Abstract
Background: The INK4a-ARF (CDKN2A) locus on chromosome 9p21 encodes two tumour suppressor proteins, p16INK4a and p14ARF, whose functions are inactivated in many human cancers.
Aims: To evaluate p14ARF and p16INK4a alterations in liver cell adenoma.
Methods: After microdissection, DNA from 25 liver cell adenomas and corresponding normal liver tissue were analysed for INK4-ARF inactivation by DNA sequence analysis, methylation specific polymerase chain reaction, restriction enzyme related-polymerase chain reaction (RE-PCR), mRNA expression, microsatellite analysis, and immunohistochemistry. In addition, microdeletion of p14ARF and p16INK4a were assessed by differential PCR.
Results: Methylation of p14ARF was found in 3/25 cases (12%) and alterations in p16INK4a occurred in 6/25 liver cell adenomas (24%) which correlated with loss of mRNA transcription. We failed to detect microdeletions or specific mutations of both exons. p16INK4a methylation appeared in the context of an unmethylated p14ARF promoter in six cases. In normal liver tissue, p14ARF or p16INK4a alterations were not observed.
Conclusions: Our data suggest that p14ARF methylation occurs independently of p16INK4a alterations in liver cell adenomas. Furthermore, methylation of p14ARF and p16INK4a may be a result of cell cycle deregulation and does not seem to be a prerequisite of malignancy.
Keywords: liver cell adenoma, p14ARF, p16INK4a, methylation, cell cycle deregulation
Liver cell adenoma (LCA) is the most important benign epithelial tumour of the liver, with an incidence of approximately 3/1 000 000 new cases per year.1 LCA are pathogenetically related to the use of oral contraceptives, androgenic steroid therapy, and have also been reported in association with glycogen storage disease. Microscopically, the neoplasm is composed of well differentiated uniform cords of proliferating hepatocytes. Normal portal tracts are absent, tumour cells are uniform in size and shape but atypical pleomorphic cells with distorted hyperchromatic nuclei may be seen.2,3 Transformation of LCA to hepatocellular carcinoma has been described but is extremely rare.4 To date, the cellular and molecular mechanisms leading to uncontrolled proliferation of hepatocytes remain unclear. Great insights will come from integrating the signals of different pathways operating at cell cycle regulation, cellular proliferation, and apoptosis.5 There is evidence that alterations in the INK4a-ARF locus, which maps to chromosome 9p21, may contribute to the development of liver tumours.6–9 The INK4a-ARF or CDKN2A locus codes for two different proteins, p16INK4a and p14ARF, both involved in cell cycle regulation.7 These two proteins are characterised by two distinct promoters and first exons spliced to a common exon 2 in different reading frames: exons 1α, 2, and 3 for p16INK4a and exons 1β, 2, and 3 for p14ARF.10
The tumour suppressor gene p16 INK4a is believed to encode a negative regulatory protein that controls the progression of eucaryotic cells through the G1 phase of the cell cycle by interacting with CDK4 and inhibiting its kinase activity.11 In the absence of functional p16 protein, CDK4 binds to cyclin D and phosphorylates pRb which stimulates entry into the S phase.6 The p16INK4a gene is inactivated by mutations, homozygous deletions, or gene methylation in many tumours of diverse origin.12 p14ARF, generated through an alternative splicing process that replaces the first exon, has been shown to function as a growth suppressor. p14ARF specifically activates the p53 pathway. p14ARF stabilises p53 by inhibiting MDM2 dependent p53 degradation, thereby inducing cell cycle arrest or apoptosis, depending on the stimulus. Data have shown that p14ARF binds to MDM2 through an NH2 terminal domain encoded by exon 1 whereas a functional domain is encoded by exon 2.13 Activation of p14ARF (in response to an oncogenic signal such as c-myc, activated ras) leads to localisation and sequestration of MDM2 in the nucleolar compartment, thereby stabilising p53 by preventing MDM2-p53 from undergoing ubiquitin mediated degradation.12–14 To date, data concerning INK4a-ARF alterations in benign tumours of the liver are lacking.
To gain insights into the role of the INK4a-ARF locus in the development of LCA, mutational and expression analyses of p16INK4a and p14ARF were performed in a large group of patients with this disease.
MATERIALS AND METHODS
Patients and tissue samples
Twenty five patients with LCA undergoing partial hepatectomy (segmental or lobar resection) between 1990 and 1999 were included in this retrospective study.
