Abstract
Background: Type A hepatitis is still a considerable problem in both underdeveloped and developed countries. Why some patients progress to fulminant type A hepatitis and others do not is unclear.
Aims: To determine if nucleotide differences in the genome of hepatitis A virus (HAV) are responsible for the range of clinical severities, we analysed the 5` non-translated region (5`NTR) of the HAV genome, which has an internal ribosomal entry site and is important for cap independent translation of the viral message.
Methods: Serum samples from 84 Japanese patients with sporadic type A hepatitis from five distant regions of Japan, comprising 12 patients with fulminant hepatitis (FH), 13 with severe acute hepatitis (AHs), and 59 with acute hepatitis (AH), were examined for HAV RNA. The fragment between nucleotides 75 and 638 of the 5`NTR was amplified by reverse transcription-polymerase chain reaction, and the nucleotide sequence was determined by direct sequencing.
Results: Comparison of sequences of the 5`NTR revealed relatively fewer nucleotide substitutions in FH and AHs patients compared with the considerable sequence variations found in strains of AH. This tendency was most prominent between nucleotides 200 and 500. Strains from FH and AHs cases had fewer nucleotide substitutions (p<0.001) in this region.
Conclusions: Nucleotide variations in the central portion of the 5`NTR of HAV may influence the severity of type A hepatitis.
Keywords: type A hepatitis, fulminant hepatitis, acute hepatitis, hepatitis A virus, 5` non-translated region
Type A hepatitis is still a major problem worldwide, not only in underdeveloped countries but also in industrialised nations. The annual incidence of hepatitis A virus (HAV) infection is 2–4 cases per 100 000 inhabitants in Nordic countries, 7–10 cases per 100 000 in Germany and the UK, and six cases per 100 000 in Italy.1 In the United States, type A hepatitis is responsible for 48% of acute viral hepatitis cases.2 Due to improvements in sanitation, type A hepatitis epidemics have not occurred recently in Japan. Although the majority of type A hepatitis cases are self limiting acute hepatitis (AH), some progress to the severe forms.3 In Japan, between 1989 and 1993, 13% of fulminant hepatitis (FH) cases were caused by HAV, and 42% of fulminant type A hepatitis patients died (Research Group of Fulminant Hepatitis, Ministry of Health and Welfare of Japan, 1994). The reason why fulminant type A hepatitis occurs in some patients and not in others remains unclear.
HAV is classified as a hepatovirus of picorna viridae. Virological studies have revealed that HAV is a positive strand RNA virus comprising approximately 7500 nucleotides and containing a 5` non-translated region (NTR), both structural and non-structural protein coding regions, and a 3`NTR.4 The HAV genotype has been determined by sequencing the viral protein 1/non-structural protein 2A (VP1/P2A) junction region.5 The 5`NTR of the virus is approximately 735 nucleotides long and has high sequence identity among the various strains reported.4,6,7 It has been shown that the 5`NTR has an internal ribosomal entry site (IRES)8,9 and is important for cap independent translation of the viral message.10–15 Therefore, nucleotide substitutions in the 5`NTR might result in significant functional changes in the virus.
Despite advances in the understanding of HAV, a correlation between HAV genomes and clinical status of type A hepatitis has not been established. We recently developed a technique for detection of HAV RNA with high frequency in the early convalescent phase of type A hepatitis.16 Polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) analysis of the central part of 5`NTR of HAV showed that the banding patterns were more similar in more severe forms of type A hepatitis than in uncomplicated self limiting AH.17 These preliminary findings may indicate that nucleotide diversity in HAV in acute type A hepatitis is greater than that in more severe forms of hepatitis although the nucleotide sequences were not analysed in that study.
Recently, we determined the full sequence of HAV from three cases of FH and three cases of AH and showed that nucleotide changes in the 5`NTR and amino acid changes in the 2B region of HAV may be more frequent between FH and AH.18 In the present study, we examined the clinicopathological features of type A hepatitis and possible correlations with nucleotide variations in the 5`NTR of the HAV genome.
METHODS
Patients
Ninety two serum samples were obtained from 92 patients with type A hepatitis recruited at six hospitals located in various regions of Japan. Serum samples were collected from patients with type A hepatitis diagnosed on the basis of a positive test for IgM anti-HAV antibody (IgM-HA) in conjunction with compatible symptoms and laboratory findings. Samples were obtained within three weeks after initial admission and were stored at −20°C. Informed consent was obtained from patients or appropriate family members. The 92 patients comprised 12 with FH, 14 with severe acute hepatitis (AHs), and 66 with AH.
