Abstract
Deamination of cytosine to uracil is the most common promutagenic change in DNA, and it is greatly increased at the elevated growth temperatures of hyperthermophilic archaea. If not repaired to cytosine prior to replication, uracil in a template strand directs incorporation of adenine, generating a G⋅C → A⋅U transition mutation in half the progeny. Surprisingly, genomic analysis of archaea has so far failed to reveal any homologues of either of the known families of uracil-DNA glycosylases responsible for initiating the base-excision repair of uracil in DNA, which is otherwise universal. Here we show that DNA polymerases from several hyperthermophilic archaea (including Vent and Pfu) specifically recognize the presence of uracil in a template strand and stall DNA synthesis before mutagenic misincorporation of adenine. A specific template-checking function in a DNA polymerase has not been observed previously, and it may represent the first step in a pathway for the repair of cytosine deamination in archaea.
Deamination of cytosine to uracil is an inherent property of the chemistry of DNA, and it produces G⋅C → A⋅T transition mutations leading to an inexorable degradation of the information content of the genome, unless corrected (1). In eubacteria, plants, animals, and some double-stranded DNA viruses, repair of cytosine deamination is achieved by a base-excision repair pathway (2) initiated by a uracil-DNA glycosylase (UDG) (3) or by a G⋅U/T mismatch-specific DNA glycosylase (TDG/MUG) (4, 5). These structurally related enzymes (6) hydrolyze the N-glycosidic bond connecting the uracil to the deoxyribose sugar and generate an abasic site, which is removed by the next enzyme in the pathway. Ultimately, repair synthesis by DNA polymerase and ligation replaces the excised nucleotide and restores the covalent integrity of the backbone (7).
Like all other DNA organisms, archaea have an absolute requirement for some type of uracil detection and repair system. However, no homologues of the two known families of uracil base excision-repair enzymes ubiquitous in the other kingdoms have been identified in archaeal genomes, so far (8–11). It has been previously noted (12) that archaeal DNA polymerases bind oligonucleotides containing deoxyuridine, which are inhibitory to PCRs utilizing these enzymes. This observation, together with the lack of obvious components for a “classical” pathway, prompted us to examine whether archaeal DNA polymerases themselves might play a role in a uracil repair system in these organisms, unlike that previously encountered.
METHODS
Source of Polymerases.
Pfu exo− was prepared from an Escherichia coli overproducing strain (BL21DE3) (Novagen) containing a pET-17b vector (Novagen) with an inserted gene coding for DNA polymerase from Pyrococcus furiosus with a mutation (D215A) abolishing exonuclease activity. Ten-milliliter cultures were grown overnight, at 37°C, in LB broth containing ampicillin (100 μg/ml) and used to inoculate 500 ml of ampicillin-containing LB broth. The large culture was incubated at 37°C until the OD600 reached ≈0.5, when isopropyl β-d-thiogalactoside (IPTG) was added (final concentration 1 mM) to induce Pfu exo− polymerase expression. After a further 4 hr, at 37°C, the cells were collected by centrifugation, suspended in 50 ml of 10 mM Tris⋅HCl, pH 7.5, containing 100 mM NaCl and 0.1 mM each phenylmethylsulfonyl fluoride and benzamidine. After cell disruption by sonication the cell debris was removed by centrifugation and the supernatant was treated with bovine pancreatic DNase I (RNase-free grade, Boehringer Mannheim; 2.5 units/ml, 37°C, 1 hr) to degrade DNA. The supernatant was then placed in an 80°C water bath for 15 min, and denatured proteins were removed by centrifugation. The supernatant was loaded onto a 40-ml DEAE-Sephacel column equilibrated to 10 mM Tris⋅HCl, pH 7.5, containing 100 mM NaCl. The polymerase does not bind to the column under these conditions and was eluted directly onto a 40-ml heparin-Sepharose Cl-6B column with 200 ml of buffer. The polymerase was eluted from the heparin column with a 400-ml gradient consisting of 100–700 mM NaCl in 10 mM Tris⋅HCl, pH 7.5. The polymerase eluted at about 350 mM salt and was concentrated in an Amicon Centriprep-50 centrifugal concentrator. Each preparation yielded between 5 and 10 mg of protein. Purity was analyzed by SDS/8% PAGE and staining with Coomassie blue. Heavily loaded samples revealed only a single band at about 90 kDa, corresponding to the polymerase. Protein thus purified can be used to grow single crystals (data not shown). All other polymerases used were recombinant from commercial suppliers: Vent exo+ and exo− and phage T4 (New England Biolabs), Pfu exo+ (Stratagene), Pwo exo+ (Boehringer Mannheim), and Taq (MBI-Fermentas, Vilnius, Lithuania).
