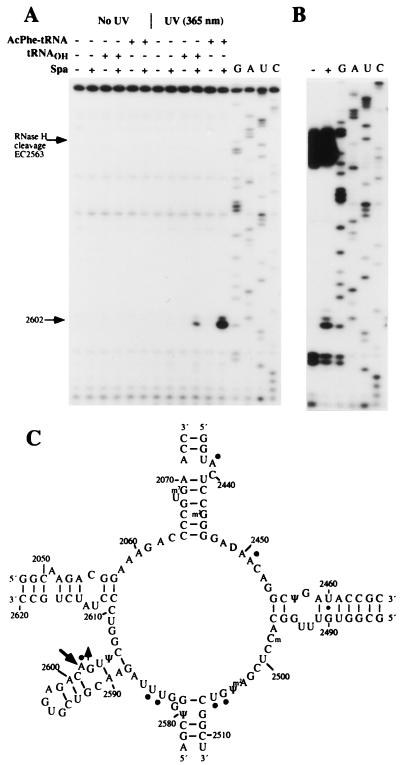
Sparsomycin was added to E. coli 70S ribosomes.poly(U) complexes in the absence of tRNA and in the presence of deacylated tRNA, or N-Ac-Phe-tRNA, bound in the P/P′-state. The resulting complexes were irradiated with UV-light at 365 nm for 15 min, and rRNA was isolated and subjected to primer extension analysis of 5S rRNA, 16S rRNA, and 23S rRNA. A single reverse transcriptase stop, which formed only in the presence of sparsomycin and P/P′-site-bound tRNA, corresponded to the universally conserved A2602 of 23S rRNA within the peptidyl transferase loop (Fig. 2 A and C). The reverse transcriptase stop was ≈6-fold stronger with N-Ac-Phe-tRNA than with deacylated tRNA, and this correlates with the observation that sparsomycin binds more strongly to ribosomes in the presence of N-Ac-Phe-tRNA (6).
Figure 2.
Primer extension analysis of sparsomycin-rRNA crosslinks. (A) The autoradiograph shows primer extension from EC2621, and the stop at A2602 is indicated. Control samples with no irradiation, and no sparsomycin, are included. 70S ribosomes (0.15 μM) and poly(U) (1 μg/pmol ribosome) were mixed with sparsomycin in the presence, or absence, of P/P′-site-bound tRNA (0.18 μM) (see Materials and Methods) and were irradiated at 365 nm for 15 min. G, A, U, and C represent sequencing tracks. (B) 70S ribosomes.poly(U).N-Ac-Phe-tRNA complexes were irradiated at 365 nm as described above, and the extracted RNA was cleaved by RNase H in the presence of oligonucleotides EC2563 and EC2654. The resulting ≈83-nt fragment was gel-purified and analyzed by primer extension from EC2621. The band at A2602 is indicated. (C) Secondary structure of the peptidyl transferase loop region of 23S rRNA from E. coli showing the crosslinked site (large arrow) and the nucleotides that display reduced (circles), or enhanced (circle plus arrow), chemical reactivities with N-Ac-Phe-tRNA in the P/P′-site (8).
The sparsomycin-induced effect at A2602 can, in principle, reflect either (i) drug-induced cleavage of the RNA backbone or (ii) a crosslink forming between one of the following: the drug and 23S rRNA, r-protein and rRNA, tRNA and rRNA, or rRNA and rRNA. To distinguish between these possibilities, complexes of 70S ribosomes, poly(U), and N-Ac-Phe-tRNA were formed and irradiated at 365 nm in the presence and absence of sparsomycin. rRNA was isolated and treated with RNase H in the presence of oligonucleotides complementary to positions 2563–2577 and 2654–2671 of E. coli 23S rRNA. The resulting ≈83-nt fragment was purified in a denaturing polyacrylamide gel and was subjected to primer extension analysis (Fig. 2B). The finding that the same sparsomycin-dependent primer extension stop occurred at position A2602 on the isolated RNase H fragment unambiguously ruled out the possibilities that the stop was caused by (i) drug-induced cleavage of the RNA backbone or (ii) drug-induced crosslinks between r-protein-rRNA, tRNA-rRNA, or rRNA-rRNA involving A2602 and a site outside of the ≈83-nt fragment. Furthermore, the finding that primer extension from an oligonucleotide located immediately 5′ to A2602 (involving screening of the 5′-region of the fragment) yielded no additional stops (data not shown) strongly supports a direct sparsomycin-A2602 crosslink within the peptidyl transferase loop region of the 70S ribosome-tRNA complex.
