Abstract
Glycosyltransferases encompass one of the largest families of enzymes found in nature. Their principle function is to catalyze the transfer of activated donor-sugar molecules to various acceptor substrates. The molecular basis that governs this specific transfer reaction, such as how a given transferase determines donor-sugar specificity, remains to be elucidated. Human α1,4-N-acteylhexosaminyltransferase (EXTL2) transfers N-acetylglucosamine and N-acetylgalactosamine but does not transfer glucose or galactose. Isothermal titration calorimetry (ITC) is a powerful technique used to characterize a variety of binding reactions, including both protein-ligand and protein-protein interactions. ITC provides the binding stoichiometry, affinity and the thermodynamic parameters free energy (ΔG), enthalpy (ΔH) and entropy (ΔS) of these binding interactions. This chapter will describe our recent ITC study demonstrating the two-step mechanism that regulates the specific binding of N-acetylhexosamines to EXTL2.
Overview
EXTL2 is a member of the exostosin (EXT)-related family of enzymes and was originally characterized as the enzyme responsible for the completion of the specific linker region of the heparan chain (Kitagawa et al., 1999). X-ray crystal structures solved in the presence of UDP-N-acetylhexosamines reveal that EXTL2 does not form any direct interaction with the signature N-acetyl group, which resides in an open space of the substrate binding cleft (Pedersen et al., 2003 and Negishi et al., 2003). This observation raises an interesting question as to how EXTL2 determines its donor substrate specificity towards the N-acetylhexosamines. Given the available structure information, ITC provides an excellent tool to explore the molecular mechanism underlying the specific donor-enzyme recognition (Sobhany et al., 2005).
ITC directly measures the heat evolved or absorbed as a result of a given binding process. An ITC instrument essentially consists of a sample cell, a reference cell and an injecting syringe. In an ITC experiment, the solution containing the protein or enzyme of interest is loaded into the sample cell of the instrument and the ligand solution is placed into the injecting syringe. A feedback control system provides thermal power constantly to maintain the exact same temperature in both the reference cell and the sample cell. When heat is generated or absorbed within the sample cell, the temperature within the cell will change and the feedback control system will alter the amount of power supplied in order to minimize the temperature imbalance (Velazquez-Campoy and Freire, 2005). The syringe injects precise amounts of the ligand into the sample cell; the syringe employs a spinning mechanism for subsequent mixing (Fig. 1). Temperature differences between the reference cell and sample cell are measured. Data analysis from a single ITC experiment yields the binding stoichiometry (n) and binding constant Ka. Analysis of the reaction heat as a function of concentration provides a complete thermodynamic characterization of a binding reaction:ΔH, ΔS and ΔG (Perozzo et al., 2004).
Figure 1. Illustration of an Isothermal Titration Calorimeter.
