Abstract
Background: Involvement of peroxisome proliferator activated receptor γ (PPARγ) in the growth response of colon cancer cells has been suggested.
Aims: To investigate the characteristics of PPARγ induced apoptosis in colon cancer cells.
Methods: The effects of ligands for each of the PPAR subtypes (α, δ, and γ) on DNA synthesis and cell viability were examined in HT-29 colon cancer cells. Modulation of apoptosis related gene expression by PPARγ ligands was screened with cDNA arrays, and the results were confirmed by quantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis.
Results: PPARα, PPARδ, and PPARγ were all expressed in HT-29 cells. PPARγ ligands, 15-deoxy-δ12,14-prostaglandin J2 (15d-PGJ2) and troglitazone (TGZ), suppressed DNA synthesis of HT-29 cells whereas ligands for PPARα and PPARδ had no significant effects. Both 15d-PGJ2 and TGZ induced HT-29 cell death in a dose dependent manner which was associated with an increase in fragmented DNA and was sensitive to a caspase inhibitor. Among several genes selected by cDNA array screening, quantitative RT-PCR analysis confirmed downregulation of c-myc expression and upregulation of c-jun and gadd153 expression by 15d-PGJ2 and TGZ. PPARγ induced apoptosis was antagonised by the presence of serum in the culture medium, and interaction between PPARγ signalling and cell survival signalling through the phosphatidylinositol 3-kinase pathway was suggested.
Conclusions: As c-myc is an important target gene of the adenomatous polyposis coli (APC)/β-catenin and/or APC/γ-catenin pathway, activation of PPARγ signalling appears to compensate for deregulated c-myc expression caused by mutated APC. The present results suggest the potential usefulness of PPARγ ligands for chemoprevention and treatment of colon cancers.
Keywords: peroxisome proliferator activated receptor, apoptosis, colon cancer
Peroxisome proliferator activated receptors (PPAR) are members of the nuclear receptor superfamily and at least three subtypes have been identified (PPARα, PPARδ, and PPARγ).1 PPARγ is expressed at a high level in adipocytes and has been shown to be an important regulator of adipocyte differentiation and fatty acid metabolism.2 PPARγ forms a heterodimer with retinoid X receptor (RXR) and regulates expression of target genes by binding to the PPAR responsive element.3 It has been demonstrated that 15-deoxy-δ12,14-prostaglandin J2 (15d-PGJ2), a metabolite of prostaglandin D2, is a potential endogenous ligand for PPARγ, and that thiazolidinediones (synthetic antidiabetic agents) such as troglitazone (TGZ) and pioglitazone are specific exogenous ligands for PPARγ.3,4 PPARγ is also expressed at high levels in colonic epithelial cells and colon cancer cells.5,6 Sarraf and colleagues7 showed that activators of PPARγ suppress the growth response of colon cancer cells. Brockman and colleagues8 reported that activation of PPARγ leads to inhibition of anchorage independent growth of colon cancer cells. Kitamura and colleagues9 also showed in HT-29 colon cancer cells that activation of PPARγ results in upregulation of differentiation marker genes such as villin and intestinal alkaline phosphatase. On the other hand, Saez and colleagues10 and Lefebvre and colleagues11 showed in APCMIN mice, an animal model of familial polyposis, that treatment with PPARγ ligands increases the frequency and size of colon polyps. Although the results of these in vitro and in vivo studies are contradictory, it is very likely that the PPARγ pathway can modulate the growth response of colon epithelial cells and colon cancer cells. As a better understanding of PPARγ signalling in colon cancer cells will benefit the development of new strategies for colon cancer prevention and treatment, we investigated the characteristics of PPARγ ligand induced growth suppression and apoptosis in colon cancer cells.
MATERIALS AND METHODS
Reagents
Wy-14643, carbaprostacyclin (cPGI), and 15d-PGJ2 were obtained from Cayman Chemical (Ann Arbor, Michigan, USA). TGZ was provided by Sankyo Pharmaceuticals (Tokyo, Japan). The caspase inhibitor (apopain inhibitor II), Z-Asp(OMe)-Glu(OMe)-Val-DL-Asp(OMe)-fmk, was obtained from Takara (Shiga, Japan). Cycloheximide (CHX), all-trans retinoic acid (AT-RA), 9-cis-retinoic acid (9c-RA), and wortmannin were purchased from Wako Pure Chemical (Osaka, Japan). NS-398 was provided by Taisho Pharmaceuticals (Tokyo, Japan).
Cell culture
HT-29 cells were obtained from Dainippon Pharmaceuticals (Tokyo, Japan). Cells were grown in Ham's F-12 culture medium (Gibco BRL, Rockville, Maryland, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco BRL). When experiments were performed in the absence of FBS, FBS was eliminated 24 hours before initiating the experiments.
Western blot analysis
HT-29 cell monolayers were washed twice with phosphate buffered saline (PBS) and scraped in a modified radioimmunoprecipitation buffer containing 50 mM Tris HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, 1 mM phenylmethylsulphonyl fluoride, 1 μg/ml aprotinin, 1 mM Na3VO4, and 1 mM NaF. Supernatants of the cell lysates (50 μg protein/lane) were separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis. After proteins were transferred to a polyvinylidene difluoride membrane (Clear blot membrane-P; Atto, Tokyo, Japan), the membrane was blocked with TTBS (10 mM Tris HCl (pH 7.6), 150 mM NaCl, and 0.1% Tween 20) containing 5% non-fat dry milk for one hour at room temperature. The membrane was then incubated with goat polyclonal anti-PPARα, anti-PPARδ, or anti-PPARγ antibody (Santa Cruz Biotechnology, California, USA) in TTBS containing 5% non-fat dry milk overnight at 4°C. After being washed with TTBS, the membrane was incubated with horseradish peroxidase conjugated antigoat IgG antibody (Santa Cruz Biotechnology) in TTBS containing 5% non-fat dry milk for one hour at room temperature. The membrane was washed again and the specific band was visualised with LumiGLO chemiluminescent reagent (New England BioLabs, Beverly, Massachusetts, USA).