Each tumour was re-evaluated with regard to typing (WHO 2000).3 In all cases, slides prepared from four different paraffin blocks of tissue, sampled from different tumour areas, were examined.
DNA samples
For each LCA sample, the histopathological lesions of interest were first identified on routinely stained slides. Parallel sections were cut with the microtome set at 6 μm, and the slides dried overnight at 37°C. Corresponding areas of interest were delineated and microdissected after rapid staining with haematoxylin and eosin. Thereafter the tissue was scraped off the slide (sections were covered by 25 μl of Tris buffer 0.05 mol) with the tip of a sealed glass pipette and then sucked into a microcapillary tube. Tissue samples were placed in Eppendorf tubes and incubated with proteinase K at 37°C overnight. Proteinase K activity was inactivated by heating to 95°C for 10 minutes. For DNA extraction, standard methods were used: after incubation with proteinase K at 37°C overnight, the tissue was extracted twice in phenol and twice in chloroform, followed by ethanol precipitation.
Methylation status of the INK4a-ARF locus
The CpG WIZ p16 methylation assay kit was used (OncorInc, Gaithersburg, Maryland, USA) according to the manufacturer's instructions. After an initial bisulphide reaction to modify the DNA, polymerase chain reaction (PCR) amplification with specific primers was performed to distinguish methylated from unmethylated DNA. Primers specific for unmethylated p16 (5-TTATTAGAGGGTGGGGTGGATTGT-3, 5-CAACCCCAAACCA CAACCATAA-3) or methylated p16 (5-TTATTAGAGGGTG GGGCGGATCGC-3, 5-GACCCCGAA CCGCGACCG TAA-3) were used. DNA (7 μg/100 μl) was denatured by 0.2 M NaOH for 10 minutes at room temperature. DNA Modification Reagent I was added, incubated for 24 hours at 50°C, and subsequently purified by DNA Modification Reagents II and III in the presence of 50 μl of water. The bisulphide modification of DNA was completed with 0.3 M NaOH treatment for five minutes followed by ethanol precipitation. For hot start PCR, the PCR mixture contained Universal PCR Buffers (1×9, 4dNTPs (1.25 nM)), and U or M primers (300 ng each per reaction). Annealing temperature was 65°C for 30 cycles. The PCR product was directly electrophoresed on a 3% agarose gel, stained with ethidium bromide, and visualised under UV illumination. Bisulphide converted DNA from corresponding normal liver tissue from each patient served as a negative control, as indicated by the presence of the unmethylated but not the methylated band. To control the efficacy of bisulphide treatment, a primer set was used for unmodified or wild-type (“w”) (5-CAGAGGGTGGGGCGGACCGA-3 and 5-CGGGCCGCG GCCGTGG-3). In the event of insufficient bisulphide modification of DNA, the wild-type primers should have been amplified.
The methylation pattern in the CpC islands of the p14ARF were determined by primers designed for either methylated or unmethylated DNA.15 The primers spanned six CpG sites within the 5` regions of the gene. The 5` positions of the sense unmethylated and methylated primers correspond to 195 and 201 bp of GenBank sequence number LA1934. The primer sequences for the unmethylated reaction were 5`TTTT TGGTGTTAAAGGGTGGTGTAGT-3` (sense) and 5`-CACAAA AACCCTCACTCACAACAA-3` (antisense), yielding a PCR product of 132 bp. The primer sequences for the methylated reaction were 5`-GTGTTAAAGGGCGGCGTAGC-3` (sense) and 5`-AAAACCCTCACTCGCGACGA-3` (antisense), which amplify a 122 bp product.12
Placental DNA treated with methyltransferase was used as a positive control for methylated alleles. The PCR products (15 μl) were electrophoresed on a 8% polyacrylamide gel, stained, and directly visualised.
In addition to methylation specific PCR (MSP), a second approach was used to determine the methylation status of p14ARF and p16INK4a, the restriction enzyme related-PCR (RE-PCR), as described by Chaubert and colleagues.9 Genomic DNA was digested with four methyl sensitive (HpaII, NaeI, EaglI, and Ksp1) and one non-methyl sensitive (MspI) restriction enzyme. After chloroform/phenol extraction and precipitation, PCR amplification of a 316 bp fragment of p14ARF exon 1 containing one HpaII and one Ksp1 site were amplified by PCR. The following primers were used: forward: GCCTGCGGGGCGGAGAT; reverse: GCGGCTGCTGCCCTAGA. For p16INK4a, a 150 bp fragment of exon 1 containing two HpaII and one Ksp1 site were amplified by PCR. The primer sets were GGGAGCAGCATGGAGCCG (forward) and CTGGATCGGCCTCCGACCGTA (reverse).