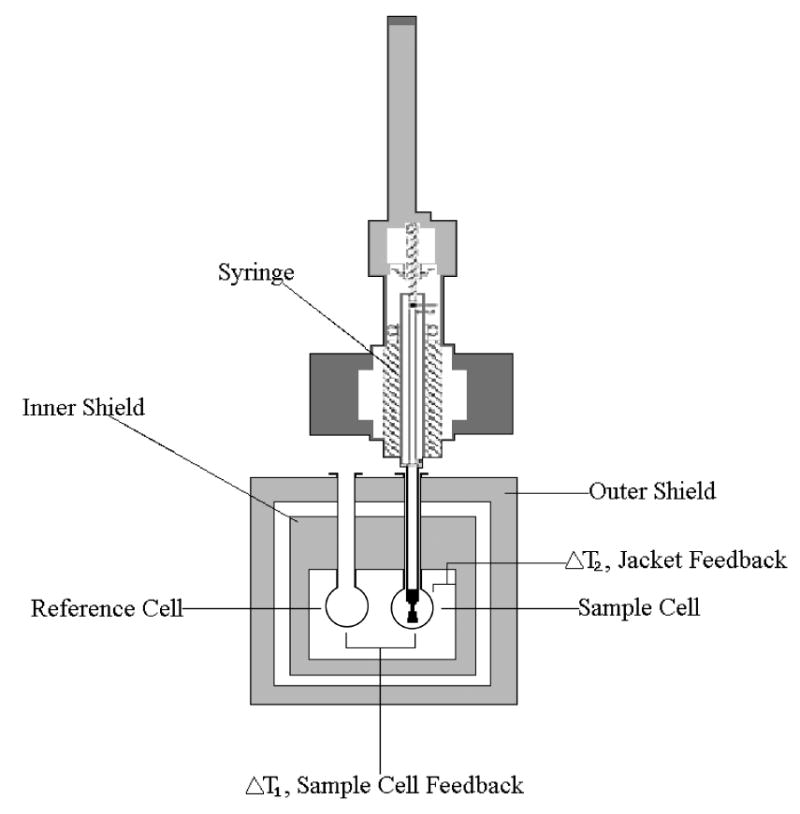
Two cells in an adiabatic environment are connected to the outside through narrow access tubes. A thermoelectric device measures the temperature difference between the two cells. A second device measures the difference in temperature between the cells and the jacket. During the process of a chemical reaction occurring in the sample cell, heat is generated or absorbed. The temperature difference between the sample and reference cells (ΔT1) is kept at zero by the addition of heat to the sample or reference cell. The integral of the power required to maintain ΔT1 = 0 over time is a measure of total heat resulting from the process being studied. A spinning syringe is used for injecting and subsequent mixing of the reactants in the experiment (VP-ITC MicroCalorimeter User’s Manual).
In general, simple reversible associations between a protein P and a ligand L, P+L↔PL are characterized by the binding constant Ka or the dissociation constant Kd: Ka = [PL]/[P][L] = 1/Kd, where [P] and [L] are the concentrations of the free reactants and [PL] is the concentration of the complex generated by the reactants (Perozzo et al., 2004 and Velazquez-Campoy et al., 2004). These values are related to the Gibbs free energy of binding (ΔG) and can be expressed in terms of the changes of enthalpy (ΔH) and entropy (ΔS) in the process: ΔG = −RTlnKa = ΔH−TΔS. R is the universal gas law constant (1.9872 cal/K·mol) and T is the absolute temperature at which the reaction took place in units Kelvin (Velazquez-Campoy et al., 2004). The perceptible response in an ITC experiment is the change in heat correlated with each injection of ligand. For every given injection of ligand, the heat generated or absorbed is directly proportional to the total amount of formed complex and can be expressed as q = V0ΔHΔ[PL]; q is the measured heat that is absorbed/generated associated with the change in complex concentration, Δ[PL], V0 is the reaction volume of the sample cell, and ΔH is the molar enthalpy of binding (Perozzo et al., 2004). As an ITC experiment progresses, the concentration of unoccupied binding sites begin to decrease and the changes in heat decrease in magnitude correspondingly as ligand is added until saturation is reached. From this titration curve, the parameters n (stoichiometry), binding constant Ka, and ΔH are directly determined. ΔS and ΔG are then calculated from these values. These calculations are performed by the software that accompanies the ITC apparatus. The software also performs computer modeling to identify the number of binding sites possessed by the protein.