Assay for cell growth response and cell viability
The effects of the PPAR ligands on DNA synthesis of cells were assessed using the cell proliferation ELISA kit (Roche Diagnostics, Tokyo, Japan). This assay is a colorimetric immunoassay for quantification of cell proliferation based on measurement of bromodeoxyuridine (BrdU) incorporation during DNA synthesis, and is a non-radioactive alternative to the [3H] thymidine incorporation assay. Cells were grown in 96 well culture dishes (104 cells/well), incubated with the PPAR ligands for 24 hours, and then labelled with BrdU for a further six hours. Incorporated BrdU was measured colorimetrically.
Cell viability was assessed with WST-1 reagent (Wako Pure Chemical). This assay is based on cleavage of the tetrazolium salt WST-1 by mitochondrial dehydrogenase of viable cells to a formazan dye. Cells grown in 96 well culture plates (104 cells/well) were incubated with test agents for 24 hours and WST-1 reagent was then added to each well. Culture plates were further incubated for two hours and cell viability was assessed colorimetrically.
Quantitative determination of fragmented DNA in cytoplasm was performed with the cell death ELISA kit (Roche Diagnostics). This ELISA detects the amount of cytoplasmic histone associated DNA fragments. Cells grown in 96 well culture plates (2×104 cells/well) were incubated with test agents for 18 hours, supernatant culture medium was then removed, and lysis buffer was added to each well. The assay was performed according to the manufacturer's manual, and the amount of fragmented DNA was measured colorimetrically.
cDNA array experiments
Changes in HT-29 cell gene expression associated with PPARγ induced apoptosis were screened with cDNA array membranes (ATLAS Human Apoptosis Array; Clontech, Palo Alto, California, USA). Cells were treated with TGZ (30 μM) or 15d-PGJ2 (10 μM) for eight hours and total RNA was extracted with Trizol reagent (Gibco BRL). Poly(A)+ RNA was further isolated with Micro-Oligo(dT) Cellulose Spin Columns (5 Prime-3 Prime, Boulder, Colorado, USA). Poly(A)+ RNA isolated from each sample (1 μg) was converted to 32P labelled first strand cDNA using [α-32P]dATP (∼110 Tbq/mmol) (Amersham-Pharmacia Biotech, Uppsala, Sweden). cDNA probes were hybridised to the ATLAS array membranes according to the manufacturer's instruction. Autoradiography and densitometric analysis were performed using a BAS2000 Bioimaging System (Fujix, Tokyo, Japan).
Real time quantitative reverse transcription-polymerase chain reaction (RT-PCR)
Real time quantitative RT-PCR analysis was performed to confirm the results of cDNA array experiments. Total RNA was isolated from cells with Trizol reagent (Gibco BRL) and first strand cDNA was made with You-Prime First Strand Beads (Amersham-Pharmacia Biotech) using oligo(dT) primers (Gibco BRL). PCR primers were designed according to the reported sequences and are summarised in table 1 ▶.
Table 1.
PCR primers used in the present study.
| c-jun (GenBank No J04111) |
| Sense 5′-TGCAAAGATGGAAACGACCTT-3′ |
| Antisense 5′-CAGGTTCAGGGTCATGCTCTG-3′ (121 bp) |
| c-myc (L16785) |
| Sense 5′-GGTGAAGTACATGAACTCAGGGC-3′ |
| Antisense 5′-GGCTTTGAATCTGCTGGATTG-3′ (111 bp) |
| gadd153 (S40706) |
| Sense 5′-TGCCTTTCTCTTCGGACACTG-3′ |
| Antisense 5′-TTTTGATTCTTCCTCTTCATTTCCA-3′ (131 bp) |
| β-actin (NM_001101) |
| Sense 5′-TTCCTGGGCATGGAGTCCT-3′ |
| Antisense 5′-AGGAGGAGCAATGATCTTGATC-3′ (204 bp) |
Real time quantitative RT-PCR analysis was performed in an ABI PRISM 7700 Sequence Detection System (PE Biosystems, Tokyo, Japan) using SYBR Green PCR reagents (PE Biosystems). In this assay, the fluorescence intensity of SYBR Green dye, which corresponds to the amount of PCR products, is monitored. After an initial 10 minutes at 95°C, the reaction was run for 40 PCR cycles of 15 seconds at 95°C and one minute at 56°C (two step PCR). β-actin mRNA quantification was also performed for standardisation. To prepare standard samples for quantitative RT-PCR, conventional RT-PCR was performed and PCR products were purified with Quiaquick PCR Purification Kits (Quiagen, Hilden, Germany). Purified PCR products were diluted and used as standard samples (6×102 to 6×107 copies) for generating a standard curve for each quantitative PCR experiment.
Statistics
Data are expressed as mean (SD). Analysis of variance (ANOVA) was performed when more than two groups were compared, and when significant (p<0.05), Scheffés multiple comparison test was applied to test for differences between individual groups. A p value <0.05 was considered to be significant.