Undigested placental and tumour DNA was used as a control. Cases were considered positive by RE-PCR when PCR amplification was obtained after digestion with one of the methyl sensitive restriction enzymes used (fig 1A, B ▶). Two CpG dinucleotides of exon 1 of p14ARF and three from p16 INK4a exon 1 were analysed. In case of a methylated p16INK4a gene, a real time quantitative MSP based on continuous optical monitoring of the progress of a fluorogenic PCR was performed. The bisulphite modified DNA was amplified using the following probes and primers: E5: 5`-CRTTATCTACTCTCCCCCTCTCC; E6: 5`-GGTTGGTTATTAGAGGGTGGGG; M probe: 5`-FAM-AACCGCCGAACGCACGC-TAMRA; and U probe: 5`-FAM-CAACCACCAAACACACACAATCCACC-TAM-RA. The sensitivity and specificity of real time PCR was determined using cloned fragments of bisulphite modified DNA as a fragment.
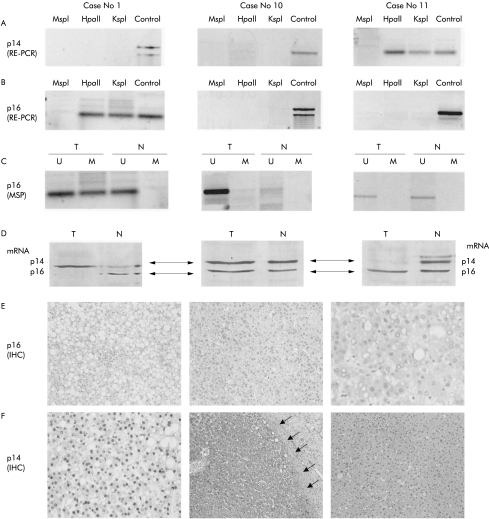
Figure 1.
Analysis of p14ARF and p16INK4a in three liver cell adenomas (case Nos 1, 10, and 11; same patients as in table 1 ▶). (A) p14ARF analysis with restriction enzyme related-polymerase chain reaction (RE-PCR). The methyl sensitive restriction enzymes used for RE-PCR are indicated (HpaII, KspI); digestion with the non-methyl sensitive enzyme MspI serves as a negative control and undigested DNA (control) serves as a positive control. The p14ARF gene is methylated in case No 11 and unmethylated in case Nos 1 and 10. (B) p16INK4a analysis with RE-PCR. Similar to (A), the methyl sensitive restriction enzymes used for RE-PCR are indicated (HpaII, KspI); digestion with the non-methyl sensitive enzyme MspI serves as a negative control and undigested DNA (control) serves as a positive control. Methylation of p16 INK4a is detected in case No 1, but not in case Nos 10 and 11. (C) p16 INK4a analysis using methylation specific polymerase chain reaction (MSP). Bisulphite treated DNA (which changes the unmethylated but not the methylated cytosines into uracil) is subjected to PCR amplification using primers designed to anneal specifically to the methylated bisulphite modified DNA. MSP results are expressed as unmethylated p16 specific bands (U) or methylated p16 specific bands (M). Bisulphite converted DNA from normal corresponding liver tissue (N) served as a negative control, as indicated by the presence of the U but not the M band. Similar to (B), methylation of p16 INK4a was detected in case No 1 but not in case Nos 10 and 11. (D) Results of multiplex reverse transcription-PCR (RT-PCR) of p14 mRNA (upper line corresponding to 200 bp) and p16 mRNA (lower line corresponding to 179 bp) for case Nos 1, 10, and 11. (E) Immunostaining of p16 INK4A protein in liver cell adenoma (LCA). Case No 1 shows methylated p16 INK4a and complete loss of p16 INK4a (LCA cells negative for p16 protein) (original magnification ×10). p16 INK4a is detectable in case Nos 10 and case 11 (dark reaction product within the cell nuclei) (original magnification ×20 and ×40). (F) Immunostaining of p14ARF protein in LCA. Case No 1 shows unmethylated p14 ARF and strong immunoreactivity of the tumour cells for p14 protein (dark reaction product within the tumour cell nuclei) (original magnification ×40). Case No 10 with unmethylated p14 ARF and strong immunoreactivity of the tumour cells for p14 protein (dark reaction product within the tumour cell nuclei). The tumour surrounding fibrous capsule (arrows) is negative (original magnification ×5). Case No 11 shows a methylated p14ARF and complete protein loss within the tumour tissue (original magnification ×20).