Chemical Characteristics of the Sparsomycin Crosslink.
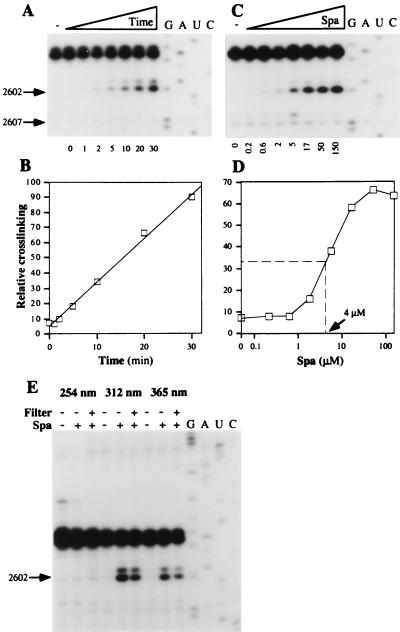
The effects of time, concentration, and UV-wavelength on the yield of the putative sparsomycin-A2602 crosslink were examined for complexes of 70S ribosomes.poly(U).N-Ac-Phe-tRNA (Fig. 3). On irradiating at 365 nm, the yield increased linearly for at least 30 min, indicating that neither the drug nor the ribosomal complex had been damaged (Fig. 3 A and B). Moreover, the dissociation constant of sparsomycin from the 70S ribosome.poly(U).N-Ac-Phe-tRNA complex was estimated at 4 μM by irradiating ribosomal complexes at 365 nm in the presence of increasing amounts of sparsomycin (Fig. 3 C and D). This dissociation constant is close to the ED50 value (8.5 μM) determined for sparsomycin in a polyphenylalanine synthesizing system of E. coli (20).
Figure 3.
Chemical characteristics of the sparsomycin crosslink. (A) Time dependence. 70S ribosomes.poly(U).N-Ac-Phe-tRNA (150 nM) and sparsomycin (50 μM) were complexed (see Materials and Methods) and irradiated at 365 nm for 1–30 min as indicated below the autoradiogram. G, A, U, and C denote sequencing tracks. (B) The reverse transcriptase stop at A2602 was quantified relative to the control band at G2607 in an Instant Imager (Packard) and was plotted as a function of the irradiation time. (C) Concentration dependence. Ribosomal complexes (30 nM) were irradiated at 365 nm for 20 min in the presence of increasing concentrations of sparsomycin (200 nM - 150 μM), indicated below the gel. (D) The reverse transcriptase stop at A2602 (in C) was quantified relative to the control band at G2607 in an Instant Imager and was plotted as a function of the sparsomycin concentration. (E) Wavelength-dependence. Ribosomal complexes (150 nM) and sparsomycin (50 μM) were mixed and irradiated at 254, 312, or 365 nm for 15 min (see Materials and Methods). When indicated (+ or −), a petri dish was used as a filter (<300 nm).
The wavelength-dependence of the sparsomycin crosslink was tested by irradiating sparsomycin and the 70S ribosome.poly(U).N-Ac-Phe-tRNA complex at 254 nm, 312 nm, and 365 nm. No sparsomycin-dependent reverse transcriptase stop was observed at 254 nm whereas irradiation at 312 nm and 365 nm generated strong and medium stops, respectively (Fig. 3E). When a petri dish was used as a filter to cut off higher energy light (<300 nm), crosslinking yields induced at 312 and 365 nm were only marginally reduced (Fig. 3E).
The rRNA Crosslinking Motif Is Universally Conserved.