General Experimental Design
Optimization of protein and ligand concentrations and the injection volume is necessary to obtain high quality data. A few preliminary experiments may need to be performed in order to establish these conditions. The protein concentration required for ITC depends on the given protein and its substrate binding affinity. For experiments that can be expected to produce a high substrate bind affinities, protein concentrations as low as 5μM can be used to successfully obtain good ITC data. However, protein-carbohydrate interactions are quite weaker than protein-protein and protein-nucleic acid interactions and higher concentrations of protein may be necessary in order to obtain observable changes in heat due to binding. For glycosyltransferases, protein concentrations of around 30μM are sufficient for most ITC experiments. Generally, the ligand concentration is ten to twenty times higher than that of the protein since several injections of small aliquots into the sample cell must take place in order to generate a sufficient number of data points for analysis. Upon completion, a titration experiment should approach or reach complete saturation of the binding sites. The time between successive injections is also an important parameter; if protein-ligand interaction is rapid, thermal equilibrium will be achieved within a short period of time (generally three-four minutes is sufficient in such cases). Conversely, heat signals of a slow process require much more time to reach thermal equilibrium and the time between injections should be adjusted accordingly. Additionally, it is absolutely critical for the solutions of the ligand and protein are pure and are exactly identical with respect to pH, buffer capacity and salt concentration; essentially it is preferable for the protein and ligand to be dissolved in the same buffer. Formation of air bubbles should be avoided, as air bubbles in the syringe can cause variation in injection volumes, and bubbles in the sample cell interfere with the thermal contact of solution and cell wall. Therefore, it is very important to thoroughly degas all solutions prior to use in the experiment.
Donor sugar binding to EXTL2
Figure 2 shows the ITC profiles of the binding of UDP-GlcNAc to EXTL2, while Table 1 summarizes the binding characteristics and the thermodynamic parameters of various donor substrates. UDP, UDP-GlcNAc and UDP-GalNAc are all found to bind to EXTL2 at a 1 to 1 ratio, thus agreeing with the X-ray crystal structure showing only one donor molecule per EXTL2 molecule (Pedersen et al., 2003). The binding affinities of the donor substrates are similar; the Kd value of UDP binding is 23 μM and those of UDP-GlcNAc and UDP-GalNAc are approximately 61 μM. UDP-Gal and UDP-Glc are found not to bind to EXTL2. These results provide direct evidence that despite having no direct interaction with the enzyme, the N-acetyl group determines the binding of UDP-N-acetylhexosamines to EXTL2.
Figure 2. Calorimetric profile of the binding of UDP-GlcNAc to EXTL2.
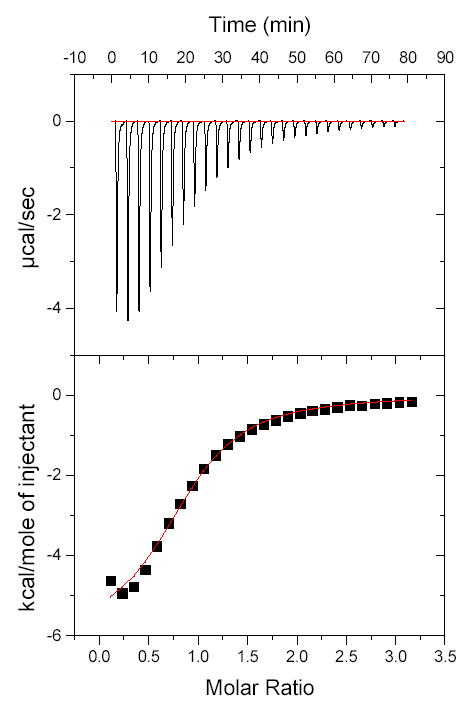
The reaction cell contained a solution of EXTL2 (1.4 ml and 165.4 μM). The syringe contained 4 mM UDP-GlcNAc dissolved in the same buffer as EXTL2. Top, raw calorimetric data obtained from the injection of 6 μl aliquots of UDP-GalNAc at three minute intervals. The lower plot shows the integrated binding isotherm with the experimental points (▪) and best fit. The final fitting parameters for the best fit, provides the values for N (number of sites), ΔH (cal/mol), ΔS (cal/mol/deg) and K (binding constant in M−1).
Table 1.