RESULTS
Expression of PPAR subtypes in HT-29 cells
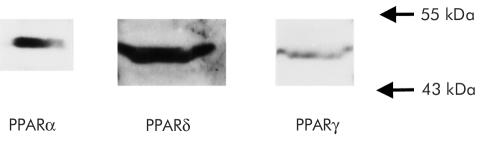
To confirm expression of PPAR subtypes (PPARα, PPARδ, and PPARγ), western blot analysis was performed using HT-29 cells. As shown in fig 1 ▶, HT-29 expressed all subtypes of PPAR.
Figure 1.
Western blot analysis showing expression of peroxisome proliferator activated receptor α (PPARα), δ (PPARδ), and γ (PPARγ) proteins in HT-29 cells.
Effects of PPAR ligands on the growth response of HT-29 cells
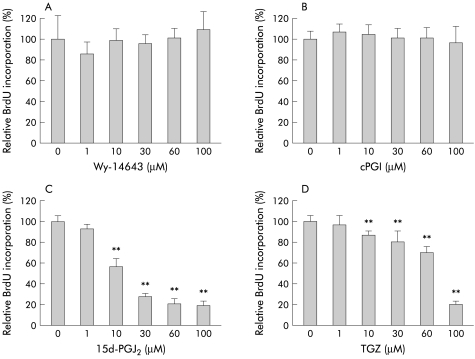
We next examined the effects of PPAR ligands on the growth response (DNA synthesis) of HT-29 cells. This series of experiments was performed in the presence of 10% FBS in the culture medium. As shown in fig 2A and B ▶, neither Wy-14643 (1–100 μM) nor cPGI (1–100 μM) affected BrdU incorporation, suggesting that PPARα and PPARδ are not critically involved in the growth response of HT-29 cells. In contrast, PPARγ ligands 15d-PGJ2 (1–100 μM) and TGZ (1–100 μM) suppressed BrdU incorporation significantly in a dose dependent manner.
Figure 2.
Effects of peroxisome proliferator activated receptor (PPAR) ligands on DNA synthesis (bromodeoxyuridine (BrdU) incorporation) of HT-29 cells (30 hours of incubation) (n=6–12). Experiments were performed in the presence of 10% fetal bovine serum. (A) Wy-14643 (PPARα ligand) (1–100 μM), (B) carbaprostacyclin (cPGI, PPARδ ligand) (1–100 μM), (C) 15-deoxy-δ12,14- prostaglandin J2 (15d-PGJ2, PPARγ ligand) (1–100 μM), and (D) troglitazone (TGZ, PPARγ ligand) (1–100 μM). **p<0.01 versus control.
Effects of PPARγ ligands on HT-29 cell viability
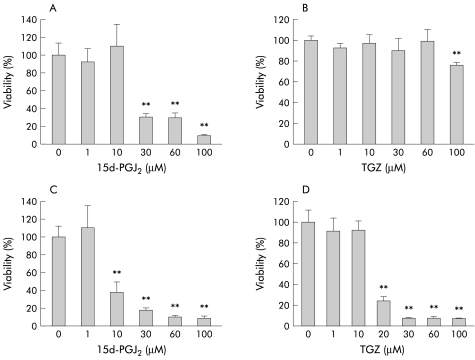
We then examined whether 15d-PGJ2 and TGZ induce cell death in HT-29 cells. Cells were treated with different concentrations of 15d-PGJ2 (1–100 μM) or TGZ (1–100 μM) for 26 hours in the presence or absence of FBS in the culture medium, and the WST-1 assay was performed. As shown in fig 3 ▶, 26 hour treatment with 15d-PGJ2 or TGZ caused a significant decrease in cell viability and FBS antagonised this effect. Neither Wy-14643 (1–100 μM) nor cPGI (1–100 μM) had significant effects on HT-29 cell viability (data not shown).
Figure 3.
Effects of different concentrations of 15-deoxy-δ12,14- prostaglandin J2 (15d-PGJ2 1–100 μM) and troglitazone (TGZ 1–100 μM) on HT-29 cell viability (26 hour incubation) in the presence (A, B) and absence (C, D) of 10% fetal bovine serum (n=8–12). **p<0.01 versus control.
Characterisation of PPARγ ligand induced HT-29 cell death
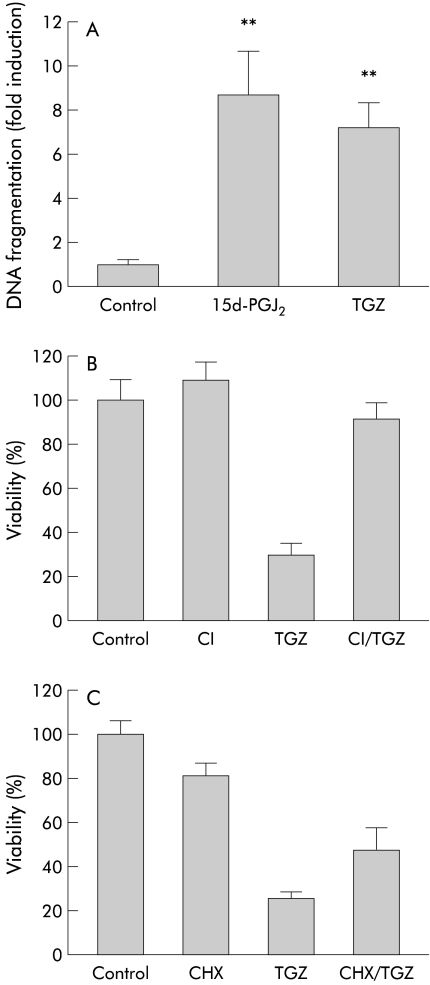
The following experiments were performed in the absence of FBS. To determine whether HT-29 cell death induced by 15d-PGJ2 or TGZ is apoptosis, we analysed the amount of fragmented DNA. Cells were incubated with 15d-PGJ2 (10 μM) or TGZ (20 μM) for 18 hours and ELISA was performed to quantify the amount of fragmented DNA in the cytoplasm. As shown in fig 4A ▶, 15d-PGJ2 (10 μM) caused a ninefold increase and TGZ (20 μM) caused a sevenfold increase in the amount of fragmented DNA, suggesting that cell death induced by these substances is apoptosis. We also examined the effect of a caspase inhibitor, Z-Asp(OMe)-Glu(OMe)-Val-DL-Asp(OMe)-fmk, on TGZ (20 μM) induced HT-29 cell death. Cells were preincubated with the caspase inhibitor (25 μM) for two hours and further incubated with TGZ (20 μM) for 24 hours in the continued presence of the caspase inhibitor. As shown in fig 4B ▶, TGZ induced cell death was significantly antagonised by the caspase inhibitor, indicating a role for caspase activation in the process.
