Abstract
The assembly of the polyketide backbone of rifamycin B on the type I rifamycin polyketide synthase (PKS), encoded by the rifA–rifE genes, is terminated by the product of the rifF gene, an amide synthase that releases the completed undecaketide as its macrocyclic lactam. Inactivation of rifF gives a rifamycin B nonproducing mutant that still accumulates a series of linear polyketides ranging from the tetra- to a decaketide, also detected in the wild type, demonstrating that the PKS operates in a processive manner. Disruptions of the rifD module 8 and rifE module 9 and module 10 genes also result in accumulation of such linear polyketides as a consequence of premature termination of polyketide assembly. Whereas the tetraketide carries an unmodified aromatic chromophore, the penta- through decaketides have undergone oxidative cyclization to the naphthoquinone, suggesting that this modification occurs during, not after, PKS assembly. The structure of one of the accumulated compounds together with 18O experiments suggests that this oxidative cyclization produces an 8-hydroxy-7,8-dihydronaphthoquinone structure that, after the stage of proansamycin X, is dehydrogenated to an 8-hydroxynaphthoquinone.
The rifamycins (1), exemplified by rifamycin B (1) (2, 3), isolated in 1959 from Amycolatopsis mediterranei (4), are a group of antibiotics of the ansamycin family (5) with pronounced antibacterial activity. Several chemically modified rifamycins, such as rifampicin, have been used since the mid-1960s for the treatment of tuberculosis and other mycobacterial infections (6). However, as in the case of many antibiotics, their effectiveness is threatened by the emergence of resistant pathogens (7), making the development of modified rifamycins not subject to these resistance mechanisms a matter of urgency. Although numerous rifamycin derivatives have been synthesized from the natural products, the structural modifications largely were confined to one region of the molecule, the C-3 position of the naphthoquinone chromophore (6, 8). More deep-seated structural alterations of the molecule are difficult to accomplish chemically and clearly call for alternative (biosynthetic) methods of structure modification.
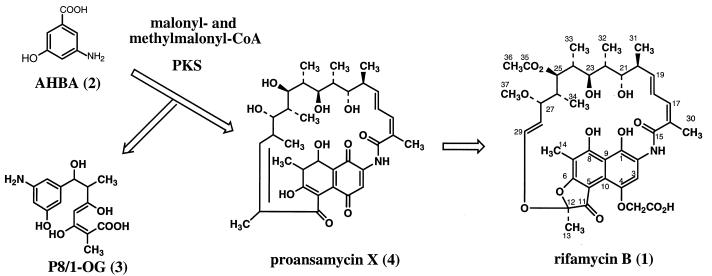
The biosynthetic assembly of the rifamycins (1) involves chain extension of an unusual starter unit, 3-amino-5-hydroxybenzoic acid (AHBA) (2) (ref. 9; ref. 10 and references therein), by propionate and acetate units on a type I polyketide synthase (PKS) (Fig. 1). Premature polyketide chain termination in a mutant strain leads to release of a tetraketide, P8/1-OG (3) (11), from the PKS and its excretion into the medium, whereas in the wild-type organism, processing continues to the stage of an undecaketide, which then is released from the enzyme as a macrocyclic lactam, presumably proansamycin X (4) (12, 13). The simplest ansamycin isolated from cultures is protorifamycin I (5, shown in Fig. 5), which has been proposed to be a precursor of the rifamycins (14). The initial cyclization product then is subject to extensive downstream processing to give the final biologically active rifamycins.
Figure 1.
Biosynthesis of rifamycin B in A. mediterranei.
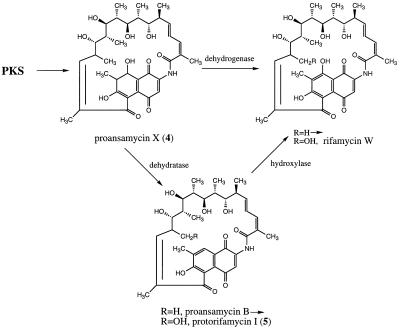
Figure 5.
Two possible routes from proansamycin X (4) to rifamycin W, the precursor of rifamycin B (1).