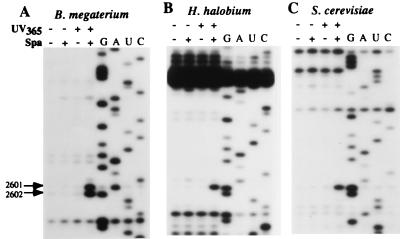
Sparsomycin is a universal drug, affecting all ribosomes, and, therefore, we tested for crosslinking to ribosomes of organisms from all three domains, including the Gram-positive bacterium B. megaterium, the haloarchaeon H. halobium, and the eukaryote S. cerevisiae, by using the same procedure as for E. coli. The sparsomycin-induced reverse transcriptase stop was observed for all three ribosomal complexes at the nucleotide corresponding to A2602 of E. coli 23S rRNA (Fig. 4 A, B, and C), although the effect at C2601 in B. megaterium 23S rRNA appears to arise from more than just stuttering of the reverse transcriptase.
Figure 4.
Crosslinking of sparsomycin to ribosome.poly(U). N-Ac-Phe-tRNA complexes from B. megaterium (A), H. halobium (B), and S. cerevisiae (C). Sparsomycin-ribosomal complexes were irradiated at 365 nm for 20 min (see Materials and Methods). Nucleotide numbering is according to E. coli. The C2601 band is only strong in A (see text). G, A, U, and C represent sequencing tracks.
Crosslinking of Sparsomycin Derivatives.
Sparsomycin has been derivatized extensively to improve its clinical properties (1). Whereas all attempts to alter the modified uracil base, except for removal of the methyl group (Fig. 1), inactivated the drug, the sulfoxy group could be modified with no loss of inhibitory activity (9). Moreover, there is a direct correlation between the degree of hydrophobicity of this part of the molecule and the inhibitory potential of the drug (1).
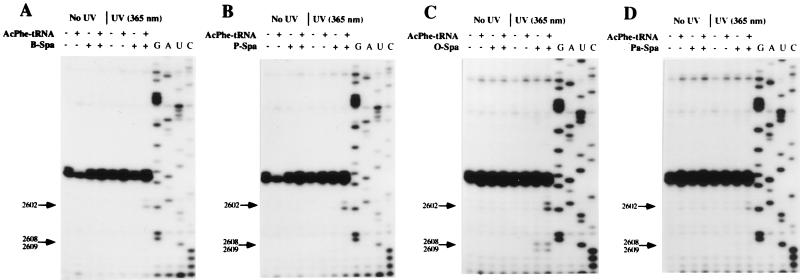
We tested the ability of some of the more potent derivatives of sparsomycin to crosslink to A2602 of E. coli ribosomes (Fig. 5). Substitution of the terminal methyl group by benzyl, pentyl, or octyl groups (Fig. 1) did not lead to decreased crosslinking yields on irradiation at 365 nm (Fig. 5 A–C). Similarly, when both the hydroxymethylene group and -CH2-S-CH3 were substituted by methyl and phenol groups, respectively, the resulting phenol-alanine-sparsomycin (Fig. 1) still crosslinked efficiently to A2602 (Fig. 5D). For each derivative, additional reverse transcriptase stops were formed at positions G2608 and U2609, and the more intense stops that occurred with octyl-sparsomycin (Fig. 5C) suggest that they depend on the chain length of the hydrophobic substituent. Importantly, these additional stops were independent of the presence of N-Ac-Phe-tRNA, suggesting that the increased hydrophobicity of the modified sulfoxy group overcomes the need for an occupied P/P′-site (Fig. 5C).
Figure 5.
Crosslinking of sparsomycin derivatives (50 μM) to E. coli 70S ribosomes.poly(U) complexes in the presence and absence of N-Ac-Phe-tRNA. (A) Benzyl-sparsomycin (B-Spa). (B) Pentyl-sparsomycin (P-Spa). (C) Octyl-sparsomycin (O-Spa). (D) Phenol-alanine-sparsomycin (Pa-Spa). The structures of the derivatives are given in Fig. 1. Complexes were formed and irradiated at 365 nm for 20 min and were analyzed by primer extension (see Material and Methods). G, A, U, and C denote sequencing tracks.