Thermodynamic parameters, ΔG (cal/mol), ΔH (cal/mol), ΔS (cal/mol/deg), and n (number of binding sites) for donor binding with wild type EXTL2 in H20 solution at 303ºK. Substrate and EXTL2 concentrations were 4 mM and 166 μM, respectively. n.d.: no detectable binding.
| Substrate | ΔG | ΔH | ΔS | Kd(μM) | n |
|---|---|---|---|---|---|
| UDP | −6443.4±193.7 | −6034±193.7 | 1.351 | 22.62 | 0.869 |
| UDP-GalNAc | −5839.1±72.14 | −2097±72.14 | 12.35 | 61.46 | 0.968 |
| UDP-GlcNAc | −5844.3±68.32 | −1975±68.32 | 12.77 | 60.80 | 1.106 |
| UDP-Gal | n.d. | n.d. | n.d. | n.d. | n.d. |
| UDP-Glc | n.d. | n.d. | n.d. | n.d. | n.d. |
ITC has previously been employed with α1,3-galactosyltransferase, which also displayed a Kd of 60 μM for its binding to UDP-Gal (Boix et al., 2002). Therefore, the interaction of glycosytransfearses with carbohydrates appeared to be around 100 μM, which is weaker than other specific interactions. Protein-protein interactions are generally quite strong; the association of a monoclonal antibody with cytochrome c yields a binding affinity of 71.43 pM (Pierce et al., 1999). Protein-nucleic acid interactions also yield robust binding affinities; for instance, the binding of Thermus aquaticus DNA polymerase to template DNA at 30°C produces a Kd of 6.2 nM (Datta and LiCatta, 2003). The binding reactions of UDP-GlcNAc and UDP-GalNAc are accompanied with an increase in ΔS. A comparison of the ΔH and TΔS values reveals that absolute values of the TΔS values are larger than those of ΔH, indicating that the binding reactions are driven by changes in entropy. Increases in entropy in systems at constant temperature are known to correlate with the loss of previously oriented waters of hydration into the bulk solvent (Sievers et al., 2004). Thus, the N-acetyl group may expel water from the area of enzyme-donor complex into the bulk solvent in order to position UDP-N-acetylhexosamine for its binding to EXTL2. To provide further evidence that water molecules may be involved in determining the specific donor binding, ITC experiments were performed in the denser and more viscous deuterium oxide (D2O). Both UDP-GlcNAc and UDP-GalNAc maintain their ability to bind to EXTL2 in D2O solution with similar affinities and ΔS values to those observed in H2O solution. The importance of water in donor binding activity was substantiated by testing the binding of UDP-Gal and UDP-Glc in D2O solution. UDP-Gal and UDP-Glc were found to be capable of binding to EXTL2 in D2O solution, although their binding affinities to EXTL2 were still 4- to 5-fold weaker than those of the UDP-N-acetylhexosamines. The bindings of UDP-Glc and UDP-Gal are accompanied with only a marginal increase in entropy, compared with the large increases in entropy observed with those of the UDP-N-acetylhexosamines in D2O solution. This data is consistent with the hypothesis that water molecules are critically involved in EXTL2 donor binding activity.
While EXTL2 forms no direct interaction with the N-acetyl group, its residues, such as Arg135, Asp246 and Arg293, interact directly with the hydroxyl groups of N-acetylhexosamine moiety (Pedersen et al., 2003; Sobhany et al., 2005). Alanine mutations of those residues result in an increase of donor binding affinity (Kd) and Km for donor hydrolysis (Sobhany et al., 2005). Moreover, the Km and Kd values are very similar in wild-type and in a given mutant of EXTL2. Thus, those residues determine the donor binding affinity and hydrolysis activity via their direct hydrogen bond interactions with the donor molecule, while the N-acetyl group determines the donor specificity by the entropy-driven positioning of UDP-N-acetylhexosamines in the active site. These experiments exemplify that, when coupled with available structural data, ITC can be a powerful tool in elucidating donor substrate recognition and catalytic mechanisms of glycosyltransferases.
Materials for Protein Expression and Purification
BL21 (DE3) Competent Cells (Stratagene, La Jolla, CA) transformed with pMAL-2c EXTL2 expression vector (New England Biolabs, Beverly, MA).