Of the 92 patients, 84 (91%) were positive for HAV RNA by nested reverse transcription (RT)-PCR, and only patients positive for HAV RNA by RT-PCR in their sera (12 FH, 13 AHs, 59 AH) were included in this study. Of these, the HAV genomes of three cases with FH (cases A1, A5, A205) and three cases with AH (cases A9, A20, A77) were sequenced completely and reported previously.18 As part of the current study, we performed amplification and sequencing of these samples again, and the results were the same. Patients with a prothrombin time of less than 40% that of controls were defined as AHs,19 and those with hepatic encephalopathy as FH. Patients who presented with a significant increase in serum blood urea nitrogen and creatinine (more than three times the upper limit of the normal range) were judged to be undergoing acute renal failure.20 IgM antibodies against hepatitis B core antigen (IgM-HBc), hepatitis B surface antigen (HBsAg), and second generation anti-HCV antibody were examined in all cases. In patients with FH and AHs, HCV RNA, IgM anti-Epstein-Barr virus antibody (IgM-EBV), IgM anti-herpes simplex virus antibody (IgM-HSV), IgM anti-cytomegalovirus antibody (IgM-CMV), antismooth muscle antibody, liver-kidney microsomal antibody 1, and antimitochondrial antibody were also examined. Histories of recent exposure to drugs and chemical agents and a history of heavy alcohol consumption (>50 g/day for >5 years) were also determined.
Laboratory data at admission were analysed homogeneneously between the FH group, AHs group, and AH group.
Histological examination was performed during the convalescent phase or after death. We performed liver biopsies in patients with FH and AHs to investigate the causes of liver injury other than type A hepatitis and to obtain additional information for the management of those patients. We do not usually perform liver biopsy in patients with self limiting acute type A hepatitis unless patients wish it to be done. Informed consent was obtained from patients or appropriate family members.
In type A hepatitis, early symptoms including fever, general malaise, fatigue, nausea, vomiting, and right upper quadrant discomfort are frequently observed, and therefore we defined the onset of these symptoms as clinical onset.
Serological markers
IgM-HA, IgM-HBc antibody, and HBsAg were measured with commercial radioimmunoassay kits (Abbott Laboratories, Chicago, Illinois, USA); second generation HCV antibody was measured with an enzyme immunoassay kit (Ortho Diagnostics, Tokyo, Japan); and HCV RNA was examined by nested RT-PCR, as described previously.21 IgM-EBV, IgM-CMV, and IgM-HSV were examined by enzyme linked immunosorbent assays. Antinuclear antibody, antismooth muscle antibody, antimitochondrial antibody, and antiliver-kidney microsomal 1 antibody were examined by a fluorescent antibody method.
Primers for PCR and direct sequencing
For amplification of HAV sequence and bidirection direct sequencing of the amplified segment, we prepared several primers for PCR and sequencing. These primers were prepared at the positions of 5`NTR based on the sequence reported by Cohen and colleagues.4
Detection of hepatitis A virus RNA in serum
RNA was extracted from sera using the acid guanidinium-phenol-chloroform method. Briefly, 50 μl of samples were mixed with 1000 μl guanidinium solution (Isogen, Nippon Gene Co., Tokyo, Japan) and 200 μl of chloroform, and precipitated with an equal volume of isopropanol. The RNA pellet was washed twice with 75% ethanol and dissolved in 11 μl of RNase free distilled water.
For reverse transcription, 11 μl of the RNA was heat denatured at 70°C for 10 minutes and then chilled rapidly on ice. RNA was mixed with 9 μl of reaction mixture on ice. The final 20 μl reaction mixture contained RNA from 50 μl of serum, 2.5 μM of antisense primer (5` AGTACCTCAGAGGCAAACAC 3`), 200 units of Superscript II reverse transcriptase (Superscript Preamplification System, Gibco BRL, Gaitherburg, Maryland, USA), 200 μM of each dNTP, 50 mM KCl, 10 mM Tris HCl (pH 8.3), 1.5 mM MgCl2, 0.001% gelatin, and 10 units of RNase inhibitor (Takara, Kyoto, Japan). The reverse transcription reaction was done by incubation at 42°C for 90 minutes, followed by heating of the mixture at 70°C for 15 minutes.
In the first round PCR reaction, 1 μl of the 20 μl of cDNA solution was used. The first round of PCR was performed in 50 μl of reaction mixture containing 25 pmol of outer antisense primer (5` AGTACCTCAGAGGCAAACAC 3`) and sense primer (5` TCTTGGAAGTCCATGGTGAG 3`), 200 μM of each dNTP, 50 mM KCl, 10 mM Tris HCl (pH 8.3), 1.5 mM MgCl2, 0.001% gelatin, and 2.5 units of Ex Taq polymerase (Takara) with proof reading activity. The final sense and antisense primer concentrations for first round PCR were 1.0 μM. Amplification conditions consisted of 35 cycles of 95°C for one minute, 50°C for one minute, and 72°C for one minute, and 1 μl of the first round product was used for the second round of PCR with the same PCR reaction mixture, except 1.0 μM each of inner sense primer (5` GGGACTTGATACCTCACCGC 3`) and antisense primer (5` CCACATAAGGCCCCAAAGAA 3`) were used. Amplification conditions for the second round were the same as those for the first round. The second round PCR products (10 μl) were analysed by 8% polyacrylamide gel electrophoresis, stained with ethidium bromide, and visualised by UV transillumination.