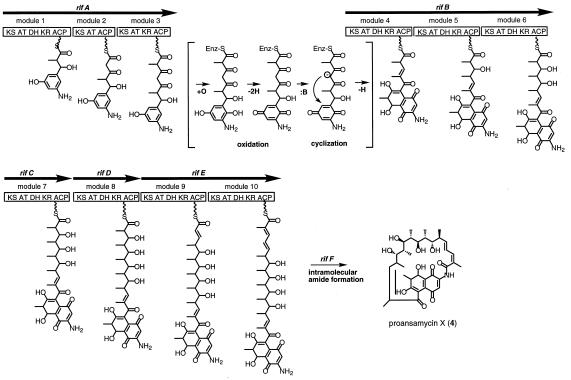
The rifamycin biosynthetic gene cluster recently has been cloned from A. mediterranei and sequenced (12, 13, 15). On one side of the cluster of about 90 kb are located about 36 kb of DNA-containing genes required for AHBA synthesis, genes involved in export and resistance, regulatory genes, and a substantial number of downstream-processing genes. The other side of this cluster consists of 52 kb of DNA encoding a type I PKS that consists of 10 modules, each with the expected functional domains, arranged colinear with the sequence of chain-extension steps that must occur in the biosynthesis of the antibiotic. These 10 modules are arranged in five genes/proteins. RifA contains the loading domain and the first three modules, rifB contains the next three modules, rifC and rifD each contain one module, and rifE contains the final two modules. An interesting feature compared with the well-characterized PKSs encoding the formation of macrolides, such as the erythromycin precursor, 6-deoxyerythronolide B (16), is the absence of a type 1 thioesterase domain (17) at the end of the last module of the rif PKS. Such a thioesterase, catalyzing the release of the fully assembled polyketide from the enzyme as the macrocyclic lactone, is characteristic of type I macrolide PKSs. A type 2 thioesterase (17), similar to one found additionally in macrolide and nonribosomal peptide biosynthetic gene clusters (18, 19) is, however, present in the rif cluster (rifR) (12). Immediately after the rifE PKS gene is located a gene, rifF, that encodes a protein with pronounced homology to mammalian arylamine:acetyl-CoA acetyltransferases (20). Based on the chemical analogy to this reaction, we have proposed (12) that this gene product catalyzes the release of the completed polyketide from the PKS by intramolecular amide formation. Here we describe an unexpected property of the rif PKS, based on the behavior of rifD, rifE, and rifF mutants, that has widespread implications for modular PKSs in general.
MATERIALS AND METHODS
Materials and General Methods.
A. mediterranei strain S699 was obtained from G. C. Lancini (Lepetit S.A., Varese, Italy); the wild type and mutants (except as described below) were cultivated as reported previously (ref. 10 and references therein). [7-13C, 18O2]-AHBA was prepared as described by Rickards and coworkers (21). Protorifamycin I was a gift from Novartis. The compounds were fed to the appropriate mutants in liquid culture (AHBA) or surface cultures grown on agar plates (protorifamycin I); the cultures then were analyzed for rifamycin B 1 by HPLC and bioassay. In the [7-13C, 18O2]-AHBA feeding, 1 was isolated by HPLC and analyzed by electrospray–MS as described (22). Liquid chromatography–MS (LC-MS) analyses were carried out with a Shimadzu LC-10AD pump connected to a Fisons Quattro II mass spectrometer (Fisons, Loughborough, U.K.).
Gene Inactivations.
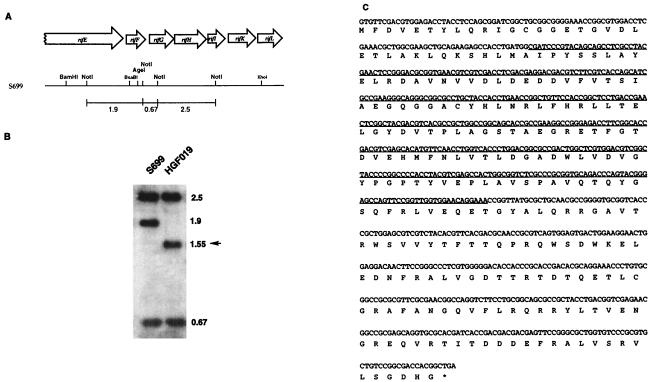
Gene disruptions were carried out essentially as described for the construction of the rifK(−) mutant (ref. 10 and references therein). For the disruption of rifF, replacement suicide vector pHGF1087 was created by insertion of a 1.75-kb BglII fragment carrying the hygromycin resistance gene (hyg) from pIJ963 (23) into BamHI-treated pHGF1086, in which the 3.0-kb BamHI–BsaBI and 3.1-kb AgeI(blunt-ended)–XhoI DNA fragments from the rif gene cluster (see Fig. 2A) were coligated into pBluescript KS(−)/(BamHI, XhoI), generating a 369-bp, in-frame deletion at the N-terminal end of rifF (see Fig. 2C). The heat-denatured pHGF1087 was introduced by electroporation into A. mediterranei S699, and six transformants resistant to hygromycin were selected for further investigation. Southern blotting of these six transformants (data not shown) demonstrated that they arose by the expected single-crossover integration either upstream or downstream of rifF. The four upstream-integrated transformants had lost the ability to produce rifamycin B, whereas the two downstream integrants produced normal rifamycin B titers. One of the downstream-integrated transformants was chosen for maintenance on synthetic agar medium (24) without hygromycin. After propagation through three generations, five colonies showed sensitivity to hygromycin and two of them also showed loss of rifamycin B production. Southern blotting with the NotI-digested genomic DNA from these hygromycin-sensitive colonies revealed that all of them had undergone a second crossover event. The three rifamycin B-producing colonies reverted back to wild type, and the two rifamycin B nonproducing mutants gave HGF019, in which the endogenous rifF was replaced with the inactive, truncated version (Fig. 2B).