Confirmation of a Sparsomycin-rRNA Crosslink.
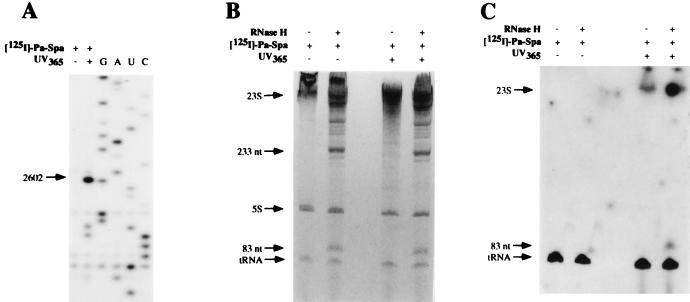
To demonstrate unambiguously that sparsomycin was crosslinked to the A2602 region of 23S rRNA, the phenol-alanine derivative of sparsomycin was radioactively labeled, by iodination at the phenol group, to produce [125I]phenol-alanine-sparsomycin (Fig. 1), which retains biological activity (6, 19). [125I]phenol-alanine-sparsomycin was complexed to 70S ribosomes.poly(U).N-Ac-[14C]Phe-tRNA, was irradiated at 365 nm, and was analyzed by primer extension. The results show that the derivative crosslinked primarily at A2602 in the same way as the parent compound phenol-alanine-sparsomycin (Fig. 6A). The rRNA was subsequently treated with RNase H, in the presence of oligonucleotides EC2563 and EC2654, to produce a radioactively labeled, ≈83-nt fragment, corresponding to positions ≈2575–2657 of E. coli 23S rRNA (see Fig. 6 B and C). In addition, neither one-dimensional SDS/PAGE of ribosomal proteins nor the use of different pairs of oligonucleotides for RNase H analyses of the three rRNAs revealed any labeling of either ribosomal proteins or other rRNA components (data not shown). This confirms that sparsomycin crosslinks exclusively with A2602 in the 23S rRNA.
Figure 6.
Crosslinking of [125I]-phenol-alanine-sparsomycin (50 μM) to E. coli 70S ribosome.poly(U) complexes by irradiating at 365 nm in the presence of N-Ac-[14C]Phe-tRNA. (A) Primer extension analysis of the isolated rRNA where the band at A2602 is indicated. G, A, U, and C represent sequencing tracks. (B) Separation of the 83- and 233-nt fragments after cleavage of the 23S rRNA by RNase H, directed by oligonucleotides EC2563 and EC2654. The treated samples were coelectrophoresed with uncleaved samples, and the gel was stained with toluidine blue. (C) Autoradiography of the gel from B. Background spots result from the long exposure.
Inhibition of the Sparsomycin Crosslink by Other Peptidyl Transferase Antibiotics.
Sparsomycin may share overlapping binding sites with other peptidyl transferase inhibitors, and, therefore, we tested the capacity of 12 of these drugs to interfere with the sparsomycin crosslink to A2602 in E. coli ribosomes. The antibiotics were added individually to a preformed complex of sparsomycin and 70S ribosome.poly(U)-N-Ac-Phe-tRNA complexes before irradiating at 365 nm and analyzing the peptidyl transferase loop region by primer extension (Fig. 7). Control experiments in which the order of addition of sparsomycin and the second drug was reversed yielded identical results (data not shown).
Figure 7.