To avoid contamination of PCR products, all reagents were UV irradiated for 10 minutes at 200 mJ/cm2 in a commercially available UV cross linker (UV Stratalinker 1800, Funakoshi Co., Tokyo, Japan), as described in the textbook by Sambrook and Russel,22 and ART aerosol resistant pipette tips (Molecular Bio-Products, Inc., San Diego, California, USA) were used. Samples from healthy subjects were always used as negative controls. In all experiments, the negative samples showed negative results for HAV RNA.
Direct sequencing of HAV cDNA fragments
For sequencing nucleotides 75–638 of the 5`NTR of HAV, 20 μl of the nested PCR products were centrifuged in microconcentrators (Ultrafree C3KT, Japan Millipore Ltd, Tokyo, Japan). The concentration of the PCR products was determined by optical densitometry (Ultrospec III, Amersham Pharmacia Biotech, Buckinghamshire, UK), and a volume of product equivalent to 56 ng was used for single sided amplification with the inner sense or antisense primers using a Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer, Foster City, California, USA) according to the manufacturer`s instructions. The dye terminator products were purified over Centri-Sep spin columns (Applied Biosystems, Foster City, California, USA). Direct sequencing was performed with an ABI 377 DNA sequencer (Applied Biosystems).
Nucleotide sequence accession numbers
The nucleotide sequence data reported in this paper will appear in DDBJ/EMBL/GenBank nucleotide sequence databases with accession numbers: AB045327 for A1, AB045328 for A10, AB045329 for A165, AB045330 for A204, AB045331 for A205, AB045332 for A206, A045333 for A413, AB045334 for A414, AB045335 for A425, AB045336 for A5, AB045337 for A502, AB045338 for A601, AB045341 for A156, AB045342 for A159, AB045343 for A16, AB045344 for A160, AB045345 for A161, AB045346 for A196, AB045347 for A197, AB045348 for A200, AB045349 for A201, AB045350 for A302, AB045351 for A6, AB045352 for A702, AB045353 for A811, AB045563 for A158, AB045564 for A168, AB045565 for A180, AB045566 for A19, AB045567 for A195, AB045568 for A20, AB045569 for A207, AB045570 for A28, AB045571 for A301, AB045572 for A303, AB045646 for A304, AB045647 for A305, AB045648 for A306, AB045649 for A307, AB045650 for A32, AB045651 for A36, AB045652 for A38, AB045653 for A39, AB045654 for A40, AB045655 for A402, AB045656 for A404, AB045657 for A406, AB045658 for A407, AB045659 for A408, AB045660 for A409, AB045661 for A422, AB045663 for A423, AB045664 for A424, AB045665 for A503, AB045666 for A51, AB045667 for A55, AB045668 for A58, AB045669 for A62, AB045670 for A65, AB045671 for A68, AB045672 for A7, AB045673 for A707, AB045674 for A71, AB045675 for A711, AB045678 for A712, AB045679 for A713, AB045680 for A75, AB045681 for A77, AB045682 for A8, AB0445683 for A80, AB045684 for A802, AB045685 for A807, AB045686 for A809, AB045687 for A812, AB045688 for A814, AB045689 for A816, AB045690 for A819, AB045691 for A85, AB045692 for A9, AB045693 for A93, and AB045694 for A97.
Phylogenetic analysis
To examine the heterogeneity of the viral sequences obtained from 84 patients, a phylogenetic tree was constructed using the neighbour joining method.23 To confirm the reliability of the phylogenetic tree, bootstrap resampling tests were performed 1000 times.24 These analyses were conducted with the Genetyx-MAC program, version 10.1 (Software Development, Tokyo, Japan).
Statistical analysis
Differences in proportions among the groups were compared by Fisher`s exact probability test, Student's t test, and Welch's t test.
RESULTS
Clinicopathological characteristics of patients analysed for HAV 5`NTR
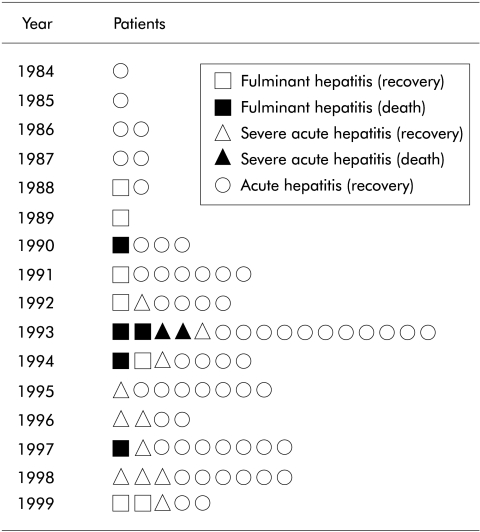
Serial changes in clinical forms of 84 patients with type A hepatitis analysed for HAV 5`NTR from 1984 to 1999 are shown in fig 1 ▶. Characteristics of these patients at admission are summarised in table 1 ▶. None of the cases was associated with an epidemic.