Figure 2.
Genetic organization and nucleotide sequence of the rifF gene in A. mediterranei S699 and HGF019. (A) A restriction map of the region of the A. mediterranei S699 chromosome encompassing the C-terminal end of the polyketide synthase (rifE) and the whole ORFs of the amide synthase (rifF), amino DHQ synthase (rifG), amino DAHP synthase (rifH), amino quinate/shikimate dehydrogenase (rifI), AHBA synthase (rifK), and oxidoreductase (rifL) genes (12). The three indicated NotI DNA fragments, 0.67, 1.9, and 2.5 kb, respectively, were used as a 32P-dCTP-labeled probe for the blot in B. (B) The autoradiograph of a Southern blot to verify the deletion of a 369-bp DNA fragment of rifF in A. mediterranei HGF019. A 0.8% agarose gel in which 5 μg of NotI-digested genomic DNA from strains S699 and HGF019 had been separated was blotted and hybridized with the three NotI DNA fragments shown above. The band corresponding to the deleted DNA fragment, 1.55 kb, of rifF in strain HGF019 is indicated by an arrow. (C) The nucleotide and deduced amino acid sequences of the rifF gene. The deleted 369-bp DNA fragment in strain HGF019 is underlined.
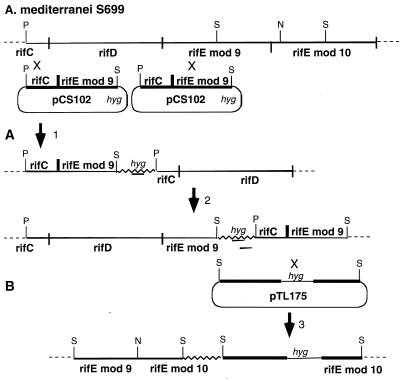
The rifE module 10 disrupted mutant was constructed by cloning a 5.1-kb SstI fragment from rifE (Fig. 3B) into pUC19 (25) to create pTL139. A 1.8-kb segment containing the hygromycin resistance gene described above was inserted at the unique NcoI site of pTL139, which is located inside the KS10 domain of rifE module 10, to form pTL175. This plasmid was introduced into the A. mediterranei S699 strain, and hygromycin-resistant transformants were selected as described above. Southern analysis of SstI-digested DNA from one of these transformants showed that it had a genotype consistent with the expected single-crossover recombination event diagrammed in Fig. 3B.
Figure 3.
Construction of rifD and rifE mutants. (A) The rifD mutant resulted from crossover 1 to produce a hygromycin-resistant strain in which the entire rifD gene has been deleted, which, in effect, produces a strain without the RifD and RifE subunits of the PKS, because transcription does not reinitiate downstream of rifC. The rifE module 9 mutant resulted from crossover 2 to produce a hygromycin-resistant strain in which module 9 for the rifE gene has been disrupted, which results in a strain without the RifE subunit of the PKS because transcription does not reinitiate downstream of rifD. (B) The rifE module 10 mutant resulted from crossover 3 to produce a hygromycin-resistant strain in which module 10 of the rifE gene has been disrupted, producing a strain with a truncated RifE protein. Thicker black lines represent the genomic DNA and the boundaries of the rif PKS genes. The thick, black line in pCS102 between rifC and rifE module 9 shows the place at which these two genes were fused. The jagged line presents the vector DNA. P, PstI; S, SstI; N, NcoI; Hyg, hygromycin resistance gene.