Figure 4.
Characterisation of peroxisome proliferator activated receptor γ (PPARγ) ligand induced HT-29 cell death. Experiments were performed in the absence of fetal bovine serum. (A) Effects of 18 hours of treatment with 15-deoxy-δ12,14- prostaglandin J2 (15d-PGJ2 10 μM) or troglitazone (TGZ 20 μM) on the amount of fragmented DNA (n=6–8). **p<0.01 versus control. (B) Effect of a caspase inhibitor (CI), Z-Asp(OMe)-Glu(OMe)-Val-DL-Asp(OMe)-fmk (25 μM), on TGZ (30 μM) induced HT-29 cell death (n=6–8). HT-29 cells were preincubated with 25 μM CI for two hours and further incubated with 20 μM TGZ for 24 hours in the continued presence of the caspase inhibitor. p<0.01 TGZ versus CI/TGZ. (C) Effect of cycloheximide (CHX 25 μM) on TGZ (20 μM) induced HT-29 cell death (n=6–8). Cells were preincubated with 25 μM CHX for two hours and further incubated with 20 μM TGZ for 24 hours in the continued presence of CHX. p<0.01 TGZ versus CHX/TGZ.
We also examined the effect of cycloheximide (CHX). Cells were preincubated with CHX (25 μM) for two hours, and further incubated with TGZ (20 μM) in the continued presence of CHX. As shown in fig 4C ▶, TGZ induced cell death was partially prevented by CHX. As CHX is an inhibitor of protein synthesis, this result suggests that the action of TGZ is mediated, at least in part, by de novo protein synthesis.
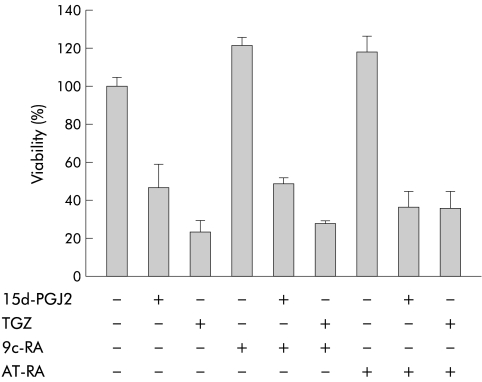
Effects of 9c-RA and AT-RA
As PPARγ forms a heterodimer with RXR and regulates expression of target genes by binding to the PPAR responsive element, we tested the effects of 9c-RA, a specific ligand for RXR, on the viability of HT-29 cells. We also tested the effects of AT-RA, a ligand for the retinoid A receptor (RAR), for comparison. Cells were incubated with 9c-RA (10 μM) or AT-RA (10 μM) in combination with 15d-PGJ2 (10 μM) or TGZ (20 μM) for 24 hours. As shown in fig 5 ▶, 9c-RA alone or AT-RA alone did not cause a significant decrease in cell viability, and HT-29 cell death induced by 15d-PGJ2 (10 μM) or TGZ (20 μM) was not affected by coincubation with 9c-RA (10 μM) or AT-RA (10 μM).
Figure 5.
Effects of 9-cis-retinoic acid (9c-RA) and all-trans retinoic acid (AT-RA) on HT-29 cell viability. Experiments were performed in the absence of fetal bovine serum. Cells were incubated with 9c-RA (10 μM) or AT-RA (10 μM) in combination with 15-deoxy-δ12,14- prostaglandin J2 (15d-PGJ2 10 μM) or troglitazone (TGZ 20 μM) for 26 hours (n=6–8).
cDNA array experiments
To examine the effects of the PPARγ ligands on expression of apoptosis related genes in HT-29 cells, we screened gene expression profiles before and after eight hours of treatment with 15d-PGJ2 (10 μM) or TGZ (30 μM) using cDNA arrays (ATLAS Human Apoptosis Arrays; Clontech). Using this screening method, we identified several genes whose expression appeared to be modulated by TGZ or 15d-PGJ2. Because quantitative assessment is difficult in cDNA array experiments, we performed real time quantitative RT-PCR analysis using an ABI PRISM 7700 sequence detection system to confirm the results of the cDNA array experiments. Data on expression of c-myc, c-jun, and gadd153 are given below.