Construction of Templates.
The 119-mer template oligonucleotide used in long-range primer-extension reactions was generated by PCR amplification of a segment of pUC19 spanning the polylinker cloning site. PCR primer sequences were A, 5′-GACGTTGTAAAACGACGGCCAGU; B, 5′-GACGTTGTAAAACGACGGCCAGT; and C, 5′-CAATTTCACACAGGAAACAGCTATGACCATG. The 119-mer oligonucleotide, incorporating either a U or T nucleotide 23 bases from the 3′ terminus of one strand, was synthesized by using Taq polymerase under standard PCR conditions, using primer C and either primer A or primer B. PCR products were purified on agarose gels and extracted by using Qiagen columns. The 44-mer template used in short-range extension reactions was synthesized directly by using a dU phosphoramidite (Cruachem, Herndon, VA) or a 1′,2′-dideoxyribose phosphoramidite (dSpacer, Glen Research, Sterling, VA) as appropriate.
Primer Extension Reactions.
For long-range primer extensions, primer C was annealed to one strand of the 119-bp PCR product by heating to 65°C in reaction buffer and cooling to room temperature. The dNTPs, [α-[35S]thio]ATP, and 5 units of DNA polymerase (Pfu, Pwo, or Taq) were added in polymerase reaction buffer (as specified by the suppliers of each polymerase) to a final volume of 20 μl, and the reaction was allowed to proceed for 60 min at 55°C. Reaction products were subjected to electrophoresis in a denaturing acrylamide gel and scanned and recorded on a Fuji FLA-2000 phosphorimager.
For short-range primer extensions, a 5′-32P-labeled 24-mer 5′-GGGGATCCTCTAGAGTCGACCTGC was annealed to the 44-mer template 5′-GGAGACAAGCTTGCATGCCTGCAGGTCGACTCTAGAGGATCCCC (the underlined region forms a duplex with the primer). We used either this uracil-free template or derivatives in which the bases at positions +1 to +7, immediately downstream (toward the 5′-direction) of the duplex region, were individually replaced by deoxyuridine. Some experiments used a template containing deoxyuridine or a stable abasic site (1′,2′-dideoxyribose) at the +10 position. The primer (2.5 nM) and template (5 nM) were mixed in the appropriate polymerase reaction buffer (as specified by the suppliers of each polymerase) and heated to 90°C for 15 min and then slowly cooled to room temperature to anneal. dNTPs (1.25 mM) and 0.5 unit of DNA polymerase (Vent exo+ or exo−, Pfu exo+ or exo−, Pwo exo+, Taq, or T4) were then added to give a final reaction volume of 20 μl. The polymerization reactions were allowed to proceed for 20 min (or the times specified in Fig. 3) at 72°C (37°C for T4). The products of the reaction were analyzed by denaturing gel electrophoresis and either autoradiography or phosphorimaging for detection.
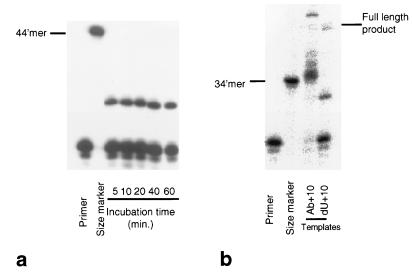
Figure 3.