The rifD and rifE module 9 disrupted mutants were made by first preparing a PCR product by using the modified splicing by overlap extension–PCR method (26). To delete the entire module 8, the first two PCRs were performed separately. One PCR mixture contained primers (a) 5′-GCGACCAGGTGGCGGCGCTGC-3′ and (b) 5′-GTCCGTGGCCATTGAACTCAACTCC CAGAACTC-3′ as primers to obtain a 0.7-kb DNA segment containing the end of module 7 attached via a 12-bp linker to the start of module 9 (underlined). The other PCR mixture used primers (c) 5′-GAGTTGAGTTCAATGGCCACGGACGAGAAACTC-3′ and (d) 5′-CAGCCGGTGCCGTCCGCGGTC-3′, to result in a product with the 0.5-kb beginning part of module 9 joined via a 12-bp linker to the end of module 7 (underlined). After the first PCR products were purified, a third PCR was carried out with these products as templates and primers (a) and (b) to assemble a final DNA segment in which all of module 8 had been deleted. From the final PCR product, a 1-kb Eco47III–XhoI fragment was purified and its sequence was confirmed, by using an Applied Biosystems model 377 sequencer and the Perkin—Elmer Amplitag Dye-terminator sequencing system. This DNA segment was joined to a 2.1-kb Eco47III–SstI fragment from module 7 and a 2.2-kb XhoI–PstI fragment from module 9 and inserted into pGEM3zF(+) (Promega) to form pCS101. A 5.3-kb BamHI–XbaI segment of pCS101 was transferred into the multiple cloning site of pANT841 (this vector was obtained from W. R. Strohl, Ohio State University, with the hyg resistance gene inserted) digested with NheI and BglII, to give pCS102 (Fig. 3A). Hygromycin-resistant transformants of the S699 strain were obtained as above and analyzed for the position of pCS102 integration by Southern blot hybridization. Two kinds of clones were obtained: in WMH-M101, the single-crossover recombination event had occurred in module 7 to produce a strain with a disrupted rifD gene (Fig. 3A-1), and in WMH-M102, the single-crossover recombination event had occurred in module 9, thereby disrupting rifE module 9 (Fig. 3A-2).
Isolation of Products from Mutants.
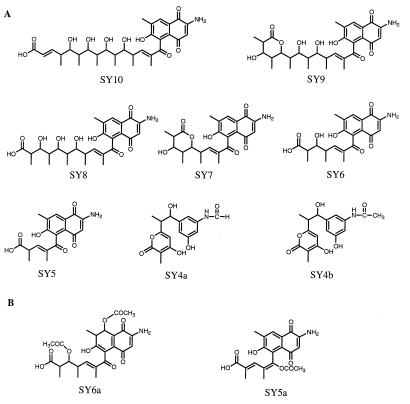
rifF(−). After 7 days of fermentation at 28°C as described (ref. 10 and references therein), the broth together with mycelia were frozen at −80°C and dried by lyophilization. The residue from 4 liters of fermentation was extracted with 500 ml of 80% aqueous methanol containing 1% formic acid. Solvent was removed under vacuum, and the residue was dissolved in 100 ml of 1% aqueous formic acid and extracted twice with 100 ml of n-butanol. After removal of the solvent, the residue from this butanol extract was partitioned between 200 ml each of ethyl acetate and 1% formic acid and the aqueous layer was extracted once more with 200 ml of ethyl acetate. The combined organic extract then was subjected to flash chromatography on C18 reversed-phase silica gel (5 μ; YMC, Kyoto), eluting with a gradient of 9–26% of CH3CN in 1% aqueous formic acid. The individual compounds were purified further by HPLC on a semipreparative C18 column (ultrasphere, 5 μ; Alltech Associates) with UV detection at 274 and 322 nm and acetonitrile/water/formic acid as the liquid phase. The approximate isolated yields of products were 4 mg of SY4a, 1 mg of SY4b, 0.75 mg of SY5, 0.6 mg of SY6, 0.75 mg of SY7, 0.75 mg of SY8, 0.5 mg of SY9, and 0.25 mg of SY10 from 1 liter of culture.
RifE(−).