Figure 1.
Serial changes in the clinical forms of the 84 patients with type A hepatitis analysed for hepatitis A virus 5`non-translated region between 1984 and 1999.
Table 1.
Clininal and laboratory data of 84 patients at admission
| FH | AHs | AH | |
| n | 12 | 13 | 59 |
| Death | 5 | 2 | 0 |
| Sex (M/F)* | 6/6 | 11/2 | 36/23 |
| Age (y)* | 43.1 (14.4) | 41.6 (12.6) | 37.4 (10.7) |
| PT (%)† | 17.1 (9.2) | 32.6 (7.7) | 73.8 (26.7) |
| ALT (IU/I)‡ | 5502 (3353) | 5025 (2101) | 2515 (1932) |
| T-Bil (mg/dl)§ | 10.1 (6.5) | 6.0 (7.0) | 5.5 (3.2) |
| IgM-HA (cut off index)* | 4.9 (2.3) | 4.6 (1.2) | 5.4 (1.6) |
Values are mean (SD) or number.
*No statistically significant between the three groups.
†Significant difference (p<0.001) between FH and AHs by Student's t test, between FH and AH by Welch's t test, and between AHs and AH by Welch's t test.
‡No significant difference between FH and AHs, statistically significant difference (p=0.001) between FH and AH by Welch's t test, and statistically significant difference (p<0.001) between AHs and AH by Student's t test.
§No significant difference between FH and AHs or between AHs and AH, statistically significant difference (p=0.03) between FH and AH by Welch's t test.
PT, prothrombin time; ALT, alanine aminotransferase; T-Bil, total bilirubin; IgM-HA, IgM anti-hepatitis A virus antibody; FH, fulminant hepatitis; AH, acute hepatitis; AHs, severe acute hepatitis.
Differences in the male to female ratio and mean age between severe (FH and AHs) and non-severe (AH) cases were not statistically significant. Serum was sampled 3–37 days after clinical onset. Mean alanine aminotransferase level was higher in FH and AHs than in AH patients (p<0.001). Due to the inclusion criteria for allocation to group FH, AHs, or AH, mean prothrombin time was prolonged in groups FH and AHs compared with group AH. Mean total bilirubin level and mean IgM-HA titre were not significantly different between severe and non-severe cases (table 1 ▶). FH and AHs cases were distributed throughout five distant regions of Japan.
Five of 12 (42%) patients with FH died of hepatic failure, and two of 13 (15%) patients with AHs died of sepsis. All 12 cases of FH required artificial liver support (plasma exchange and haemodiafiltration). Eight (10%) patients comprising three (25%) with FH, one (8%) with AHs, and four (7%) with AH had acute renal failure and were treated with haemodiafiltration.
Two patients with AH were positive for HBsAg and anti-HBc antibody, and one patient with AH was positive for antinuclear antibody but showed a typical type A hepatitis course. HCV RNA was negative in all cases of FH and AHs. One patient with FH was positive for HCV antibody on admission, and he had a history of tattooing 18 years earlier, although data for previous liver function tests were not available and he may have been a HCV carrier.
IgM-EBV, IgM-HSV, IgM-CMV, antismooth muscle antibody, liver-kidney microsomal antibody 1, and antimitochondrial antibody were negative in all cases of FH and AHs. Two patients with FH and AHs had histories of heavy alcohol consumption. Significant histories of recent exposure to drugs or chemical agents thought to contribute to the clinical course were not observed.
Histological examination was performed in all 12 FH cases, 10/13 AHs cases, and 10/59 AH cases. This was performed 47 (50) (mean (SD)) days after admission in the FH group, 46 (11) days in the AHs group, and 22 (11) days in the AH group.
In the FH cases, liver histology revealed massive necrosis in three patients, submassive necrosis in three, and residual phase of acute hepatitis in six. One patient with a history of heavy alcohol consumption who recovered also showed pericellular fibrosis, which is consistent with alcoholic liver disease.
In the 10 AHs cases, liver histology revealed submassive necrosis in two patients who died of sepsis and residual phase of acute hepatitis in eight. One of the eight patients who recovered had a history of heavy alcohol consumption and showed pericellular fibrosis.
In all 10 AH cases, liver histology showed subsiding acute hepatitis.
Sequence analysis of HAV strains from FH, AHs, and AH
The sequence between nucleotides 75 and 638 of the 5`NTR was determined in 84 patients. The nucleotide sequence was first compared with the sequence of HM-175, a representative HAV sequence reported by Cohen and colleagues.4 A large number of nucleotide variations were clustered between nucleotides 75 and 199 and between nucleotides 501 and 638 in all cases of FH, AHs, and AH, and there were no obvious sequence differences between severe (FH and AHs) and non-severe (AH) cases in these regions. In contrast, fewer nucleotide variations were found between nucleotides 200 and 500 of the 5`NTR in cases of FH and AHs than in cases of AH.