Time course of short-range primer extension and effect of an abasic site in the template. (a) Denaturing polyacrylamide gels showing the short-range primer extension products produced by Pfu (exo−) on a template containing a single uracil positioned 10 bases from the end of the primer-template duplex region. The time of the incubation was varied as shown and in all cases a truncated product is produced. The first lane (primer) contains just the primer strand as a marker. The second lane (no dU) shows the maximum length 44-mer product. (b) Denaturing polyacrylamide gels showing the short-range primer extension products produced by Pfu (exo−) on a template containing either a stable abasic site (Ab+10) or a single uracil (dU+10) positioned 10 bases from the end of the primer-template duplex region. The first two lanes contain markers 24 (primer size) and 34 bases in length. With Ab+10 two terminated products, 35 and 36 bases long, are produced. These correspond to incorporation of a dNTP at the abasic site followed by the random non-template-directed addition of two further nucleotides. As the polymerase used lacks a functional 3′-5′ exonuclease, these nontemplate additions persist, so that the full-length product is longer than the 44-mer (marker, and small amount of full-length 44-mer produced in the dU+10 case). With the dU+10 template, the terminated product is shorter, clearly indicating a stop preceding the dU rather than in the vicinity of the dU.
Sequencing and Termination-Site Mapping.
A sequencing ladder for pUC19 was generated by using a T7 Sequenase v2.0, DNA chain-termination sequencing kit (Amersham Life Sciences) in accordance with the manufacturers instructions, using primer C (see above). The sequencing reaction products, and the products of the long-range primer extension reaction with Pfu exo+ were run alongside each other on a denaturing gel (Stratagene CastAway, 42 × 18 cm), and visualized by autoradiography.
Surface Plasmon Resonance.
Surface plasmon resonance measurements of Pfu binding to various DNAs were performed on a Biacore Biosensor (Stevenage, U.K.) under essentially the same conditions as previously described (13), with biotinylated oligonucleotides immobilized on a streptavidin-coated SA5 Biosensor chip. Single-stranded (ss) oligonucleotides were 35-mers of sequence 5′-biotin-CCGAATCAGTTCACTTCNAGCCGAGGTATTTAGCC for ssC and ssU, where N is C in ssC and N is U in ssU. An immobilized double-stranded oligonucleotide containing a single central U⋅G mispair was generated by annealing a nonbiotinylated oligonucleotide 5′-GGCTAAATACCTCGGCTGGAAGTGAACTGATTCGG to the immobilized ssU oligonucleotide in situ, as previously described (13). Immobilized single-stranded oligonucleotide containing a single central abasic site, and a duplex containing a widowed guanine opposite an abasic site, were generated by passing an excess of recombinant herpes simplex virus 1 uracil-DNA glycosylase over surfaces containing immobilized ssU and U⋅G duplex, respectively, as previously described (6, 13).
RESULTS
We examined the ability of the DNA polymerases from the thermophilic eubacterium Thermus aquaticus (Taq) and from the thermophilic archaea Pyrococcus furiosus (Pfu) and Pyrococcus woesei (Pwo) to extend a primer across a template containing a single deoxyuridine. The eubacterial Taq DNA polymerase was able to extend the primer to the full length defined by the template. In contrast, the major primer-extension product generated by the archaeal DNA polymerases was significantly shorter (Fig. 1a). With the equivalent template in which the deoxyuridine was replaced by thymidine, all the DNA polymerases produced full-length products with no premature termination. To determine the position at which primer-extension by the archaeal DNA polymerases had halted on the deoxyuridine template, the products of the reactions were analyzed by gel electrophoresis, alongside sequencing reactions of the same template. The major products generated by the archaeal DNA polymerases corresponded to termination of primer extension around 4–6 nucleotides upstream of the position of the uracil in the template strand (Fig. 1b).
Figure 1.