The mycelium of the rifE mutant, which was stocked in 20% glycerol at −80°C, was transferred into three 250-ml baffled flasks, each containing 50 ml of seed medium (4 g of yeast extract, 10 g of malt extract, and 4 g of glucose in 1 liter of water), hygromycin (250 μg/ml), and 4 g glass beads and incubated for 3 days on a shaker (250 rpm, 30°C). The seed culture then was inoculated into three 2-liter baffled flasks, each containing 500 ml of medium (5 g of meat extract, 5 g of peptone, 5 g of yeast extract, 2.5 g of enzyme hydrolysate of casein, 20 g of glucose, and 1.5 g of NaCl in 1 liter of water), and fermented for 7 days. The cultures were combined and adjusted to pH 4.5 with 15% aqueous trichloroacetic acid. The mycelium was removed by centrifugation, and the broth was extracted with ethyl acetate (two times with same volume as the broth). The ethyl acetate-soluble fraction was evaporated under reduced pressure (<30°C), and the resulting residue was passed through a Sep-Pak C18 cartridge (Waters), eluting with 40% CH3CN. Acetic anhydride (1.5 ml) and pyridine (3 ml) were added to the material (300 mg), and the mixture was stirred for 20 h at room temperature. The solution was evaporated and the residue was separated by silica gel column chromatography (1.0 × 30 cm; Aldrich), eluting with toluene/MeOH from 95:5 to 1:1 (vol/vol). The material eluting from 95:5 to 90:10 toluene/MeOH (233 mg) was subjected to silica gel column chromatography, eluting with hexane/ethyl acetate from 90:10 to 10:90, and a 73-mg portion of the material recovered from 50:50 to 40:60 hexane-ethyl acetate was passed through a Sep-Pak C18 cartridge. The fraction eluting with 35% CH3CN finally was purified by RP-HPLC [Nova-Pak C18; Waters; 10 × 300 mm; flow rate, 2.5 ml/min; detection by UV at 320 nm; eluant CH3CN plus 0.05% trifluoroacetic acid/H2O (35:65, vol/vol)] to yield SY6a (tret, 25.5 min; 31.6 mg) and SY5a (tret, 12.6 min; 23.6 mg).
The constitutional structures of the isolated compounds were determined by high-resolution fast atom bombardment MS and by one-dimensional (1H, 13C, difference nuclear Overhauser effect) and two-dimensional (heteronuclear multiple bond correlation) NMR spectroscopy. Rotation measurements showed them to be optically active, but their stereochemistries, assumed to correspond to those of the natural rifamycins, were not determined independently. The structure elucidation of these compounds will be reported in a forthcoming paper.
RESULTS
Disruption of rifF.
To probe the function of the proposed off-loading enzyme RifF, we constructed a mutant of A. mediterranei S699 in which part of the rifF gene had been deleted from the genome (Fig. 2). The mutant construction provided evidence that there is no endogenous promoter located on the 2.9-kb C-terminal end of rifE. Taken together with the deduced translational coupling between rifE and rifF, this shows that the PKS products (RifA–E), or at least the one from rifE, and the product from rifF should share the same transcription unit. It also revealed that there might be a promoter located downstream of rifF that directly controls the downstream genes such as rifG, rifH, and rifI, which are thought to be involved in the formation of the starter unit AHBA (12). The rifF(−) mutant no longer synthesizes rifamycin B, the main product of this fermentation, but HPLC analysis of the organic extract revealed a very complex product pattern. Isolated and fully characterized by HR-MS and one- and two-dimensional NMR was a series of linear polyketides, from the pentaketide SY5 to the decaketide SY10, all carrying a naphthoquinone chromophore without a hydroxyl group at C-8 (Fig. 4). The heptaketide and the nonaketide were isolated as the carboxyl-terminal lactones, but LC-MS also indicated the presence of the corresponding open-chain acids. The corresponding undecaketide, if present at all, is not a prominent metabolite as judged from LC-MS evidence. Also isolated and structurally characterized were N-formyl and N-acetyl derivatives of the tetraketide, SY4a and SY4b, lactone analogs of the previously reported compound 3 (11), carrying a 3-amino-5-hydroxybenzene chromophore, and a hydroxyl group at the position corresponding to C-8 in the rifamycins (Fig. 4). LC-MS evidence additionally suggests the presence of the corresponding free amino compound P8/1-OG lactone. The tetra- and pentaketides are the most prevalent compounds; the amounts of metabolite obtained decrease with increasing chain length, but the di- and triketides, if present at all, also are formed only in trace amounts. The sum of the accumulated metabolites adds up to approximately 10–20% of the amount of rifamycin B formed in a wild-type fermentation. Original indications based on feeding experiments with 13C-AHBA and NMR analysis were that these compounds are not formed by the wild-type organism. However, subsequent LC-MS analysis with selective ion monitoring and comparison with the authentic samples revealed that the same compounds also are present in extracts of wild-type A. mediterranei S699.
Figure 4.
Structures of compounds isolated and characterized from the rifF(−) mutant (A) and the rifE disrupted mutant (B).
Disruption of PKS Genes.