We then performed phylogenetic analysis of the region between nucleotides 200 and 500 and classified the virus strains into three groups (fig 2 ▶). Bootstrap analysis to evaluate the statistical reliability of the tree revealed 1000/1000 (100%) reliability. The majority of strains, including all strains from FH and AHs cases, belonged to one group. One sequence in this group (A10) had the highest homology with wild-type HM-175, and this sequence was tentatively referred to as the HAV prototype (HAVPT).
Figure 2.
Phylogenetic tree of the sequence between nucleotides 200 and 500 of the 5`non-translated region of the hepatitis A virus genome, as determined by the neighbour joining method. To confirm the statistical reliability of the tree, bootstrap analysis was performed 1000 times. Numbers beside the phylogenetic roots are the results of bootstrap analyses. Horizontal lines indicate the nucleotide sequence distance between the sequences.
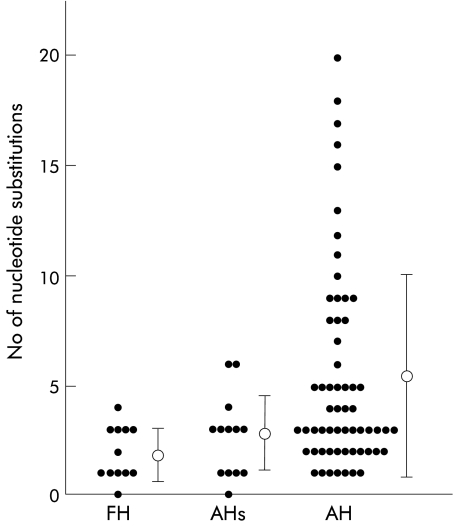
Sequences between nucleotides 200 and 500 were then compared with the HAVPT sequence. The nucleotide sequence homologies for the 84 sequences were between 93.4% and 100% in this region. The distribution of nucleotide variations is shown in fig 3 ▶. Sequences from cases of FH and AHs were most similar to that of HAVPT. In contrast, considerable sequence variations were found in cases of AH. The number of nucleotide substitutions is shown in fig 4 ▶. The average number of nucleotide substitutions between nucleotides 200 and 500 was 1.8 (1.2) (mean (SD)) per case in FH, 2.8 (1.7) in AHs, and 5.4 (4.6) in AH. Differences between FH and AH, AHs and AH, and severe cases (FH and AHs) and non-severe cases (AH) were statistically significant (p<0.001, p=0.001, and p<0.001, respectively). Differences between FH and AHs were not statistically significant (p=0.13) (fig 4 ▶).
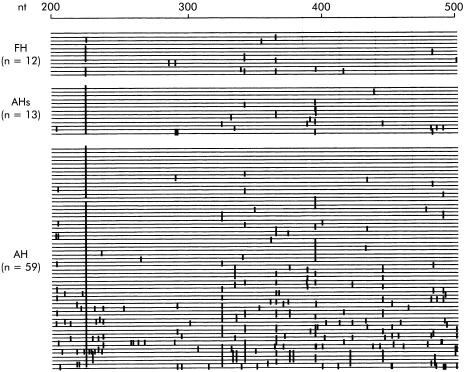
Figure 3.
Nucleotide sequence of the central portion of the 5`non-translated region of the hepatitis A virus between nucleotides 200 and 500. Horizontal lines indicate identical nucleotides between the individual strains and hepatitis A virus prototype (HAVPT). Vertical lines indicate positions where nucleotide changes occurred in the individual strains compared with HAVPT. FH cases were A10, A1, A413, A425, A502, A601, A204, A5, A206, A414, A165, and A205, respectively. AHs cases were A16, A159, A197, A811, A156, A196, A6, A200, A201, A702, A161, A302, and A160, respectively. AH cases were A141, A180, A404, A406, A713, A809, A819, A19, A39, A62, A168, A301, A304, A402, A711, A802, A814, A8, A32, A36, A38, A51, A55, A68, A303, A306, A407, A422, A424, A807, A816, A97, A157, A195, A207, A503, A9, A162, A707, A305, A307, A409, A75, A812, A7, A40, A65, A71, A80, A712, A93, A28, A20, A77, A85, A158, A423, A408, and A58, respectively.
Figure 4.
Number of nucleotide substitutions between nucleotides 200 and 500. Bars represent mean (SD). FH, fulminant hepatitis; AH, acute hepatitis; AHs, severe acute hepatitis.