Real time quantitative RT-PCR analysis
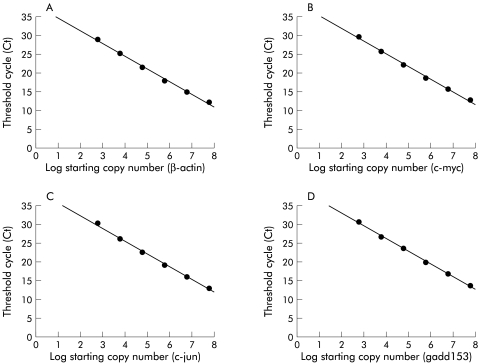
The real time quantitative RT-PCR system employed is based on measurement of fluorescence signal and fig 6 ▶ shows representative standard curves plotting the log starting target copy number of c-myc, c-jun, gadd153, and β-actin mRNA and threshold cycles (Ct, PCR cycle number at which the fluorescence signal reached above baseline). β-actin mRNA measurement was performed in all samples for standardisation of the data.
Figure 6.
Representative standard curves for quantitative reverse transcription-polymerase chain reaction. (A) For β-actin, the threshold cycles (Ct)—that is, the polymerase chain reaction cycle number at which the fluorescence signal reached above baseline—was −3.36 log (starting copy number) + 38.07, r2=0.996. (B) For c-myc, Ct=−3.38 log (starting copy number) + 38.63, r2=0.997. (C) For c-jun, Ct=−3.45 log (starting copy number) + 39.44, r2=0.997. (D) For gadd153, Ct=−3.42 log (starting copy number) + 39.69, r2=0.999.
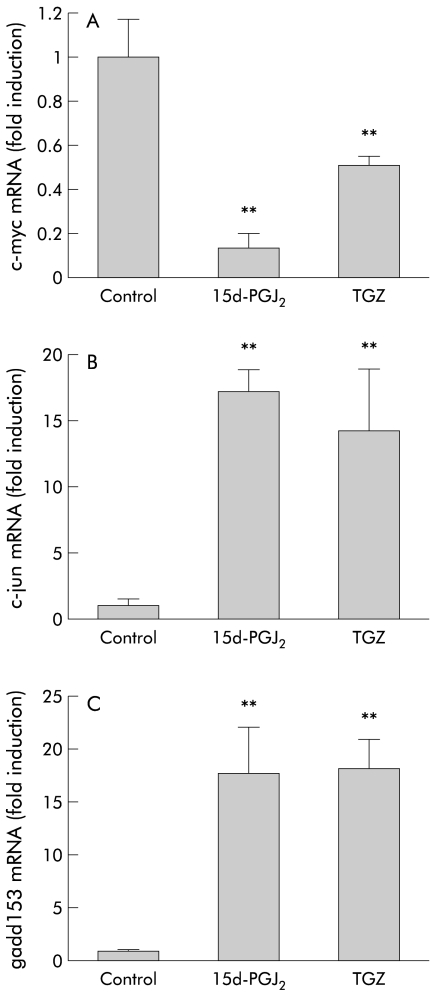
Figure 7A ▶ shows the effects of the PPARγ ligands on expression of c-myc mRNA in HT-29 cells. As seen in fig 7 ▶, eight hours of treatment with 15d-PGJ2 (10 μM) caused approximately 90% suppression of c-myc mRNA expression and eight hours of treatment with TGZ (30 μM) suppressed it by 50%. In contrast, eight hours of treatment with 15d-PGJ2 or TGZ caused an approximate 17-fold and 14-fold increase in expression of c-jun mRNA, respectively (fig 7B ▶). PPARγ ligands also significantly upregulated expression of gadd153 mRNA, as shown in fig 7C ▶, in which eight hours of treatment with 15d-PGJ2 or TGZ caused an approximate 17-fold increase.
Figure 7.
Quantitative reverse transcription-polymerase chain reaction analysis showing the effects of eight hours of treatment with 15-deoxy-δ12,14- prostaglandin J2 (15d-PGJ2 10 μM) or troglitazone (TGZ 30 μM) (n=6). Experiments were performed in the absence of fetal bovine serum. Data were standardised by the expression level of β-actin. (A) Expression of c-myc. (B) Expression of c-jun. (C) Expression of gadd153. **p<0.01 versus control.
Interaction between cell survival signalling and PPARγ signalling
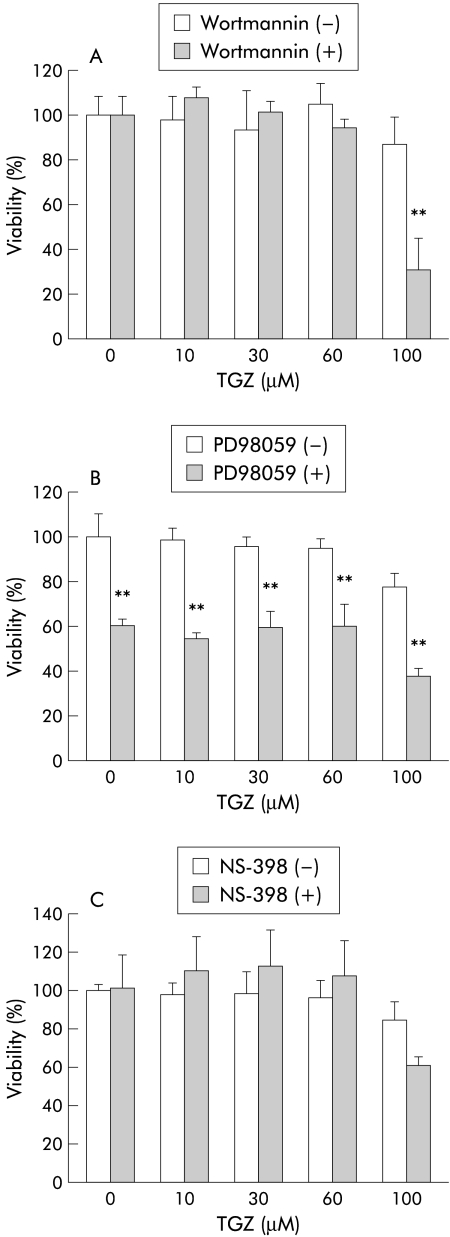
As already shown in fig 3 ▶, 15d-PGJ2 and TGZ induced HT-29 cell death was partially antagonised in the presence of FBS. This may be due to the presence of cell protective substances contained in FBS, such as peptide growth factors. Growth factor linked cell survival signalling has been reported to be mediated by the phosphatidylinositol 3-kinase (PI3-kinase)/Akt pathway.12 Cyclooxygenase 2 (COX-2) is also assumed to be important in antiapoptotic signalling in colon cancer cells.13 Thus we examined the effects of inhibitors of PI3-kinase (wortmannin) and COX-2 (NS-398) on TGZ induced HT-29 cell death in the presence of 10% FBS. We also tested the effect of a mitogen activated protein (MAP) kinase inhibitor (PD98059) because interaction between the MAP kinase pathway and PPARγ has been suggested.14 Figure 8A ▶ shows the effects of wortmannin (10 μM). Although wortmannin alone did not affect HT-29 cell viability, TGZ at 100 μM caused significant cell death in the presence of wortmannin compared with that in the absence of wortmannin. Figure 8B ▶ shows the effect of PD98059 (20 μM) on TGZ induced HT-29 cell death. PD98059 alone was found to decrease cell viability and thus interpretation of the results shown in fig 8B ▶ is rather difficult. The effect of NS-398 (20 μM) is shown in fig 8C ▶. NS-398 alone did not have a significant effect on cell viability. TGZ at 100 μM appeared to cause more HT-29 cell death in the presence of NS-398 although the effect was not significant.