Multiplex RT-PCR
To compare relative levels of p16INK4a and p14ARF mRNA, multiplex reverse transcription-PCR (RT-PCR) was performed. Total RNA was extracted from 30 μg of microdissected LCA tissue by TRIzol reagents (Gibco BRL, Rockville, Maryland, USA). After ethanol washing and drying, RNA was suspended in 60 μl of diethyl pyrocarbonate treated water. After concentration determination, 2 μg of total RNA were subjected to a reverse transcription reaction using random oligonucleotide primers and superscript II reverse transcriptase (Gibco BRL) in a 20 μl reaction volume for 60 minutes at 42°C. The RT reaction product (1 μl) was then amplified by PCR using the forward primers of exons 1α and 1β and the reverse primer for exon 2 of the p16INK4a-p14ARF gene. The primers were as follows: forward exon 1α (sense 1): 5`-GCTGCCCACGCACCGAATA-3; exon 1β (sense 2): 5`CCCTCGTGCTGATGCTACTGA-3`; and reverse primer (antisense) 5`ACCACCAGCGTGTCCAGGAA-3`. Hot start PCR was performed for 35 cycles (95°C for 45 seconds, 57°C for 45 seconds, and 72°C for 60 seconds). The sizes of the products were 179 bp for p16INK4a and 200 bp for p14ARF, respectively. PCR products were electrophoresed on a 2% agarose gel and stained. β-actin amplification was performed to show RNA quality.
Allelic dosage analysis of loss of heterozygosity and homozygous deletion, and DNA sequencing for the INK4a-ARF (CDKN2A) locus
Allelic dosage analysis of the p14ARF and p16INK4a genes was performed using differential PCR. DNA fragments were amplified in exon 1β of p14ARF, exon 3 of p16INK4a, and exon 2 using the following primers: p14arf exon 1b: ARF2F 5`-CTCGTGCTGATGCTACTAGAG-3` and ARF2R 5`-AAGTCGTT GTAACCCGAATG-3`; p16 exon 3: p16ex3F: 5`-CGATTGAA AGAACCAGAGAG-3` and p16ex3R 5`-ATGGACATTTACGG TAGTGG-3`; interferon γ: INFGF2dF 5`-GCAGGTCATTCAGA TGTAGC-3` and INFG2RdR 5`-AGAGCACAAACAGAGGATGA-3`. As a negative control, a glioblastoma cell line (LNZ343) with a known deletion of the INK4a-ARF locus was used. For positive controls, the hepatocellular carcinoma cell line HepG2 with an intact INK4-ARF locus was analysed. The ratio of DNA fragment intensity in HepG2 between exon 1b or exon 3 and the internal control interferon γ was used to normalise the results. Hemizygous deletion was diagnosed if the ratio of the tumour sample was 50% of the one found in HepG2. If the ratio was less than 40%, the tumour sample was considered to harbour a homozygous deletion (fig 2 ▶). To control the data for gene dosage analysis, microsatellite analysis using nine microsatellites of chromosome 9p21 was performed, as described previously.16 The markers used were D9S161, D9S126, D9S171, D9S1752, D9S1748, D9S1747, D9S1749, D9S1751, and IFNA, and were obtained from Research Genetics (Hubtsville Alabama, USA) (fig 3 ▶).
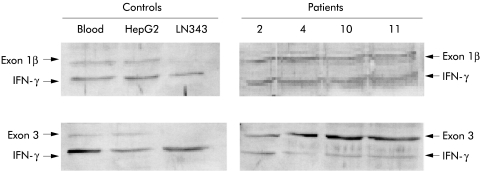
Figure 2.
Allelic dosage analysis of p14ARF and p16INK4a. Results of differential polymerase chain reaction technique, as described in the text. Negative control: LNZ343 with known deletion of the INK4a-ARF locus. Positive control: HepG2 with an intact INK4-ARF locus. The ratio of DNA fragment intensity in HepG2 between exon 1β or exon 3 and the internal control interferon γ (IFN-γ) was used to normalise the results. In patient Nos 2, 4, 10, and 11, the ratio between exon 1β or exon 3 and IFN-γ was 70–100% of the ratio found in HepG2, suggesting that there was no loss at the INK4a-ARF locus.