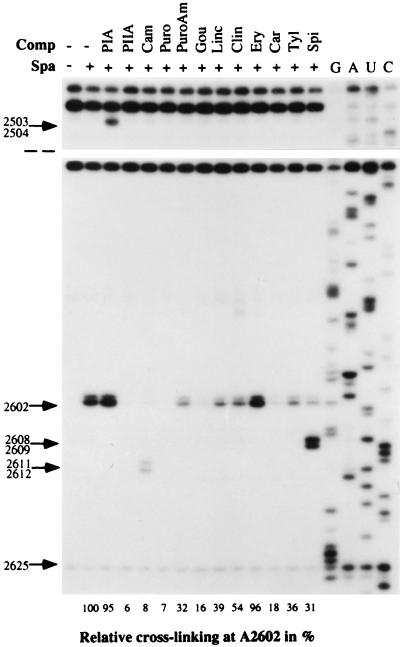
Interference of peptidyl transferase antibiotics with the sparsomycin crosslink at A2602. Sparsomycin (50 μM) was complexed to E. coli 70S ribosome.poly(U) complexes in the presence of N-Ac-Phe-tRNA and was incubated at 37°C for 5 min. The second drug was added, and, after 15 min of incubation, the samples were irradiated for 15 min at 365 nm. Primer extension analyses were performed by using EC2654. Drug concentrations were pristinamycin IA (PIA, 200 μM), pristinamycin IIA (PIIA, 200 μM), chloramphenicol (Cam, 1.2 mM), puromycin (Puro, 1 mM), puromycin aminonucleoside (PuroAm, 1 mM), gougerotin (Gou, 200 μM), lincomycin (Linc, 200 μM), clindamycin (Clin, 200 μM), erythromycin (Ery, 200 μM), carbomycin (Carb, 200 μM), tylosin (Tyl, 200 μM), and spiramycin (Spi, 200 μM). The primer extension stop at A2602 was quantified in an Instant Imager, using the stop at 2625 as a reference, and the results are given as a percentage below the gel, relative to a control sample in the absence of competitor. Control experiments, performed in the absence of sparsomycin, established that the observed effects depended on the presence of sparsomycin, except for pristinamycin IA and chloramphenicol (see text). G, A, U, and C denote sequencing tracks.
Strong reductions in crosslinking yields were observed with pristinamycin IIA (streptogramin A), puromycin, chloramphenicol, and gougerotin whereas puromycin aminonucleoside, lincomycin, clindamycin, and the macrolides carbomycin, tylosin, and spiramycin caused smaller decreases. Only pristinamycin IA (streptogramin B) and erythromycin produced no effect (Fig. 7). Additional primer extension stops were observed in the peptidyl transferase loop with pristinamycin IA (2503/2504), chloramphenicol (2611/2612), and spiramycin (2608/2609). Although the stops produced by pristinamycin IA also probably derive from a drug-rRNA crosslink (18), the spiramycin effect entirely depends on the presence of sparsomycin. This may reflect a small spiramycin-induced displacement of the crosslinking moiety of sparsomycin toward G2608/U2609, similar to the effects produced by the hydrophobic moieties of the sparsomycin derivatives (Fig. 5). This would, in turn, imply that A2602 and G2608/U2609 are juxtaposed structurally. The chloramphenicol effects were independent of the presence of sparsomycin and are currently being investigated.
DISCUSSION
Sparsomycin inhibits peptide bond formation universally, a rare property that it shares with the 4-amino hexose cytosine drugs that include amicetin, blasticidin S, and gougerotin (21). This provides a basis for its antitumor activity, although its selectivity for some human tumors must depend on the preferential membrane permeability of the active sparsomycin derivatives (1). The identification of a ribosomal binding site for sparsomycin on 23S-like rRNA provides an important insight into a conserved feature of the catalytic center of peptide bond formation. By using a mild UV irradiation method, primer extension with reverse transcriptase, and the radioactively labeled derivative [125I]phenol-alanine sparsomycin, we could show unambiguously that sparsomycin generates a crosslink at the universally conserved A2602 of 23S rRNA in E. coli ribosomes. Moreover, no other crosslinks were observed to either rRNA or ribosomal proteins.
The finding that sparsomycin crosslinked efficiently to the corresponding position in the 23S-like rRNAs of another bacterium, an archaeon, and a eukaryote underlines the universal potency of sparsomycin and reinforces the idea that A2602 is located within a unique and highly conserved ribosomal motif. Moreover, the failure to isolate haloarchaeal rRNA mutants that exhibit high resistance to sparsomycin and carry alterations at A2602 probably reflects the idea that mutations at this position produce dominant lethal phenotypes that escape selection, as has, indeed, been shown for E. coli (K. Lieberman and A. Dahlberg, personal communication).