Substitution sites in the proposed secondary structure of 5`NTR of HAV
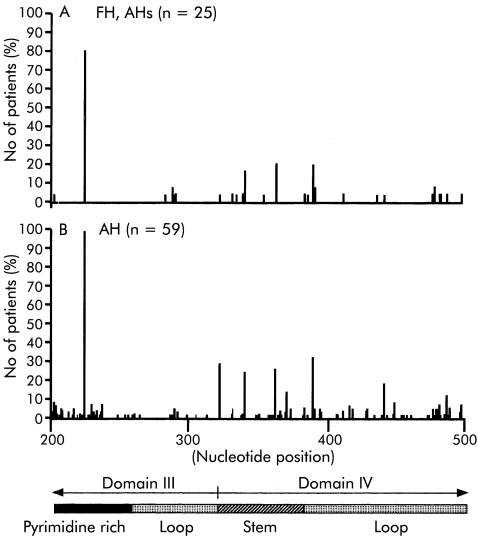
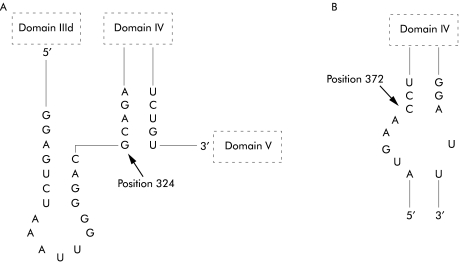
The 5`NTR of HAV contains a 5` terminal hairpin, two pseudoknots, a pyrimidine rich tract (pY1), and an internal ribosomal entry site (IRES).8 There are six domains in the IRES, which is located between nucleotides 151 and 734. Portions of domains III and IV are present between nucleotides 200 and 500 (fig 5 ▶). Domain III is located between nucleotides 99 and 323, and domain IV is located between nucleotides 324 and 586. The region between nucleotides 203 and 250 is particularly pyrimidine rich.
Figure 5.
Distribution of nucleotide substitutions between nucleotides 200 and 500 of the 5`non-translated region. Bars indicate the percentage of cases that have substitutions at each nucleotide position. FH, fulminant hepatitis; AH, acute hepatitis; AHs, severe acute hepatitis.
Substitutions that occurred in more than 10% of cases were found at nucleotides 225 (80%), 342 (16%), 364 (20%), and 391 (20%) in FH and AHs and at nucleotides 225 (98%), 324 (29%), 342 (24%), 364 (25%), 372 (14%), 391 (32%), 443 (19%), and 489 (12%) in AH (fig 5 ▶). Comparison of the differences between severe (FH and AHs) and non-severe (AH) cases revealed that nucleotide changes occurred more frequently at nucleotides 324 and 372 in AH than in FH and AHs (fig 5 ▶).
Portions of the secondary structure of the 5`NTR of HAV proposed by Brown and colleagues8 are shown in fig 6 ▶. A change of G to A at nucleotide 324 was found in 29% of AH cases in contrast with 4% of FH and AHs cases. This nucleotide is positioned at the beginning of the stem structure of domain IV, and the change is predicted to stabilise the structure. A change of C to T or G at nucleotide 372 was found in 14% of AH cases in contrast with 0% of FH and AHs cases. This position is in the middle of the stem structure of domain IV, and the change from C to T is predicted to stabilise the structure.
Figure 6.
Portions of the proposed secondary structure of the 5`non-translated region of hepatitis A virus.5 Arrows indicate positions of nucleotide substitutions. (A) Nucleotide 324 and (B) nucleotide 372 where substitutions are found more frequently in acute hepatitis than in fulminant hepatitis and severe acute hepatitis.
DISCUSSION
Although the severity of acute type A hepatitis varies, it is not clear why it is more severe in some patients than in others. It is thought that disease severity may be dependent on the characteristics of individual patients. Muraoka25 reported that aging and underlying chronic liver disease could be factors that increase the severity of type A hepatitis. Willner and colleagues26 reported that type A hepatitis caused serious illness and death during an urban epidemic in the USA and that complications were more frequent in patients 40 years of age and older, although young healthy subjects were also at risk for severe complications. Vento and colleagues27 reported that patients with chronic hepatitis C had a substantial risk of FH and death associated with HAV superinfection.
In the past several years, increased numbers of patients with sporadic type A hepatitis, especially the more severe forms, have visited our hospital, and our analysis of factors that could contribute to the severity of the disease revealed no significant difference in patient characteristics, including age,28 suggesting that viral factors might determine the severity of the disease. Therefore, to identify possible differences in non-translates with respect to different types of hepatitis, we analysed the HAV genome in sera from type A hepatitis patients with various clinicopathological features.
The 5`NTR of HAV is thought to be conserved between the reported strains7,9,10 and contains an IRES and pyrimidine rich tract.5,11 The HAV IRES is located downstream of nucleotide 151 and extends to within 40 nucleotides of the first initiator AUG, which is located at nucleotide 734. The pyrimidine rich tract is located between nucleotides 96 and 154.5 They form a highly ordered secondary structure and contain elements necessary for RNA translation and thus viral replication.5,11
In a previous study, we performed PCR-SSCP of the region between nucleotides 278 and 551 of the 5`NTR region of HAV from various forms of type A hepatitis and found that there might be differences between HAV sequences from patients with AH and those from patients with more severe forms of type A hepatitis, although we did not analyse the sequences themselves in that study.17 In the present study, we found that the central portions of the 5`NTR sequence of HAV obtained from patients with fulminant or severe type A hepatitis were relatively similar. In contrast, HAV sequences from patients with ordinary self limiting AH showed a wide variety of substitutions in this region. Thus the current findings support our hypothesis based on our previous study. These data suggest that viruses with fewer nucleotide substitutions in this region may be more virulent in comparison with strains with more nucleotide substitutions.