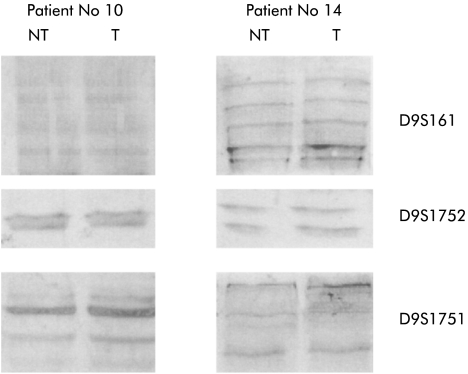
Figure 3.
Microsatellite analysis of tumour (T) and non-tumour (NT) tissue of patient Nos 10 and 14. Three different microsatellite markers were used (D9S161, D9S1751, and D9S1752).
Single strand conformational polymorphism (SSCP) analysis is a technique for the detection of mutations based on the three dimensional conformation taken by a single strand of DNA in a non-denaturing environment. Coding sequences and flanking intronic sequences of exons 1α, β, and 2 of the INK4a-ARF gene were analysed by PCR-SSCP. Primer sequences for exons 1α, β, and 2 have been described previously.16 Exon 1β was analysed through two overlapping PCR products generated with the primer pairs P14F1 (5` TCAGGGAAGGGCGGGTGCG 3`) and P14R1 (5` GCCGCGGGATGTGAACCA 3`), which generated a 245 bp product, and the primer pair P14F2 (5` GCCGCGAGTGAGGGTTTT 3`) and P14R2 (5` CACCGCGGTTATCTCCTC 3`), which generated a 257 bp product. The primers were labelled with 32P-ATP and each sample was subjected to PCR analysis (denaturing for 30 seconds, annealing for 45 seconds, extension for 30 seconds at 94°C, 55–60°C, and 72°C, respectively). The PCR products were electrophoresed, and the gels dried and autoradiographed. Variant SSCP bands were cut out from the gel and the DNA eluted. Variant bands and 3 μl of the eluted DNA were used as templates for unlabelled PCR. After purification of the PCR products, sequencing analysis was performed using the DNA Sequenase Kit (Amersham, Germany) and an automatic sequencing analyser (ABI 373; Applied Biosystems-Perkin-Elmer, Germany). All mutations found were confirmed by direct sequencing of the amplified tumour and corresponding non-tumorous DNA to identify germline mutations and polymorphisms.
Immunohistochemical analysis and assessment
Immunohistochemical analysis was performed as described previously.16 In all cases tumour and non-neoplastic liver tissue was examined.
The following antibodies were used: p16 (polyclonal; rabbit, dilution 1:500; Pharmingen, San Diego, California, USA), and p14 (polyclonal; rabbit, dilution 1:100; Zymed Laboratories, South San Francisco, California, USA).
Sections known to stain positively were included in each batch and negative controls were also performed by replacing the primary antibody with mouse or goat ascites fluid (Sigma-Aldrich Biochemicals, St Louis, Missouri, USA).
RESULTS
Analysis of INK4a-ARF deletions and mutations
Twenty five normal/tumour pairs were interpreted for allelic dosage analysis (table 1 ▶, fig 2 ▶). The allelic balance of the two genes was determined using the interferon γ gene as an internal control (fig 2 ▶). The two genes, p14ARF and p16INK4a, were expressed in all cases examined; deletions were not observed. No exclusive loss of either p16INK4a or p14ARF was found in our tumours. Loss of heterozygosity analysis revealed an identical status of the microsatellite markers used in paired samples of LCA and corresponding liver (fig 3 ▶).
Table 1.