The most important functional part of the sparsomycin structure is the modified uracil ring because its derivatization produces drug inactivation. In contrast, the sulfoxy group of sparsomycin can be altered without major loss of activity (Fig. 1). Thus, we infer that the modified uracil ring is likely to be located at A2602, and this is consistent with both the wavelength dependence of the crosslinking and with the observation that the sparsomycin derivatives that were modified at the sulfoxy group crosslinked to A2602 (Fig. 5). The putative interaction between the modified uracil ring of sparsomycin and A2602 is likely to involve stacking of the bases. Moreover, the recent observation that the 3′-terminus (A76) of P/P′-site-bound tRNA and A2602 are juxtaposed (22) suggests that A76 may stabilize the stacking interaction and thereby provide a rationale for the interdependence of tRNA and sparsomycin binding.
The sparsomycin crosslink at A2602 also depends on the presence of P/P′-site-bound tRNA. Moreover, the observation that N-Ac-Phe-tRNA stimulates the crosslinking yield 6-fold more that deacylated tRNA, whereas no crosslinking occurs to the free ribosome, correlates with an earlier finding that P/P′-site-bound N-Ac-Phe-tRNA, but not deacylated tRNA, strongly enhances the chemical reactivity of A2602 (8). This implies, in turn, that the N-Ac-Phe-tRNA-induced conformational switch at A2602 is of functional importance. The hypothesis is reinforced by the observation that ribosomal binding of the ternary complex of aminoacyl-tRNA.EF-Tu.GTP (or a non-hydrolyzable analogue), in the presence of deacylated tRNA, renders A2602 chemically unreactive (8) and, moreover, that the resultant complex is destabilized by sparsomycin (23).
The results are consistent with A2602 being one of a few key nucleotides lying at the peptidyl transferase center that are proximal to the 3′-ends of both donor and acceptor tRNA substrates on the ribosome. Thus, all of the tested antibiotics that are known to directly inhibit peptide bond formation, in vitro (21, 24), reduced the yield of the sparsomycin crosslink, to some degree (Fig. 7). Only erythromycin and a streptogramin B, which primarily affect the nascent peptide and not peptide bond formation, had no effect. Further support comes from the observation that P/P′-site bound azido-tRNAs, derivatized at A76, can crosslink to A2602 (22). Thus, we can conclude that sparsomycin inhibits protein synthesis by blocking the 3′-terminus of P/P′-site-bound tRNA at A2602 on the ribosome. This may lead to distortion of the positioning of the 3′-end of the aminoacyl-tRNA and to inhibition of peptide bond formation.
Acknowledgments
We are grateful to Dr. V. I. Makhno at Petersburg Nuclear Physics Institute for preparation of ribosomal particles and to Prof. J. P. G. Ballesta for helpful counsel on sparsomycin. The research was supported by the Danish RNA Regulation Centre and by a grant from the Novo Nordisk Foundation to B.T.P.