Disruption of the rifD module 8 and rifE modules 9 and 10 genes, as shown in Fig. 3, also led to the production of linear polyketides. Acetylation of the crude material obtained from the rifE module 10 mutant before further purification resulted in isolation of SY6a, the diacetate of the 8-hydroxy-7,8-dihydronaphthoquinone analog of hexaketide SY6, plus SY5a, the monoacetate of the enol of pentaketide SY5 (Fig. 4). The structure of SY6a was fully consistent with its FAB-MS and one- and two-dimensional NMR data. Moreover, tetraketide 3 to decaketide SY10 were detected by HPLC analysis of the rifE module 10 mutant (which cannot make the undecaketide), whereas the rifD mutant produced the tetraketide-to-octaketide products (heptaketide SY7 and octaketide SY8 were the main products); the rifE module 9 mutant accumulated a similar set of compounds (octaketide and nonaketide were the main products). Peaks with a UV spectrum characteristic of a dihydronaphthoquinone ring, as in SY6a, were seen in these extracts as well, but not in the rifF(−) extract.
Origin of C-8 Oxygen in Rifamycins.
The isolation of a series of polyketides mostly containing a naphthoquinone chromophore without the 8-hydroxy group raises a question about the pathway of rifamycin formation that is unsettled in the literature. Clearly, the structure of the chromophore initially generated on the enzyme, and presumably present in the first cyclic product released, proposed to be 4, must be that of an 8-hydroxylated 7,8-dihydronaphthoquinone. This follows from the structure of the first PKS module, which has a functional ketoreductase domain but an inactive dehydratase domain and is supported by the structures of 3, SY4a, SY4b, and, particularly, SY6a. The question, however, is whether this 8-hydroxy-7,8-dihydronaphthoquinone is converted to the 8-hydroxynaphthoquinone chromophore of the rifamycins by (i) dehydration followed by hydroxylation at C-8 or (ii) by a direct dehydrogenation to introduce the 7,8-double bond (Fig. 5). The first pathway has been proposed by Ghisalba et al. (14) based on the structure of 5 and their finding of its partial conversion into rifamycins. On the other hand, Rickards and coworkers (21) fed AHBA carrying 18O in the carboxyl group and reported incorporation of 18O into the 8-hydroxy group of rifamycins, implying that route ii must be operating. Because the two reports are in conflict, we reexamined both of these experiments. Feeding of 5 to a mutant, HGF010 (unpublished data), of A. mediterranei carrying a deletion from the rif gene cluster covering part of the last PKS module through rifI, i.e., a mutant that can neither synthesize AHBA nor assemble a complete rifamycin polyketide, but that contains all the downstream-processing genes, resulted in no detectable formation of rifamycin B (although small amounts of an unidentified yellow pigment were produced). Although one cannot rule out the possibility that the precursor does not penetrate the cell membrane, this result provides no support for the proposed precursor role of 5 in rifamycin biosynthesis. Conversely, [7-13C,18O2]AHBA (85% 18O, 89% 13C) was fed to a rifK(−) mutant that depended on external AHBA for 1 synthesis. The resulting rifamycin B from this mutant contained 82% 18O in addition to the 13C, as demonstrated by electrospray ionization–MS, indicating that the C-8 oxygen is derived from the carboxyl group of AHBA. These results thus strongly support a pathway of rifamycin biosynthesis involving direct dehydrogenation of 4. Hence, both 5 and the naphthoquinone compounds SY5 through SY10 isolated in this work are either shunt metabolites or artifacts of the work-up process arising from the true intermediate structures carrying an 8-hydroxy-7,8-dihydronaphthoquinone chromophore, as in SY6a.
DISCUSSION
Because inactivation of rifF abolished the synthesis of 1 but not that of the open-chain polyketides, the results of this study support the proposed role of RifF as the chain-termination enzyme in rifamycin B 1 biosynthesis. This represents another mode of release of an assembled polyketide chain from a type I PKS, an alternative to the well-known chain-termination catalyzed by thioesterase domains, as exemplified by the erythromycin PKS system. The data also provide very clear evidence that the rif PKS assembles polyketide chains in a processive manner as illustrated in Fig. 6. It seems plausible, and is consistent with the data, that multiple polyketide chains are synthesized simultaneously on the same enzyme molecule, but an alternative model in which ketides of different chain length are released from different protein molecules cannot be ruled out. Evidence for the processive nature of polyketide assembly on type I PKSs comes primarily from feeding experiments demonstrating the intact incorporation of partially assembled precursor oligoketides, which first were carried out in the erythromycin and tylosin systems (27, 28). Only a few studies have provided evidence for the accumulation of such intermediates during either the normal or impaired function of a type I PKS. Working on the mycinamicin producer, Micromonospora griseorubida, Kinoshita et al. (29) isolated from a nonproducing mutant a triketide containing the starter and first two chain-extension units and, from the wild type, the corresponding tetraketide and the decarboxylation product of the pentaketide. Tetra- and decarboxylated pentaketides were isolated from nonproducing mutants of the tylosin producer Streptomyces fradiae (30). However, noncyclized polyketide intermediates have not been observed during extensive work with 6-deoxyerythronolide B synthase (31) in vivo and in vitro (R. McDaniel and C. Khosla, personal communication). The present work, showing the accumulation of a whole series of ketides of different chain length, represents perhaps the most direct support for the widely accepted notion that type I PKSs assemble polyketide chains processively.