Pathohistological data and INK4a-ARF alterations
| p14 alterations | p16 alterations | ||||||||
| Patient No | Sex (M/F) | Tumour size diameter (cm) | RE-PCR | MSP | EXP | RE-PCR | MSP | EXP | Allelic status |
| 1 | F | 2.5 | — | — | +++ | +++ | +++ | — | ND |
| 2 | F | 4.9 | — | — | +++ | — | — | +++ | ND |
| 3 | F | 10.5 | — | — | +++ | — | — | +++ | ND |
| 4 | M | 5.5 | — | — | +++ | — | — | +++ | ND |
| 5 | F | 6.5 | — | — | +++ | — | — | +++ | ND |
| 6 | F | 3.0 | — | — | +++ | — | — | +++ | ND |
| 7 | F | 12.5 | — | — | +++ | — | — | +++ | ND |
| 8 | F | 7.6 | — | — | +++ | — | — | +++ | ND |
| 9 | M | 8.0 | — | — | +++ | — | — | +++ | NI |
| 10 | F | 8.5 | — | — | +++ | — | — | +++ | ND |
| 11 | F | 10.8 | +++ | +++ | — | — | — | +++ | ND |
| 12 | F | 4.5 | — | — | +++ | — | — | +++ | ND |
| 13 | F | 14.0 | — | — | +++ | — | — | +++ | NI |
| 14 | F | 10.5 | — | — | +++ | — | — | +++ | ND |
| 15 | F | 9.5 | — | — | +++ | — | — | +++ | ND |
| 16 | F | 8.2 | +++ | +++ | — | — | — | +++ | ND |
| 17 | F | 7.5 | — | — | +++ | — | — | +++ | ND |
| 18 | F | 9.6 | — | — | +++ | +++ | +++ | — | ND |
| 19 | M | 8.3 | — | — | +++ | — | — | +++ | ND |
| 20 | F | 7.8 | — | — | +++ | — | — | +++ | ND |
| 21 | F | 3.9 | — | — | +++ | +++ | +++ | — | ND |
| 22 | F | 9.0 | — | — | +++ | +++ | +++ | — | ND |
| 23 | F | 6.5 | — | — | +++ | +++ | +++ | — | ND |
| 24 | F | 10.2 | +++ | +++ | — | — | — | +++ | ND |
| 25 | F | 7.8 | — | — | +++ | +++ | +++ | — | ND |
RE-PCR, restriction enzyme related-polymerase chain reaction; MSP, methylation specific PCR; EXP, gene mRNA expression analysed by reverse transcription PCR; ND, not detected (wild-type, both allels expressed as defined by multiplex PCR); NI, not informative.
Mutations of exons 1 and 2 were analysed by SSCP-PCR followed by direct sequencing of the cases with anomalous migrating bands. In nine cases, abnormal bands were visible. However, we failed to detect specific mutations within both exons. In one case, a polymorphism was identified in normal liver but not within LCA tissue (c442G >A; A148T).
Methylation status of the p14ARF and p16INK4A genes
Promoter methylation of p14ARF was present in 3/25 cases (12%). In all patients, corresponding non-neoplastic liver tissue was also analysed; no p14ARF promoter methylation was observed in any case. Analysis of the methylation status of the adjacent p16INK4a gene revealed that 6/25 LCA (24%) examined showed aberrant methylation at the 5`CpG island. Despite microdissection, amplification of unmethylated templates was also detected to some degree, probably because of contaminated normal intratumorous tissue (fibroblasts, endothelial cells, inflammatory cells). In normal LCA surrounding liver tissue, methylation of p14ARF or p16INK4a was not observed.
All six LCA with methylated p16INK4a exhibited an unmethylated p14ARF promoter. A coincidence of both p14ARF and p16INK4a methylation was not found. Thus the methylation status of p14ARF and p16INK4a promoters does not seem to be directly related.
Real time PCR of those samples with a methylated p16INK4a gene showed a level of methylation of approximately 75%.
All six cases with aberrant methylation of the p16INK4a or p14ARF gene showed complete loss of immunoreactvity (fig 1E, F ▶) within the tumour tissue. In the 19 cases shown to lack p16INK4a promoter methylation, nuclear staining of p16INK4a protein was observed in nearly all LCA cells with a moderate to strong intensity of immunoreactivity. In normal liver tissue, p16INK4a protein was detected in all cases (fig 1E, F ▶). Three LCA with a methylated p14ARF promoter lacked specific p14ARF immunostaining (fig 1E, F ▶).
Multiplex RT-PCR for p16INK4A and p14ARF mrna
Using specific sense primers for exon 1α and exon 1β, and a common reverse primer for exon 2, both transcripts were simultaneously amplified in a single reaction. p16INK4a mRNA was amplified in 19/25 cases and p14ARF transcripts were detected in 22/25 tumours (fig 1D ▶). Among the tumours with downregulated p16INK4a or p14ARF mRNA, methylation of the corresponding promoters was observed in six and three cases, respectively.
DISCUSSION
Recently, aberrant methylation of the p16INK4a promoter has been reported not only in various types of carcinomas but also in early preneoplastic lesions in the lung, stomach, oesophagus, and pancreas.17–21 Ours is the first study to examine alterations in the INK4a-ARF (also termed CDKN2A) locus on chromosome 9p21 in LCA, the most important benign epithelial tumour of the liver. We examined the status of p14ARF and p16INK4a simultaneously to answer the question of whether alterations in these genes may function as cooperative or alternative mechanisms in the pathogenesis of these tumours.