Long-range primer extension reactions on normal and single-uracil templates. (a) Denaturing polyacrylamide gel showing products of reactions in which a 31-mer primer was extended against a 119-nucleotide template (see Methods) containing either a deoxyuridine (U) or a deoxythymidine (T) 23 nucleotides from the 5′ end. Reactions were performed with DNA polymerases from Pyrococcus furiosus (Pfu), Pyrococcus woesei (Pwo), and Thermus aquaticus (Taq). All three polymerases produce full-length products on T templates, but the archaeal enzymes produce smaller major products on U templates. (b) Polyacrylamide gel showing the position of the major premature termination product from the primer extension reaction against the U template with Pyrococcus furiosus DNA polymerase (Pfu exo+), relative to a sequencing ladder of the segment of pUC19 used in the long-range primer extension reaction (see Methods). The sequence for the template strand is given, reading 3′→5′ from the bottom (shorter primer extension products) to the top (longer primer extension products). The position of the deoxyuridine (U) is indicated. The major Pfu products correspond to termination of the polymerase reaction 4–6 bases upstream of the template deoxyuridine. The position of the full-length product obtained from primer-extension against the T template (not shown) was consistent with termination at the end of the 119-base template.
The spacing of 4–6 nucleotides between the last nucleotide incorporated into the primer and the position of the uracil in the template is suggestive of a uracil-sensor “leading” the polymerase activity in the archaeal enzymes. To further analyze the significance of this spacing, we constructed a series of primer-template duplexes in which a deoxyuridine was placed in the template at between +1 and +7 bases downstream of the duplex junction, and we measured the ability of Vent, Pfu, Pwo, Taq, and T4 DNA polymerases to initiate primer extension reactions on these substrates. With deoxyuridine present in the template in the first four positions downstream of the primer-template duplex, minimal primer extension reaction was observed with the archaeal DNA polymerases. At spacings of +5, +6, and +7 between the duplex junction and the deoxyuridine, some primer extension products were observed with the archaeal enzymes, corresponding to addition of 1, 2, and 3 nucleotides respectively, with virtually no full-length product produced (Fig. 2). In contrast, Taq and T4 produced a full-length major product with all of these templates. Again these results are consistent with archaeal DNA polymerases stalling DNA synthesis 4–6 nucleotides upstream of a template strand deoxyuridine. Polymerase arrest was very persistent; incubation of a template with a deoxyuridine in the +10 position with Pfu polymerase for up to 1 hr did not result in any significant readthrough (Fig. 3a). The lack of termination seen with Taq suggests this property is not universally present in thermophilic polymerases. Similarly, the negative results with T4 confirm that deoxyuridine-directed stalling is not a general property of the polymerase α family [the archaeal polymerases and T4 belong to the polymerase α group, whereas Taq is a member of the polymerase I family (14, 15)]. A similar pattern of primer extension was observed with the exo+ or exo− versions of the Vent and Pfu polymerases, which contain or lack the 3′-5′ proofreading exonuclease activity. However, products shorter than the original primer were observed with the exo+ enzymes and also with Pwo polymerase, for which only the exo+ form was available. These observations suggest that, unlike the 5′-3′ polymerase activity, the 3′-5′ exonuclease “proofreading” activity of the archaeal enzymes is insensitive to the presence of deoxyuridine and is not involved in detection and stalling at template deoxyuridine.
Figure 2.
Short-range primer extensions. Denaturing polyacrylamide gels showing short-range primer extension products on otherwise identical templates containing either no uracil (no dU) or a single uracil positioned 1–7 bases (dU+n) from the end of the primer-template duplex region. The first lane (primer) contains just the primer strand as a marker. Gels are for the following: Vent exo+ and exo− (a), Pfu exo+ and exo− (b), Pwo exo+ (c), Taq (d), and T4 (e). In all cases the “no dU” lane shows the maximum-length 44-mer product. All the archaeal enzymes fail to produce full-length product on templates containing uracil, but they are able to extend the primer slightly when the uracil is 5 or more bases beyond the end of the duplex region. Products shorter than the original primer are present in reactions involving enzymes with functional 3′→5′ exonuclease activity (exo+).