Several regions of the 5`NTR, including the pyrimidine rich tract and IRES, have been examined for possible correlations with replication of HAV RNA in vitro.10–12 It has been reported that HAV strains adapted to cell culture systems have mutations in the 5`NTR and P2 region.29,30 Schultz and colleagues15 reported recently that mutations within the 5`NTR of cell adapted HAV enhance cap independent translation directed by HAV IRES in a cell type specific fashion. Day and colleagues13 reported that mutations in the 5`NTR significantly enhance growth of the virus in a cell culture system. Thus nucleotide variations in the 5`NTR may influence replication of the virus and thereby affect virulence.
Although our study demonstrates a possible association between the severity of type A hepatitis and nucleotide variations in the 5`NTR of HAV, it is unclear if the nucleotide sequence of the 5`NTR of HAV is related directly to virulence of the virus. The role of viral factors will need to be confirmed by examination of IRES function in viruses from patients with FH and from those with self limiting AH. We are planning to examine the differences in IRES function in these viruses. We are also planning to examine the correlation between the nucleotide variations in the 5`NTR and levels of virus in such cases. Such studies might reveal the functional implications of the nucleotide variations found in this study.
However, some patients with FH had virus with significant numbers of substitutions and some patients with AH had virus with only a small number of mutation(s) in this region; therefore, the number of substitutions does not appear to be the sole factor that decides the outcome of HAV infection. Additionally, unlike epidemics of fulminant type B hepatitis, epidemics of fulminant type A hepatitis have not been reported. These findings suggest that viral factors may not play a direct role in HAV infection in contrast with hepatitis B viral infection, and that there may be additional factors, such as underlying liver disease, that influence the outcome of HAV infection.21
Our current study suggests that both viral and host factors should be considered and examined when discussing the mechanisms responsible for the severity of type A hepatitis. Although we analysed the 5`NTR of HAV in the current study, there are reports that the pathogenicity of HAV may be related to mutations within the 5`NTR, P2, and P3 segments.31–33 Our recent study of the entire sequence of HAV showed that amino acid changes in the 2B region (part of P2 segment) may differ between FH and AH.18 Therefore, to better understand the characteristic features of HAV responsible for the more severe forms of hepatitis, it will be interesting to examine other portions of HAV, including the 2B region, in specimens obtained from patients with severe and non-severe forms of type A hepatitis.
Abbreviations
HAV, hepatitis A virus
HAVPT, HAV prototype
FH, fulminant hepatitis
AH, acute hepatitis
AHs, severe acute hepatitis
NTR, non-translated region
IRES, internal ribosomal entry site
PCR-SSCP, polymerase chain reaction-single strand conformation polymorphism
RT-PCR, reverse transcription-polymerase chain reaction
anti-HBc, antibodies against hepatitis B core antigen
HBsAg, hepatitis B surface antigen
EBV, Epstein-Barr virus
HSV, herpes simplex virus
CMV, cytomegalovirus
REFERENCES
- 1.Nordenfelt E. Current epidemiological trends of viral hepatitis in northern Europe. In: Rizzetto M, Purcell RH, Gerin JL, et al. eds. Viral hepatitis and liver disease. Turin: Edizioni Minerva Medica, 1997:545–50.
- 2.Alter MJ, Gallagher M, Morris TT, et al. Acute non-A-E hepatitis in the United States and the role of hepatitis G virus infection. N Engl J Med 1997;336:741–6. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi Y, Okuda K. Fulminant and subfulminant hepatitis in Japan. Indian J Gastroenterol 1993;12:19–21. [PubMed] [Google Scholar]
- 4.Cohen JI, Ticehurst JR, Purcell RH, et al. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picorna viruses. J Virol 1987;61:50–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson BH, Jansen RW, Khanna B, et al. Genetic relatedness of hepatitis A virus strains recovered from different geographic regions. J Gen Virol 1992;73:1365–77. [DOI] [PubMed] [Google Scholar]
- 6.Robertson BH, Khanna B, Nainan OV, et al. Epidemiologic patterns of wild-type hepatitis A virus determined by genetic variation. J Infect Dis 1991;163:286–92. [DOI] [PubMed] [Google Scholar]
- 7.Jansen RW, Newbold JE, Lemon SM. Complete nucleotide sequence of a cell culture-adapted variant of hepatitis A virus: comparison with wild-type of a neutralization-resistant antigenic variant of hepatitis A virus. J Infect Dis 1990;161:7–13.1688601 [Google Scholar]