Our study showed that the p14ARF promoter was inactivated in 12% of cases. In 24% of all LCA examined, promoter methylation of the neighbouring gene, p16INK4a, was observed. We failed to detect simultaneous methylation of both genes and conclude that p14ARF methylation is independent of p16INK4a. Thus the p14ARF promoter demonstrates selective epigenetic silencing independent of that of p16INK4a. The strong correlation between promoter methylation and transcriptional inactivation, as examined by multiplex RT-PCR, indicates that aberrant methylation is a major mechanism of inactivation of the INK4a-ARF locus in LCA.
In concordance with data reported for cell lines, we failed to detect specific mutations of the p14ARF or p16INK4a gene.14 p14ARF can also be lost by (homozygous) deletion but this loss also targets p16INK4a in the vast majority of cases.19,22 Only a few examples currently exist of specific p14ARF deletions that spare the remainder p16INK4a coding region: a melanoma cell line and a glioma xenograft.23
In human cells, transcriptional silencing usually involves methylation of CpG rich sequences (CpG islands) in the promoters of affected genes. Such silencing is clonal and thought to be physiologically irreversible in somatic cells. Neoplastic cells often display aberrant methylation of multiple genes, including genes that regulate critical processes such as cell cycle control, DNA repair, and angiogenesis.12,18,24 The cause(s) of aberrant promoter methylation in neoplastic cells remains to be elucidated. It has been proposed that age related methylation identifies and contributes to an acquired predisposition to neoplasia (for example, colon cancer) because it parallels an age related increased cancer incidence and has the potential to alter the physiology of aging cells and tissues.25,26 This hypothesis predicts that higher levels of age related methylation may be present in conditions of rapid cell turnover that mimic premature aging. In LCA, an increase in cellular proliferation is often visible histologically. The proliferative activity of the neoplastic hepatocytes is significantly higher than in adenoma surrounding non-neoplastic liver tissue.2,3,27 Therefore, we hypothesise that methylation and consecutive silencing of the p16INK4a and p14ARF promoter may cause induction of increased cell turnover via affecting the G1/S phase transition of the cell cycle. In contrast with Rashid et al who found aberrant methylation of p16INK4a in approximately 73% of tubulovillous colon adenoma,28 a clear precancerous lesion, we detected aberrant methylation only in 24% of LCA. Together with the observation that altered methylation is also observed in liver cirrhosis,24 our data favour the hypothesis that methylation is a phenomenon of increased cellular proliferation and immortalisation rather than a conditio-sine-qua-non of malignant transformation.
Abbreviations
LCA, liver cell adenoma
MSP, methylation specific polymerase chain reaction
RE-PCR, restriction enzyme-related polymerase chain reaction
RT-PCR, reverse transcription-polymerase chain reaction
SSCP, single strand conformational polymorphism
REFERENCES
- 1.Wittekind Ch. Hepatocellular carcinoma and cholangiocarcinoma. In: Hermanek P, Gospodarowicz MK, Henson DE, et al, eds. Prognostic factors in cancer. Berlin: Springer, 1995:88–93.
- 2.Nagorney DM. Benign hepatic tumors: focal nodular hyperplasia and hepatocellular adenoma. World J Surg 1995;19:13–18. [DOI] [PubMed] [Google Scholar]
- 3.WHO. Pathology and genetics. Hamilton SR, Aaltonen LA, eds. Lyon: IARC Press, 2000:173–180.