Figure 8.
Interaction between cell survival signalling and peroxisome proliferator activated receptor γ (PPARγ) signalling. Experiments were performed in the presence of 10% fetal bovine serum (FBS). (A) Effects of wortmannin (10 μM), an inhibitor of phosphatidylinositol 3-kinase (PI3-kinase), in combination with troglitazone (TGZ) on HT-29 cell viability in the presence of 10% FBS. Cells were treated with different concentrations of TGZ (1–100 μM) in the presence and absence of wortmannin for 24 hours (n=5–6). **p<0.01 wortmannin (−) versus wortmannin (+). (B) Effects of PD98059 (20 μM), an inhibitor of mitogen activated protein kinase, in combination with TGZ on HT-29 cell viability in the presence of 10% FBS. Cells were treated with different concentrations of TGZ (1–100 μM) in the presence and absence of PD98059 for 24 hours (n=6–8). **p<0.01 PD98059 (−) versus PD98059 (+). (C) Effects of NS-398 (20 μM), an inhibitor of cyclooxygenase 2, in combination with TGZ on HT-29 cell viability in the presence of 10% FBS. Cells were treated with different concentrations of TGZ (1–100 μM) in the presence and absence of NS-398 for 24 hours (n=6–8).
DISCUSSION
The major findings of the present study were: (1) PPARγ ligands suppressed the growth response of colon cancer cells whereas ligands for PPARα and PPARδ did not. (2) PPARγ ligand induced apoptosis in HT-29 cells was associated with downregulation of c-myc expression and upregulation of c-jun and gadd153 expression. (3) PPARγ ligand induced apoptosis in HT-29 cells was antagonised by signalling mediated through PI3-kinase.
Induction of apoptosis by PPARγ ligands has been reported in several cell types, including macrophages,15 endothelial cells,16 breast cancer cells,17 and choriocarcinoma cells.18 In the present study, we showed that PPARγ ligands induced apoptosis in HT-29 cells and that cell death induced by PPARγ ligands was antagonised when serum was present in the culture medium. Bishop-Bailey16 observed the protective effects of serum against PPARγ ligand induced apoptosis in endothelial cells. This may be explained by the presence of polypeptide growth factors and other cell protective substances contained in serum. Consistent with this, we found that wortmannin, an inhibitor of PI3-kinase, antagonised the protective effect of serum, suggesting a possible interaction between PPARγ signalling and PI3-kinase mediated cell survival signalling.12 Further studies are required to confirm this finding. Although the molecular mechanisms of PPARγ induced apoptosis are not clear, Chinetti and colleagues15 showed that, in macrophages, PPARγ inhibits the transcriptional activity of the nuclear factor κB p65/RelA subunit, and speculated that PPARγ activators induce macrophage apoptosis by negatively interfering with the antiapoptotic nuclear factor κB signalling pathway. Elstner and colleagues17 showed that apoptosis induced by the combination of TGZ and AT-RA was associated with downregulation of bcl-2 in MCF-7 breast cancer cells. We screened for the effects of the PPARγ ligands on expression of apoptosis related genes using cDNA arrays. We did not find visible changes in mRNA levels of bcl family genes after incubation with 15d-PGJ2 or TGZ (data not shown). We selected several genes whose expression appeared to be modulated by PPARγ ligands prior to induction of apoptosis, and confirmed downregulation of c-myc and upregulation of c-jun and gadd153 by quantitative RT-PCR analysis.