References
- 1.Ottenheijm H C J, van den Broek L A G M, Ballesta J P G, Zylicz Z. Prog Med Chem. 1986;23:220–268. doi: 10.1016/s0079-6468(08)70344-8. [DOI] [PubMed] [Google Scholar]
- 2.Garrett R A, Rodriguez-Fonseca C. In: Ribosomal RNA: Structure, Evolution, Processing and Function in Protein Biosynthesis. Zimmermann R A, Dahlberg A E, editors. Boca Raton, FL: CRC; 1995. pp. 327–355. [Google Scholar]
- 3.Lieberman K R, Dahlberg A E. Prog Nucl Acid Res Mol Biol. 1995;50:1–23. [PubMed] [Google Scholar]
- 4.Moazed D, Noller H F. Proc Natl Acad Sci USA. 1991;88:3725–3728. doi: 10.1073/pnas.88.9.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Fonseca C, Amils R, Garrett R A. J Mol Biol. 1995;247:224–235. doi: 10.1006/jmbi.1994.0135. [DOI] [PubMed] [Google Scholar]
- 6.Lázaro E, van den Broek L A G M, Felix A S, Ottenheijm H C J, Ballesta J P G. Biochemistry. 1991;30:9642–9648. doi: 10.1021/bi00104a011. [DOI] [PubMed] [Google Scholar]
- 7.Monro R E, Celma M L, Vázquez D. Nature (London) 1969;222:356–358. doi: 10.1038/222356a0. [DOI] [PubMed] [Google Scholar]
- 8.Moazed D, Noller H F. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 9.Lázaro E, van den Broek L A G M, Felix A S, Ottenheijm H C J, Ballesta J P G. Biochimie. 1991;73:1137–1143. doi: 10.1016/0300-9084(91)90157-v. [DOI] [PubMed] [Google Scholar]
- 10.Porse B T, Rodriguez-Fonseca C, Leviev I, Garrett R A. Biochem Cell Biol. 1995;73:877–885. doi: 10.1139/o95-095. [DOI] [PubMed] [Google Scholar]
- 11.Tan G T, DeBlasio A, Mankin A S. J Mol Biol. 1996;261:222–230. doi: 10.1006/jmbi.1996.0454. [DOI] [PubMed] [Google Scholar]
- 12.Lázaro E, Rodriguez-Fonseca C, Porse B, Ureña D, Garrett R A, Ballesta J P G. J Mol Biol. 1996;261:231–238. doi: 10.1006/jmbi.1996.0455. [DOI] [PubMed] [Google Scholar]
- 13.Makhno V I, Peshin N N, Semenkov, Yu. P, Kirillov S V. Mol Biol. 1988;22:528–536. [PubMed] [Google Scholar]
- 14.Porse B T, Garrett R A. J Mol Biol. 1999;286:375–387. doi: 10.1006/jmbi.1998.2509. [DOI] [PubMed] [Google Scholar]
- 15.Leviev I G, Rodriguez-Fonseca C, Phan H, Garrett R A, Heilek G, Noller H F, Mankin A S. EMBO J. 1994;13:1682–1686. doi: 10.1002/j.1460-2075.1994.tb06432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirillov S V, Makhno V I, Semenkov, Yu. P. Eur J Biochem. 1978;89:297–304. doi: 10.1111/j.1432-1033.1978.tb20927.x. [DOI] [PubMed] [Google Scholar]
- 17.Christiansen J, Egebjerg J, Larsen N, Garrett R A. In: Ribosomes and Protein Synthesis: A Practical Approach. Spedding G, editor. Oxford: Oxford Univ. Press; 1990. pp. 229–252. [Google Scholar]
- 18.Porse B T, Kirillov S V, Awayez M J, Garrett R A. RNA. 1999;5:585–595. doi: 10.1017/s135583829998202x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tejedor F, Ballesta J P G. Anal Biochem. 1982;127:143–149. doi: 10.1016/0003-2697(82)90156-7. [DOI] [PubMed] [Google Scholar]
- 20.van den Broek L A G M, Lázaro E, Zylicz Z, Fennis P J, Missler F A N, Lelievel P, Garzotto M, Wagener D J T, Ballesta J P G, Ottenheijm H C J. J Med Chem. 1989;32:2022–2015. doi: 10.1021/jm00128a051. [DOI] [PubMed] [Google Scholar]
- 21.Vázquez D. Inhibitors of Protein Synthesis. New York: Springer; 1979. [Google Scholar]
- 22.Kirillov, S. V., Porse, B. T. & Garrett, R. A. (1999) RNA5, in press. [DOI] [PMC free article] [PubMed]
- 23.Hornig H, Woolley P, Luhrmann R. Biochimie. 1987;69:803–813. doi: 10.1016/0300-9084(87)90207-0. [DOI] [PubMed] [Google Scholar]
- 24.Gale E F, Cundliffe E, Reynolds P E, Richmond R A, Waring M J. The Molecular Basis of Antibiotic Action. London: Wiley; 1981. pp. 402–457. [Google Scholar]