- 4.Foster JH, Berman MM. The malignant transformation of liver cell adenomas. Arch Surg 1994;129:712–17. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler JM, Bodmer WF, Mortensen NJ. DNA mismatch repair genes and colorectal cancer. Gut 2000;47:148–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roussel MF. The INK4 family of cell cycle inhibitors in cancer. Oncogene 1999;18:5311–17. [DOI] [PubMed] [Google Scholar]
- 7.Liew CT, Li HM, Lo KW, et al. High frequency of p16INK4A gene alterations in hepatocellular carcinoma. Oncogene 1999;18:789–95. [DOI] [PubMed] [Google Scholar]
- 8.Matsuda Y, Ichida T, Matsuzawa J, et al. p16(INK4) is inactivated by extensive CpG methylation in human hepatocellular carcinoma. Gastroenterology 1999;116:394–400. [DOI] [PubMed] [Google Scholar]
- 9.Chaubert P, Gayer R, Zimmermann A, et al. Germ-line mutations of the p16INK4(MTS1) gene occur in a subset of patients with hepatocellular carcinoma. Hepatology 1997;25:1376–81. [DOI] [PubMed] [Google Scholar]
- 10.Bradley G, Irish J, MacMillan C, et al. Abnormalities of the ARF-p53 pathway in oral squamous cell carcinoma. Oncogene 2001;20:654–8. [DOI] [PubMed] [Google Scholar]
- 11.Serrano M, Lin AW, McCurrach ME, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997;88:593–602. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M, Tortola S, Toyota M, et al. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res 2000;60:129–33. [PubMed] [Google Scholar]
- 13.Karayan L, Riou JF, Seite P, et al. Human ARF protein interacts with topoisomerase I and stimulates its activity. Oncogene 2001;20:836–48. [DOI] [PubMed] [Google Scholar]
- 14.Fulci G, Labuhn M, Maier D, et al. p53 gene mutation and ink4a-arf deletion appear to be two mutually exclusive events in human glioblastoma. Oncogene 2000;19:3816–22. [DOI] [PubMed] [Google Scholar]
- 15.Lo DYM, Wong IHN, Zhang J, et al. Quantitative analysis of aberrant p16 methylation using real-time quantitative methylation-specific polymerase chain reaction. Cancer Res 1999;59:3899–903. [PubMed] [Google Scholar]
- 16.Tannapfel A, Benicke M, Katalinic A, et al. Frequency of p16INK4A alterations and k-ras mutations in intrahepatic cholangiocarcinoma of the liver. Gut 2000;47:721–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahrendt SA, Eisenberger CF, Yip L, et al. Chromosome 9p21 loss and p16 inactivation in primary sclerosing cholangitis-associated cholangiocarcinoma. J Surg Res 1999;84:88–93. [DOI] [PubMed] [Google Scholar]
- 18.Esteller M. Epigenetic lesions causing genetic lesions in human cancer. Promoter hypermethylation of DNA repair genes. Eur J Cancer 2000;36:2294–300. [DOI] [PubMed] [Google Scholar]
- 19.Soufir N, Daya-Grosjean L, de La Salmoniere P, et al. Association between INK4a-ARF and p53 mutations in skin carcinomas of xeroderma pigmentosum patients. J Natl Cancer Inst 2000;92:1841–7. [DOI] [PubMed] [Google Scholar]
- 20.Pinyol M, Hernandez L, Martinez A, et al. INK4a/ARF locus alterations in human non-Hodgkin's lymphomas mainly occur in tumors with wild-type p53 gene. Am J Pathol 2000;56:1987–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin A, Baran-Marzak F, El Mansouri S, et al. Expression of p16/INK4a in posttransplantation lymphoproliferative disorders. Am J Pathol 2000;156:1573–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labuhn M, Jones G, Speel EJ, et al. Quantitative real-time PCR does not show selective targeting of p14(ARF) but concomitant inactivation of both p16(INK4A) and p14(ARF) in 105 human primary gliomas. Oncogene 2001;20:1103–9. [DOI] [PubMed] [Google Scholar]
- 23.Jen J, Harper JW, Bigner SH, et al. Deletion of p16 and p15 genes in brain tumors. Cancer Res 1994;54:6353–8. [PubMed] [Google Scholar]
- 24.Kaneto H, Sasaki S, Yamamoto H, et al. Detection of hypermethylation of the p16(INK4A) gene promoter in chronic hepatitis and cirrhosis associated with hepatitis B or C virus. Gut 2001;48:372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang QS, Papanikolaou A, Nambiar PR, et al. Differential expression of p16(INK4a) in azoxymethane-induced mouse colon tumorigenesis. Mol Carcinog 2000;28:139–47. [DOI] [PubMed] [Google Scholar]
- 26.Awad MM, Sanders JA, Gruppuso PA. A potential role of p15(ink4b) and p57(kip2) in liver development. FEBS Lett 2000;483:160–4. [DOI] [PubMed] [Google Scholar]
- 27.WHO. Histological typing of tumours of the liver (International histological classification of tumours), 2nd edn. Ishak T, Anthony PP, Sobin LH, et al, eds. Berlin: Springer, 1994.
- 28.Rashid A, Shen L, Morris JS, et al. CpG island methylation in colorectal adenomas. Am J Pathol 2001;159:1129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]