A series of genetic alterations are associated with the development of colon cancers.19 Most colorectal tumours are initiated by inactivating mutations in the adenomatous polyposis coli (APC) tumour suppressor gene, which is located on 5q21.19 APC is a cytoplasmic protein and the tumour suppressive effect of APC is believed to be mediated by its ability to bind to β-catenin.20 Mutations of β-catenin have also been identified in colon cancer cells which lack APC mutations.21 β-catenin binds to the T cell factor/lymphoid enhancer factor (Tcf/Lef) transcription factors and activates expression of genes containing Tcf/Lef binding sites in their regulatory regions.22,23 Wild-type APC binds to β-catenin and promotes the degradation of β-catenin which results in inhibition of β-catenin-Tcf/Lef regulated gene transcription.24 As mutated APC (or mutated β-catenin) is deficient in this ability, increased β-catenin-Tcf/Lef mediated transcription is closely associated with colonic tumorigenesis. Since Kolligs and colleagues25 recently showed that γ-catenin is also regulated by APC and that γ-catenin activates the transcription of Tcf/Lef target genes, γ-catenin may also play an important role in colonic tumorigenesis.
Although the target genes of β-catenin-Tcf/Lef are not fully understood, He and colleagues26 identified the c-myc gene as a target of this signalling. It is also reported that c-myc expression is greatly enhanced by γ-catenin-Tcf/Lef. 25 c-Myc protein is an oncogenic transcription factor and expression of c-myc mRNA and c-Myc protein are frequently altered in various cancers.27 Although c-myc is reported to be overexpressed in most colon cancers,28 rearrangement and amplification of this gene are rare in colon cancers.29 Thus deregulation of the APC/β-catenin pathway may be responsible for overexpression of c-myc in colon cancers. Consistent with this, He and colleagues26 showed that introduction of wild-type APC to HT-29 cells results in downregulation of c-myc expression. The present study demonstrated that PPARγ ligands, 15d-PGJ2 and TGZ, both significantly downregulated expression of c-myc prior to induction of apoptosis in HT-29 cells. Thus it is very likely that in colon cancer cells, activation of PPARγ signalling can compensate for deregulation of c-myc expression induced by altered APC/β-catenin and/or APC/γ-catenin signalling.
Mann and colleagues30 have reported that c-jun and fra-1 are also targets of β-catenin-Tcf/Lef. c-jun and fra-1 proto oncogenes are components of AP-1 transcription factor and regulate expression of many genes. As the present results showed that PPARγ ligands upregulate expression of c-jun, PPARγ signalling does not appear to counteract the altered APC/β-catenin signalling in the case of c-jun expression. However, in certain cell types,31,32 c-jun has also been reported to be associated with cell cycle arrest and apoptosis. Further studies may be required to evaluate the significance of c-jun in the growth regulation of colon cancer cells. We also identified in the present study that PPARγ ligands upregulate expression of gadd153 prior to induction of apoptosis. Expression of gadd153 is induced by a variety of DNA damaging and growth arrest inducing agents and is also associated with apoptosis in various cell types.33,34 He and colleagues35 recently showed that PPARδ is a target of APC/β-catenin signalling, suggesting the involvement of PPARδ in colonic tumorigenesis. However, in the present study, cPGI, a ligand for PPARδ, did not affect the growth response of HT-29 cells. Thus further studies are required to understand the significance of PPARδ signalling and its interaction with PAPRγ signalling.
In conclusion, the present results, together with reports by other investigators,7–9 suggest a potential usefulness of PPARγ ligands for chemoprevention and treatment of colon cancers. Further basic as well as clinical studies are required to develop new strategies to fight colon cancers using PPARγ ligands.
Acknowledgments
This work was supported in part by a grant in aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture in Japan.
Abbreviations
PPAR, peroxisome proliferator activated receptor
RT-PCR, reverse transcription-polymerase chain reaction
TGZ, troglitazone
APC, adenomatous polyposis coli
RXR, retinoid X receptor
cPGI, carbaprostacyclin
AT-RA, all-trans retinoic acid
9c-RA, 9-cis-retinoic acid
FBS, fetal bovine serum
CHX, cycloheximide
RAR, retinoid A receptor
COX-2, cyclooxygenase 2
PI3-kinase, phosphatidylinositol 3-kinase
Tcf/Lef, T cell factor/lymphoid enhancer factor
PBS, phosphate buffered saline
BrdU, bromodeoxyuridine
MAP, mitogen activated protein
REFERENCES
- 1.Vamecq J, Latruffe N. Medical significance of peroxisome proliferator-activated receptors. Lancet 1999;354:141–8. [DOI] [PubMed] [Google Scholar]
- 2.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994;79:1147–56. [DOI] [PubMed] [Google Scholar]
- 3.Forman BM, Tontonoz P, Chen J, et al. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell 1995;83:803–12. [DOI] [PubMed] [Google Scholar]
- 4.Kliewer SA, Lenhard JM, Willson TM, et al. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor gamma and promotes adipocyte differentiation. Cell 1995;83:813–19. [DOI] [PubMed] [Google Scholar]
- 5.Mansen A, Guardiola-Diaz H, Rafter J, et al. Expression of the peroxisome proliferator-activated receptor (PPAR) in the mouse colonic mucosa. Biochem Biophys Res Commun 1996;222:844–51. [DOI] [PubMed] [Google Scholar]
- 6.Fajas L, Auboeuf D, Raspe E, et al. The organization, promoter analysis, and expression of the human PPARgamma gene. J Biol Chem 1997;272:18779–89. [DOI] [PubMed] [Google Scholar]