The premature halt generated by the presence of a template-strand uracil was not 100%, and a small proportion of full-length product was sometimes observed with the archaeal enzymes. Scaling-up the reactions produced enough of this full-length product to perform Maxam–Gilbert sequencing, which confirmed the incorporation of adenine opposite the template uracil, as expected from Watson–Crick base pairing. Furthermore, analysis of the template strand, after such a treatment, revealed it to be unchanged. Thus, in response to a template deoxyuridine the archaeal polymerases do not nick the template strand, hydrolyze the deoxyuridine glycosidic bond to give an abasic site, or replace the deoxyuridine with another base (data not shown).
Bulky lesions such as pyrimidine photodimers (16) and aminofluorene adducts (17), or abasic sites (18), will stall or at least pause DNA synthesis by a wide variety of polymerases. However, these cause arrest at the site of the lesion, presumably because of poor base pairing interactions available for an incoming nucleoside triphosphate. We observe a similar effect with Pfu, which stalls and misincorporates nucleotides on a template containing a stable abasic site (1′,2′-dideoxyribose) (Fig. 3b) 10 nucleotide downstream of the primer-template duplex. In marked contrast, the equivalent template with a deoxyuridine at that position results in a shorter product (Fig. 3b) consistent with arrest 4–6 nucleotides upstream of the template strand uracil, and in agreement with the data presented in Figs. 1 and 2. Finally, Pfu polymerase binds tightly and specifically to uracil-DNA, only in a single-stranded context, and not to abasic sites (Fig. 4). Thus the specific arrest of archaeal DNA synthesis elicited by template-strand uracil, characterized by stalling 4–6 bases upstream of the uracil and tight binding of single-stranded uracil, is qualitatively different from the nonspecific arrest of DNA polymerases in general at abasic sites. To determine whether upstream stalling was a general response to deaminated bases, a template containing inosine (the deamination product of guanine) in place of uracil was used in similar primer extension reactions; it elicited no stalling and produced only full-length product (data not shown).
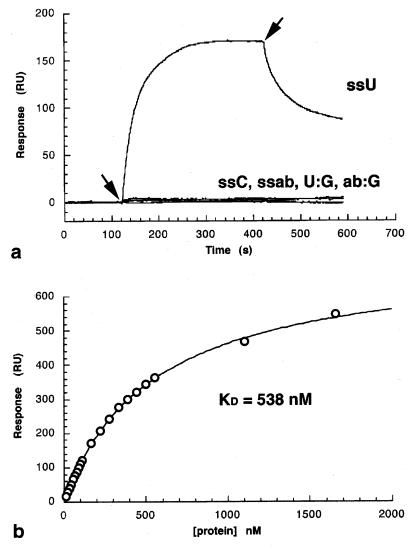
Figure 4.
DNA-binding specificity of Pfu DNA polymerase. (a) Surface plasmon resonance measurements of Pfu DNA polymerase interactions with DNA. The signal in response units (RU) is proportional to the mass of enzyme bound to the oligonucleotides immobilized on the surface of the Biacore chip. The beginning and end of injection of Pfu DNA polymerase over the chip surface are indicated by the arrows. Interactions were measured with single-stranded 35-mer oligonucleotides containing either a single uracil (ssU), a single abasic site (ssab), or no modification (ssC), and with double-stranded 35-mers containing a single U⋅G mispair (U:G) or a guanine opposite and abasic site (ab:G). Pfu polymerase interacts strongly with ssU but no other oligonucleotides. (b) Dose–response curve for binding of Pfu polymerase to ssU. Experimental data (○) were fit to the equation R = Rmax⋅C/(Kd + C), where C is the protein concentration and Rmax is the maximal response, giving an estimated Kd = 0.54 μM.