Figure 6.
Proposed mechanism of polyketide assembly, dihydronaphthoquinone ring closure, and macrolactam ring closure on the type I rifamycin polyketide synthase of A. mediterranei.
Because the ketides isolated from the rifF(−) mutant also are found in the wild-type extract, although their presence is masked by large amounts of rifamycin, their formation is not the result of the mutation to eliminate the off-loading mechanism. It is noteworthy that all of the isolated ketides, with the exception of SY6a, which comes from a PKS-truncated mutant, have undergone a structural modification in the aromatic moiety, either a dehydration from the 8-hydroxy-7,8-dihydronaphthaline to the naphthalene ring structure in SY5-SY10 or an acylation of the nitrogen in SY4a and SY4b. If these modifications occurred on the enzyme-bound ketides, either spontaneously or by the aberrant action of an enzyme, they may prevent the further processing of the modified ketide, with consequent inactivation of the PKS. The release of these products then may reflect the operation of a repair mechanism, which removes the aberrant product from the enzyme. Such a repair process may be an inherent function of the PKS itself, or it may be catalyzed by an auxiliary enzyme, such as the type 2 thioesterase encoded by rifR (12), as has been suggested for this type of enzyme in macrolide (19) and nonribosomal peptide biosynthesis (18). Alternatively, however, the structural modifications may be introduced only after release of the incomplete polyketides from the PKS, and the accumulated ketides simply may represent normal shunt metabolites of this fermentation.
Our data indicate that the ring closure from the benzenoid to the naphthoquinoid system may not be a postsynthetic modification of the polyketide produced by the rifA–F genes, but could occur either during its assembly or before the ansamacrolide ring is closed by the RifF enzyme, to form 4 (Fig. 6). Mechanistically, this transformation presumably requires enzymatic oxidation to the benzoquinone followed by Michael addition of a C-5 carbanion, which may or may not be enzyme-controlled, driven by reoxidation of the resulting naphthohydroquinone to the quinone (Fig. 6). This modification most likely occurs between the tetraketide and pentaketide stage, coincident with the handover of the substrate from the last module of RifA to the first module of RifB. The absence of uncyclized pentaketide among the products isolated from the rifF mutant implies that oxidative cyclization is less likely to occur after formation of the pentaketide by the first module of RifB. The fact that 3 does not seem to be a free intermediate in rifamycin biosynthesis (C. Khosla, personal communication) suggests that the cyclization may occur on a PKS-bound tetraketide chain. (Cyclization after release would require that the enzymes involved have an unusually broad substrate specificity, encompassing the pentaketide-to-undecaketide substrates but ignoring the tetraketide.) Thus, the enzyme(s) catalyzing it either must be part of the PKS or must dock to the PKS, and module 4 of RifB may only chain-extend a (dihydro)naphthoquinoid, i.e., cyclized tetraketide. Downstream recognition of modifications in the starter unit had been demonstrated earlier by the observation that 3-hydroxy- and 3,5-dihydroxybenzoic acid were used as starter units but resulted only in the formation of the desamino or 3-hydroxy analogs of 3, respectively (32). The cumulative evidence points to module 4 as the site of this discrimination. This obviously has implications for future efforts to produce modified rifamycins by genetic engineering of the biosynthetic process.
Acknowledgments
We are indebted to Dr. Ross Lawrence of the University of Washington Mass Spectrometry Center for numerous mass spectral analyses. This work was supported by National Institutes of Health Grants AI 20264 to H.G.F and AI 38947 to C.R.H.