- 7.Sarraf P, Mueller E, Jones D, et al. Differentiation and reversal of malignant changes in colon cancer through PPARgamma. Nat Med 1998;4:1046–52. [DOI] [PubMed] [Google Scholar]
- 8.Brockman JA, Gupta RA, Dubois RN. Activation of PPARgamma leads to inhibition of anchorage-independent growth of human colorectal cancer cells. Gastroenterology 1998;115:1049–55. [DOI] [PubMed] [Google Scholar]
- 9.Kitamura S, Miyazaki Y, Shinomura Y, et al. Peroxisome proliferator-activated receptor gamma induces growth arrest and differentiation markers of human colon cancer cells. Jpn J Cancer Res 1999;90:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saez E, Tontonoz P, Nelson MC, et al. Activators of the nuclear receptor PPARgamma enhance colon polyp formation. Nat Med 1998;4:1058–61. [DOI] [PubMed] [Google Scholar]
- 11.Lefebvre AM, Chen I, Desreumaux P, et al. Activation of the peroxisome proliferator-activated receptor gamma promotes the development of colon tumors in C57BL/6J-APCMin/+ mice. Nat Med 1998;4:1053–7. [DOI] [PubMed] [Google Scholar]
- 12.Yao R, Cooper GM. Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science 1995;267:2003–6. [DOI] [PubMed] [Google Scholar]
- 13.Tsujii M, DuBois RN. Alterations in cellular adhesion and apoptosis in epithelial cells overexpressing prostaglandin endoperoxide synthase 2. Cell 1995;83:493–501. [DOI] [PubMed] [Google Scholar]
- 14.Hu E, Kim JB, Sarraf P, et al. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARgamma. Science 1996;274:2100–3. [DOI] [PubMed] [Google Scholar]
- 15.Chinetti G, Griglio S, Antonucci M, et al. Activation of proliferator-activated receptors alpha and gamma induces apoptosis of human monocyte-derived macrophages. J Biol Chem 1998;273:25573–80. [DOI] [PubMed] [Google Scholar]
- 16.Bishop-Bailey DHT. Endothelial cell apoptosis induced by the peroxisome proliferator-activated receptor (PPAR) ligand 15-deoxy-delta12, 14-prostaglandin J2. J Biol Chem 1999;274:17042–8. [DOI] [PubMed] [Google Scholar]
- 17.Elstner E, Muller C, Koshizuka K, et al. Ligands for peroxisome proliferator-activated receptor gamma and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in BNX mice. Proc Natl Acad Sci USA 1998;95:8806–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keelan JA, Sato TA, Marvin KW, et al. 15-Deoxy-delta(12,14)-prostaglandin J(2), a ligand for peroxisome proliferator-activated receptor-gamma, induces apoptosis in JEG3 choriocarcinoma cells. Biochem Biophys Res Commun 1999;262:579–85. [DOI] [PubMed] [Google Scholar]
- 19.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell 1996;87:159–70. [DOI] [PubMed] [Google Scholar]
- 20.Rubinfeld B, Souza B, Albert I, et al. Association of the APC gene product with beta-catenin. Science 1993;262:1731–4. [DOI] [PubMed] [Google Scholar]
- 21.Morin PJ, Sparks AB, Korinek V, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 1997;275:1787–90. [DOI] [PubMed] [Google Scholar]
- 22.Behrens J, von Kries JP, Kuhl M, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 1996;382:638–42. [DOI] [PubMed] [Google Scholar]
- 23.Molenaar M, van de Wetering M, Oosterwegel M, et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 1996;86:391–9. [DOI] [PubMed] [Google Scholar]
- 24.Korinek V, Barker N, Morin PJ, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science 1997;275:1784–7. [DOI] [PubMed] [Google Scholar]
- 25.Kolligs FT, Kolligs B, Hajra KM, et al. Gamma-catenin is regulated by the APC tumor suppressor and its oncogenic activity is distinct from that of beta-catenin. Genes Dev 2000;14:1319–31. [PMC free article] [PubMed] [Google Scholar]
- 26.He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science 1998;281:1509–12. [DOI] [PubMed] [Google Scholar]
- 27.Dang CV, Resar LM, Emison E, et al. Function of the c-Myc oncogenic transcription factor. Exp Cell Res 1999;253:63–77. [DOI] [PubMed] [Google Scholar]
- 28.Erisman MD, Scott JK, Watt RA, et al. The c-myc protein is constitutively expressed at elevated levels in colorectal carcinoma cell lines. Oncogene 1988;2:367–78. [PubMed] [Google Scholar]
- 29.Erisman MD, Rothberg PG, Diehl RE, et al. Deregulation of c-myc gene expression in human colon carcinoma is not accompanied by amplification or rearrangement of the gene. Mol Cell Biol 1985;5:1969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mann B, Gelos M, Siedow A, et al. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci USA 1999;96:1603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kharbanda S, Datta R, Kufe D. Regulation of c-jun gene expression in HL-60 leukemia cells by 1-beta-D-arabinofuranosylcytosine. Potential involvement of a protein kinase C dependent mechanism. Biochemistry 1991;30:7947–52. [DOI] [PubMed] [Google Scholar]
- 32.Colotta F, Polentarutti N, Sironi M, et al. Expression and involvement of c-fos and c-jun protooncogenes in programmed cell death induced by growth factor deprivation in lymphoid cell lines. J Biol Chem 1992;267:18278–83. [PubMed] [Google Scholar]
- 33.Matsumoto M, Minami M, Takeda K, et al. Ectopic expression of CHOP (GADD153) induces apoptosis in M1 myeloblastic leukemia cells. FEBS Lett 1996;395:143–7. [DOI] [PubMed] [Google Scholar]
- 34.Eymin B, Dubrez L, Allouche M, et al. Increased gadd153 messenger RNA level is associated with apoptosis in human leukemic cells treated with etoposide. Cancer Res 1997;57:686–95. [PubMed] [Google Scholar]
- 35.He TC, Chan TA, Vogelstein B, et al. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell 1999;99:335–45. [DOI] [PMC free article] [PubMed] [Google Scholar]