- 8.Brown EA, Day SP, Jansen RW, et al. The 5` nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J Virol 1991;65:5828–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown EA, Zajac AJ, Lemon SM. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5` nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J Virol 1994;68:1066–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whetter LE, Day SP, Elroy-Stein O, et al. Low efficiency of the 5` nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J Virol 1994;68:5253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carneiro JS, Equestre M, Pagnotti P, et al. 5` UTR of hepatitis A virus RNA: mutations in the 5`-most pyrimidine-rich tract reduce its ability to direct internal initiation of translation. J Gen Virol 1995;76:1189–96. [DOI] [PubMed] [Google Scholar]
- 12.Shaffer DR, Emerson SU, Murphy PC, et al. A hepatitis A virus deletion mutant which lacks the first pyrimidine-rich tract of the 5` nontranslated RNA remains virulent in primates after direct intrahepatic nucleic acid transfection. J Virol 1995;69:6600–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day SP, Murphy P, Brown EA, et al. Mutations within the 5` nontranslated region of hepatitis A virus RNA which enhance replication in BS-C cells. J Virol 1992;66:6533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaffer DR, Lemon SM. Temperature-sensitive hepatitis A virus mutants with deletions downstream of the first pyrimidine-rich tract of the 5` nontranslated RNA are impaired in RNA synthesis. J Virol 1995;69:6498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz DE, Honda M, Whetter LE, et al. Mutations within the 5` nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey kidney cells. J Virol 1996;70:1041–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiwara K, Yokosuka O, Ehata T, et al. Frequent detection of hepatitis A viral RNA in serum during the early convalescent phase of acute hepatitis A. Hepatology 1997;26:1634–9. [DOI] [PubMed] [Google Scholar]
- 17.Fujiwara K, Yokosuka O, Ehata T, et al. PCR-SSCP analysis of 5` nontranslated region of hepatitis A viral RNA: comparison with clinicopathological features of hepatitis A. Dig Dis Sci 2000;45:2422–7. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara K, Yokosuka O, Fukai K, et al. Analysis of full-length hepatitis A virus genome in sera from patients with fulminant and self-limited acute type A hepatitis. J Hepatol 2001;35:112–19. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi Y, Shimizu M. Aetiology and prognosis of fulminant viral hepatitis in Japan: a multicentre study. The Study Group of Fulminant Hepatitis. J Gastroenterol Hepatol 1991;6:159–64. [DOI] [PubMed] [Google Scholar]
- 20.Brenner BM, Lazarus JM. Acute renal failure, 3rd Edn. New York: Churchill Livingstone, 1993.
- 21.Okamoto H, Okada S, Sugiyama Y, et al. Detection of hepatitis C virus RNA by a two-step polymerase chain reaction with two pairs of primers deduced from the 5`-noncoding region. Jpn J Exp Med 1990;60:215–22. [PubMed] [Google Scholar]
- 22.Sambrook J, Russel DW. Molecular cloning. A laboratory manual, 3rd Edn. New York: Cold Spring Harbor Laboratory Press, 2001.
- 23.Gojobori T, Ishii K, Nei M. Estimation of average number of nucleotide substitutions when the rate of substitution varies. J Mol Evol 1982;28:414–23. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985;39:783–91. [DOI] [PubMed] [Google Scholar]
- 25.Muraoka H. Clinical and epidemiological study on factors of serious development of viral hepatitis type A. Nippon Shokakibyo Gakkai Zasshi 1990;87:1383–91. [PubMed] [Google Scholar]
- 26.Willner IR, Uhl MD, Howard SC, et al. Serious hepatitis A: An analysis of patients hospitalized during an urban epidemic in the United States. Ann Intern Med 1998;128:111–14. [DOI] [PubMed] [Google Scholar]
- 27.Vento S, Garofano T, Renzini C, et al. Fulminant hepatitis associated with hepatitis A virus superinfection in patients with chronic hepatitis C. N Engl J Med 1998;338:286–90. [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara K, Ehata T, Yokosuka O, et al. The recent increase of severe type A hepatitis in Chiba area. Int Hepatol Commun 1995;3:S37. [Google Scholar]
- 29.Cohen JI, Rosenblum B, Ticehurst JR, et al. Complete nucleotide sequences of an attenuated hepatitis A virus: comparison with wild-type virus. Proc Natl Acad Sci USA 1987;84:2497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graff J, Kasang C, Normann A, et al. Mutational events in consecutive passages of hepatitis A virus strain GBM during cell culture adaptation. Virology 1994;204:60–8. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Chao S-F, Ping LH, et al. An infectious cDNA clone of a cytopathic hepatitis A virus: genomic regions associated with rapid replication and cytopathic effect. Virology 1995;212:686–97. [DOI] [PubMed] [Google Scholar]
- 32.Raychaudhuri G, Govindarajan S, Shapiro M, et al. Utilization of chimeras between human (HM-175) and simian (AGM-27) strains of hepatitis A virus to study the molecular basis of virulence. J Virol 1998;72:7467–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brack K, Frings W, Dotzauer A, et al. A cytopathic, apoptosis-inducing variant of hepatitis A virus. J Virol 1998;72:3370–6. [DOI] [PMC free article] [PubMed] [Google Scholar]