ABBREVIATIONS
- PKS
polyketide synthase
- AHBA
3-amino-5-hydroxybenzoic acid
- rif
rifamycin
- LC-MS
liquid chromatography–MS
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Lancini G, Cavalleri B. In: Biotechnology of Antibiotics. 2nd Ed. Strohl W R, editor. New York: Dekker; 1997. pp. 521–549. [Google Scholar]
- 2.Brufani M, Fedeli W, Giacomello G, Vaciago A. Experientia. 1964;20:339–342. doi: 10.1007/BF02171085. [DOI] [PubMed] [Google Scholar]
- 3.Oppolzer W, Prelog V. Helv Chim Acta. 1973;56:2287–2314. doi: 10.1002/hlca.19730560717. [DOI] [PubMed] [Google Scholar]
- 4.Sensi P, Margalith P, Timbal M T. Farmaco Ed Sci. 1959;14:146–147. [PubMed] [Google Scholar]
- 5.Rinehart K L, Jr, Shields L S. Fortsch Chem Org Naturst. 1976;33:231–307. doi: 10.1007/978-3-7091-3262-3_3. [DOI] [PubMed] [Google Scholar]
- 6.Sepkowitz K A, Rafalli J, Riley L, Kiehn T E, Armstrong D. Clin Microbiol Rev. 1995;8:180–199. doi: 10.1128/cmr.8.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Service R F. Science. 1995;270:724–727. doi: 10.1126/science.270.5237.724. [DOI] [PubMed] [Google Scholar]
- 8.Lancini G C, Zanichelli W. In: Structure-Activity Relationships Among the Semisynthetic Antibiotics. Perlman D, editor. New York: Academic; 1977. pp. 537–600. [Google Scholar]
- 9.Ghisalba O. Chimia. 1985;39:79–88. [Google Scholar]
- 10.Kim C-G, Yu T-W, Fryhle C B, Handa S, Floss H G. J Biol Chem. 1998;273:6030–6040. doi: 10.1074/jbc.273.11.6030. [DOI] [PubMed] [Google Scholar]
- 11.Ghisalba O, Fuhrer H, Richter W, Moss S. J Antibiot. 1981;34:58–63. doi: 10.7164/antibiotics.34.58. [DOI] [PubMed] [Google Scholar]
- 12.August P R, Tang L, Yoon Y J, Ning S, Müller R, Yu T-W, Taylor M, Hoffmann D, Kim C-G, Zhang X, et al. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 13.Tang L, Yoon Y J, Choi C Y, Hutchinson C R. Gene. 1998;216:255–265. doi: 10.1016/s0378-1119(98)00338-2. [DOI] [PubMed] [Google Scholar]
- 14.Ghisalba O, Traxler P, Nüesch J. J Antibiot. 1978;31:1124–1131. doi: 10.7164/antibiotics.31.1124. [DOI] [PubMed] [Google Scholar]
- 15.Schupp T, Toupet C, Engel N, Goff S. FEMS Microbiol Lett. 1998;159:201–207. doi: 10.1111/j.1574-6968.1998.tb12861.x. [DOI] [PubMed] [Google Scholar]
- 16.Donadio S, Staver M J, McAlpine J B, Swanson S J, Katz L. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 17.Magnuson S, Rock C O, Cronin J E., Jr Microbiol Revs. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider A, Marahiel M A. Arch Microbiol. 1998;169:404–410. doi: 10.1007/s002030050590. [DOI] [PubMed] [Google Scholar]
- 19.Xue X, Zhao L, Liu H-w, Sherman D H. Proc Natl Acad Sci USA. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohsako S, Ohtomi M, Sakamoto Y, Uyemura K, Deguchi T. J Biol Chem. 1988;263:7534–7538. [PubMed] [Google Scholar]
- 21.Anderson, M. G., Monypenny, D. & Rickards, R. W. (1989) J. Chem. Soc. Chem. Commun., 311–313.
- 22.Kim C-G, Kirschning A, Bergon P, Zhou P, Su E, Sauerbrei B, Ning S, Ahn Y, Breuer M, Leistner E, et al. J Am Chem Soc. 1996;118:7486–7491. [Google Scholar]
- 23.Lydiate D J, Malpartida F, Hopwood D A. Gene. 1985;35:223–235. doi: 10.1016/0378-1119(85)90001-0. [DOI] [PubMed] [Google Scholar]
- 24.Lancini G. In: Biotechnology. Rehm H-J, Reed G, editors. Vol. 4. New York: VCH; 1986. pp. 431–469. [Google Scholar]
- 25.Yanish-Perron C, Vieira J, Messing J. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 26.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 27.Yue S, Duncan J S, Yamamoto Y, Hutchinson C R. J Am Chem Soc. 1987;109:1253–1255. [Google Scholar]
- 28.Cane D E, Yang C C. J Am Chem Soc. 1987;109:1255–1257. [Google Scholar]
- 29.Kinoshita, K., Takenaka, S. & Hayashi, M. (1988) J. Chem. Soc. Chem. Commun., 943–945.
- 30.Huber M L B, Paschal J W, Leeds J P, Kirst H A, Wind J A, Miller F D, Turner J R. Antimicrob Agents Chemother. 1990;34:1535–1541. doi: 10.1128/aac.34.8.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cane D E, Walsh C T, Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 32.Hunziker D, Yu T-W, Hutchinson C R, Floss H G, Khosla C. J Am Chem Soc. 1998;120:1092–1093. [Google Scholar]