DISCUSSION
Hydrolytic deamination of cytosine to uracil is a very subtle change in terms of the structure of the incorrect base thus produced, and recognition by known uracil-DNA repair enzymes (19,20) involves highly specific interactions to allow it to be distinguished from the normal DNA pyrimidines, cytosine and thymine. Unlike some other lesions, uracil is entirely compatible with Watson–Crick base-paired double-stranded DNA, and it is efficiently utilized as a template for the incorporation of adenine by all other classes of DNA polymerases, including the nonarchaeal members of the polymerase α family. Given the range of covalent modifications that can occur as a result of DNA damage in general, we cannot dismiss the possibility that other lesions, particularly those involving bulky adducts, might also cause upstream stalling by archaeal, and indeed other, DNA polymerases. However, in terms of the most biologically important bases from which uracil must be distinguished, the phenomenon we describe is highly specific, with the archaeal DNA polymerases, but not other polymerase α family members, stalling in response to uracil, but not to the natural DNA pyrimidines or to deaminated bases in general.
The behavior of the archaeal DNA polymerases on deoxyuridine-templates is fully consistent with the presence of a “read-ahead” function within the polymerase, scanning the template before its utilization in directing primer extension, and stopping the 5′-3′ polymerase activity whenever a deoxyuridine is encountered. We know of no previous observation of any base-specific template-checking function in a DNA polymerase. Unlike the 3′-5′ proofreading exonuclease activity present in many DNA polymerases, this read-ahead function appears to be passive, and we observe no changes in the chemical structures of the template-strand deoxyuridine as a result of its exposure to the polymerase.
Given the high affinity of archaeal DNA polymerases for single-stranded uracil-DNA, an alternative explanation might be possible, at least for the long-range primer extensions, in which one polymerase molecule actively extending the primer is blocked by a second polymerase bound to the deoxyuridine in the single-stranded template downstream (Fig. 5a). This “blocking” model is unlikely for two reasons. First, structural studies of polymerase I family eubacterial DNA polymerases show that at least 10–12 template nucleotides are bound within the body of the enzyme (21, 22), and the “footprint” of the α polymerases (23), which include the archaeal enzymes, is certainly not significantly smaller. As two polymerases cannot approach closer than the “footprint” width along the same DNA, blocking of a running polymerase by a polymerase stalled on a downstream deoxyuridine would predict a gap of at least the “footprint” width between the last nucleotide incorporated and the template-deoxyuridine, rather than the significantly smaller gap that is actually observed (Figs. 1 and 2). Second, with native exo+ enzymes there is clear 3′-5′ exonuclease “trimming” of the input primer, even in experiments in which 5′-3′ polymerase activity is blocked. It is very difficult to understand how a blocking polymerase molecule bound to the template deoxyuridine might prevent access by a second enzyme for primer extension, while still allowing access for exonuclease trimming, given that both reactions initially require full access to the primer-template junction. On the other hand, these data are fully consistent with a simple read-ahead model in which the DNA polymerase that performs the 3′-5′ exonuclease and 5′-3′ polymerase activities is the same molecule whose 5′-3′ progress is prevented by specific interaction with a deoxyuridine upstream in the single-stranded segment of the template (Fig. 5b).
Figure 5.
Read-ahead uracil detector. (a) Blocking of a running polymerase actively extending a primer, by a second polymerase bound to deoxyuridine in the single-stranded segment of the template, would give a gap between the last incorporated nucleotide (arrowed) and the template deoxyuridine (U), larger than the “footprint” of the polymerase. (b) In the read-ahead model, specific stalling of a running polymerase at a deoxyuridine upstream in the single-stranded segment of the template would give a gap between the last incorporated nucleotide (arrowed) and the template deoxyuridine (U), smaller than the “footprint” of the polymerase, as is observed.