Luria-Bertani Media: 10 g of Bacto-Tryptone, 5 g of Bacto-Yeast Extract, 10 g of NaCl, dissolved in distilled water, final volume one liter with pH adjusted to 7.5. Working media solution modified with 100 μg/mL ampicillin.
2X YT Media: 16 g Bacto-Tryptone, 10 g of Bacto-Yeast Extract, 10 g of NaCl, dissolved in dissolved in distilled water, final volume one liter with pH adjusted to 7.5. Working media solution modified with 100 μg/mL ampicillin.
DU 640 Spectrophotometer (Beckman Coulter, Fullerton, CA).
Isopropylthio-β-D-galactoside (IPTG) (Invitrogen, Carslbad, CA).
FRENCH Pressure Cell and Press (Thermo IEC, Needham Heights, MA).
J6-MI Centrifuge and Opitma L-90K ultracentrifuge (Beckman Coulter, Fullerton, CA).
Innova 44 Shaking Incubator (New Brunswick Scientific, Edison, NJ).
Amylose Resin (New England Biolabs, Beverly, MA).
Molecularporous membrane tubing (Spectrum Laboratories, Inc., Rancho Dominguez, CA).
Bio-Rad Protein Assay (Bio-Rad, Hercules, CA).
Solutions
Lysis Buffer: 30 mM Tris-Cl (pH 7.5), 350 mM NaCl, 5 mM dithiothreitol (DTT) (ICN Biomedicals, Inc., Aurora OH).
Elution Buffer: 30 mM Tris-Cl, 350 mM NaCl, 5 mM dithiothreitol (DTT) modified with 20 mM Maltose.
Reaction Buffer: 25 mM HEPES (pH 7.5), 100 mM NaCl, 20 mM MnCl2, 1 mM.
Method for Protein Expression and Purification
One milliliter of BL21 (DE3) cells transformed with pMAL-2c EXTL2 expression vector was added to 100 mLs of Luria-Bertani media modified with 100 μg/mL ampicillin and placed on a shaker at 210 rpm at 37ºC for an overnight culture.
10 mLs of pre-culture each was added to ten 2-liter flasks containing one liter of 2X YT media modified with 100 μg/mL ampicillin.
These flasks were placed in the Innova 44 Shaking Incubator set to shake at 210 rpm at 37ºC. When A550 reached 0.8, isopropylthio-β -D-galactoside was added to each flask, final concentration of 0.2 mM.
The temperature of the incubator was then set to 23ºC and the flasks were allowed to shake overnight.
The cells were then harvested by spinning at 4000 rpm for twenty-five minutes and then resuspended in lysis buffer.
The cells were disrupted in a French Press at room temperature under pressures varying between 8,000 and 24,000 pounds per square inch.
Following disruption, the cells were spun down at 38,000 rpm for forty-five minutes.
The soluble fraction obtained was then allowed to bind to amylose resin at 4ºC for 2 hours with gentle agitation.
The resin was then spun down at 4000 rpm for five minutes. The supernatant was poured off and the resin was washed with the lysis buffer. Resin was pelleted and washed a total of three times.
Protein was then eluted with the elution buffer.
Following elution, the protein solution was placed into Molecularporous membrane tubing and dialyzed overnight into reaction buffer. Following dialysis, protein concentration was determined using Bio-Rad Protein Assay.
Materials for Isothermal Titration Calorimetry
Uridine Diphosphate N-acteylglucosamine (Sigma-Aldrich, St. Louis, MO).
ThermoVac Sample Degasser and Thermostat (MicroCal, LLC., Northhampton, MA).
VP-ITC MicroCalorimeter (MicroCal, LLC., Northhampton, MA).
Origin Scientific Graphing and Analysis Software (OriginLab, Northhampton, MA).
Solutions
Reaction Buffer: 25 mM HEPES (pH7.5), 100 mM NaCl, 20 mM MnCl2, 1 mM CaCl2.