We have observed this uracil-detecting read-ahead function in three DNA polymerases available to us from archaea. The enzymes used were recombinant proteins expressed in E. coli, either commercially or from our own expression system, so that no other archaeal proteins were present in the reactions. Template-strand uracil detection is thus an inherent function of the DNA polymerases themselves. Sensitivity to inhibition by uracil-containing oligonucleotides in PCRs, which would seem to be a necessary corollary of the uracil-detection function we describe here, has been observed in DNA polymerases from two other archaeal sources (12), including the crenarchaeon Desulfurococcus. Thus, although we cannot be certain that an ability to detect template-strand uracil is a universal property of archaeal DNA polymerases, its presence in enzymes from two distinct archaeal kingdoms would suggest that it is at least widespread. That a wide variety of nonarchaeal polymerases, including mesophilic and thermophilic eubacterial enzymes, and polymerase α family members show no particular affinity for uracil-DNA strengthens the concept that uracil detection may be a unique attribute of archaeal enzymes.
A promutagenic deoxyuridine in a template strand becomes a fixed mutation only once DNA polymerase has incorporated an adenine opposite it on the daughter strand. The DNA polymerase itself is therefore presented with the last opportunity to recognize the presence of the promutagenic base in the template strand. Incorporation of a template-strand uracil-detection function within the polymerase is thus an efficient strategy. With notable exceptions (24, 25), the recognition and repair of DNA damage involves multiple steps in ordered pathways, frequently involving multiprotein complexes (26). As uracil detection by the archaeal DNA polymerases appears to be passive and merely stops the polymerase upstream of the lesion, it can represent only the first step in such a pathway. In eukaryotes, single-stranded DNA can act as a primary signal for initiation of DNA repair (27). Thus, the stalled polymerase and exposed template might act as the signal for the recruitment, and possibly the organization, of the protein(s) that perform the subsequent steps. However, the nature of those subsequent steps is not at all obvious. Full repair would minimally require the removal of the uracil base (or nucleotide) and the restoration of deoxycytidine. An excision-repair process (base or nucleotide) comparable to the processes in eubacteria or eukaryotes, seems unlikely, as these depend absolutely on the presence of an intact complementary strand to direct resynthesis and facilitate ligation after the excision step(s).
Maintenance of genomic integrity in archaea requires that uracils are ultimately replaced by cytosine, although the enzyme systems that accomplish this remain to be discovered. However, the block to replication that would result from stalling of archaeal DNA polymerases might be relieved without uracil excision, by a postreplicative repair pathway (28). In eubacteria, gaps in daughter strands resulting from polymerases stalled at unrepaired bulky damage or abasic sites can be repaired by means of homologous recombination with the complementary strand from a sister duplex (29), facilitated by RecA (30). While this damage-tolerant mechanism does not repair the original template-strand lesion, it at least allows the completion of DNA synthesis. With the daughter-strand gaps generated by persistent arrest of archaeal DNA polymerases upstream of template-strand uracils, a homologous recombination mechanism would regenerate a G⋅U mismatch, maintaining the promutagenic status of the uracil, but avoiding the unrepairable formation of a mutated A⋅U base pair, and allowing replication to continue. Unlike the various nucleotide excision systems and uracil-DNA glycosylases, which are widespread and strongly conserved among and between eubacteria and eukaryota but apparently absent from archaea, clear archaeal homologues of RecA can be identified. At least in principle, therefore, a homologous recombination mechanism analogous to daughter-strand gap repair could also operate in archaea. Whether the ability of archaeal DNA polymerases to detect template-strand uracil provides the first step in a prereplicative repair mechanism remains to be seen. However, given the possibility of daughter-strand gap repair in archaea, it at least provides a means of avoiding the fixation of a promutagenic G⋅U mispair into an A⋅U mutation.
Acknowledgments
We are grateful to Maria Davis, Shauna Brummet, Barbera Kaboord, Don Cowan, and Renos Savva for useful discussion and to Pauline Heslop for expert technical assistance. This work was supported by a LINK Grant from the U.K. Medical Research Council and Nycomed Amersham, PLC, to B.A.C. and L.H.P.
ABBREVIATION
- exo
exonuclease activity
Note Added in Proof
While this manuscript was in review, a uracil-DNA glycosylase activity very distantly related to MUG (5, 6) was described in a thermophilic eubacterium (31). Homologues of this enzyme identifiable in some archaeal genomes would then provide the first step in a conventional excision-repair pathway required for the essential replacement of uracil by cytosine in these organisms. Whether the polymerase-based uracil-detection system we describe here directly invokes this pathway, or provides a damage-tolerance mechanism allowing postreplicative excision repair, remains to be determined.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Seeberg E, Eide L, Bjørås M. Trends Biochem Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. Proc Natl Acad Sci USA. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nedderman P, Jiricny J. J Biol Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 5.Gallinari P, Jiricny J. Nature (London) 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 6.Barrett T E, Savva R, Panayotou G, Barlow T, Brown T, Jiricny J, Pearl L H. Cell. 1998;92:117–129. doi: 10.1016/s0092-8674(00)80904-6. [DOI] [PubMed] [Google Scholar]
- 7.Dianov G, Lindahl T. Curr Biol. 1994;4:1069–1076. doi: 10.1016/s0960-9822(00)00245-1. [DOI] [PubMed] [Google Scholar]
- 8.Bult C J, White O, Olsen G J, Zhou L X, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–378. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 10.Smith D R, Doucette Stamm L A, Deloughery C, Lee H M, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 12.Lasken R S, Schuster D M, Rashtchian A. J Biol Chem. 1996;271:17692–17696. doi: 10.1074/jbc.271.30.17692. [DOI] [PubMed] [Google Scholar]
- 13.Panayotou G, Brown T, Barlow T, Pearl L H, Savva R. J Biol Chem. 1998;273:45–50. doi: 10.1074/jbc.273.1.45. [DOI] [PubMed] [Google Scholar]
- 14.Joyce C M, Steitz T A. Annu Rev Biochem. 1994;63:777–882. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 15.Delarue M, Poch O, Tordo N, Moras D, Argos P. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 16.Taylor J-S, O’Day C L. Biochemistry. 1990;29:1624–1632. doi: 10.1021/bi00458a038. [DOI] [PubMed] [Google Scholar]
- 17.Moore P D, Rabkin S D, Strauss B S. Nucleic Acids Res. 1980;8:4473–4484. doi: 10.1093/nar/8.19.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajagopalan M, Lu C, Woodgate R, O’Donnell M, Goodman M F, Echols H. Proc Natl Acad Sci USA. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savva R, McAuley-Hecht K, Brown T, Pearl L H. Nature (London) 1995;373:487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]
- 20.Mol C D, Arvai A S, Slupphaug G, Kavli B, Alseth I, Krokan H E, Tainer J A. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 21.Doublie S, Tabor S, Long A M, Richardson C C, Ellenberger T. Nature (London) 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 22.Kiefer J R, Mao C, Braman J C, Beese L S. Nature (London) 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Sattar A, Wang C C, Karam J D, Konigsberg W H, Steitz T A. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 24.Olsson M, Lindahl T. J Biol Chem. 1980;255:10569–10571. [PubMed] [Google Scholar]
- 25.Sancar A. Biochemistry. 1994;33:2–9. doi: 10.1021/bi00167a001. [DOI] [PubMed] [Google Scholar]
- 26.Svejstrup J Q, Wang Z G, Feaver W J, Wu X H, Bushnell D A, Donahue T F, Friedberg E C, Kornberg R D. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- 27.Garvik B, Carson M, Hartwell L. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 407–464. [Google Scholar]
- 29.Shinohara A, Ogawa T. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 30.West S C, Cassuto E, Howard-Flanders P. Nature (London) 1981;294:659–662. doi: 10.1038/294659a0. [DOI] [PubMed] [Google Scholar]
- 31.Sandigursky M, Franklin W A. Curr Biol. 1999;9:531–534. doi: 10.1016/s0960-9822(99)80237-1. [DOI] [PubMed] [Google Scholar]