Method for Isothermal Titration Calorimetry
Degass all protein samples, buffers and donor substrate solutions were thoroughly prior to testing.
Load EXTL2 sample into the sample cell of the VP-ITC MicroCalorimeter.
Load reaction buffer into the reference cell.
Dissolve Uridine Diphosphate N-acteylglucosamine (UDP-GlcNAc) in the reaction buffer at a concentration 10–20 times that of the EXTL2. Then load the UDP-GlcNAc solution into the syringe.
Set the VP-ITC Microcalorimeter to perform twenty-six injections of 6 μl at 180 sec intervals. Run program; data acquisition and analysis will be completed by the Origin software package.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, and NIEHS.
References
- Boix E, Zhang Y, Swaminathan J, Brew K, Achaya KR. Structural basis of ordered binding of donor and acceptor substrates to the retaining glycosyltransferase, alpha-1,3-galactosyltransferase. J Biol Chem. 2002;277:28310–28318. doi: 10.1074/jbc.M202631200. [DOI] [PubMed] [Google Scholar]
- Datta K, LiCata VJ. Thermodynamics of the binding of Thermus Aquaticus DNA polymerase to primed-template DNA. Nucleic Acids Res. 2003;31:5590–5597. doi: 10.1093/nar/gkg774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa H, Shimakawa H, Sugahara K. The Tumor Suppressor EXT-like Gene EXTL2 Encodes an α1, 4-N-Acetylhexosaminyltransferase That Transfers N-Acetylgalactosamine and N-Acetylglucosamine to the Common Glycosaminoglycan-Protein Linkage Region. J Biol Chem. 1999;274:13933–13937. doi: 10.1074/jbc.274.20.13933. [DOI] [PubMed] [Google Scholar]
- Negishi M, Dong J, Darden TA, Pedersen LG, Pedersen LC. Glucosaminylglycan biosynthesis: what we can learn from the X-ray crystal structures of glycosyltransferases GlcAT1 and EXTL2. Biochem Biophys Res Commu. 2003;303:393–398. doi: 10.1016/s0006-291x(03)00356-5. [DOI] [PubMed] [Google Scholar]
- Pedersen LC, Dong J, Taniguchi F, Kitagawa H, Krahn JM, Pedersen LG, Sugahara K, Negishi M. Crystal structure of an alpha 1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis. J Biol Chem. 2003;278:14420–14428. doi: 10.1074/jbc.M210532200. [DOI] [PubMed] [Google Scholar]
- Perozzo R, Folkers G, Scapozza L. Thermodynamics of protein-ligand interactions: history, presence, and future aspects. J Recept Sig Transd. 2004;24:1–52. doi: 10.1081/rrs-120037896. [DOI] [PubMed] [Google Scholar]
- Pierce MM, Raman CS, Nall BT. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19:213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- Sievers A, Beringer M, Rodina MV, Wolfenden R. The ribosome as an entropy trap. Proc Natl Acad Sci USA. 2004;101:7897–7901. doi: 10.1073/pnas.0402488101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhany M, Dong J, Negishi M. Two-step mechanism that determines the donor binding specificity of human UDP-N-acetylhexosaminyltransferase. J Biol Chem. 2005;280:23441–23445. doi: 10.1074/jbc.M413379200. [DOI] [PubMed] [Google Scholar]
- Velazquez-Campoy A, Leavitt SA, Freire E. Characterization of protein-protein interactions by isothermal titration calorimetry. Methods Mol Biol. 2004;261:35–54. doi: 10.1385/1-59259-762-9:035. [DOI] [PubMed] [Google Scholar]
- Velazquez-Campoy A, Freire E. ITC in the post-genomic era…? Priceless. Biophys Chem. 2005;115:115–124. doi: 10.1016/j.bpc.2004.12.015. [DOI] [PubMed] [Google Scholar]
- VP-ITC MicroCalorimeter User’s Manual. MicroCal, LLC.; Northampton, MA: [Google Scholar]


