Abstract
Background and aim: Complement receptor type 1 (CR1) is a transmembrane protein, and human erythrocyte CR1 (E-CR1) is involved in the transport of circulating immune complexes (IC) from the circulation to the reticuloendothelial system, including the liver and spleen. In chronic viral hepatitis, increased levels of IC containing viral particles and an association with various extrahepatic manifestations have been reported. However, regulatory mechanisms for IC levels are not fully understood.
Patients/subjects and methods: We analysed IC, E-CR1, and quantitative polymorphism of the CR1 gene in 149 patients with chronic viral liver diseases and in 64 normal blood donors using an enzyme linked immunosorbent assay, radioimmunoassay, and polymerase chain reaction-restriction fragment length polymorphism, respectively. We also analysed the effect of CR1 gene polymorphism on IC binding to E-CR1 using molecular methods.
Results: E-CR1 levels in patients with chronic hepatitis and chronic viral liver diseases as a whole correlated inversely with increased levels of IC. Moreover, significantly high levels of IC were observed in patients with chronic hepatitis C (CH-C) who were homozygous for the E-CR1 low density allele. We also found low levels of E-CR1 in liver cirrhosis and CH-C but not in CH-B. Low levels of E-CR1 in CH-C were observed, even after considering the polymorphism of the CR1 gene. Finally, we demonstrated CR1 gene polymorphism dependent binding of hepatitis virus containing IC.
Conclusions: Our results emphasise the important role of E-CR1 in clearance of IC from the circulation and the acquired, rather than inherited, decrease in E-CR1 in chronic viral liver diseases, especially of type C.
Keywords: complement receptor type 1, erythrocytes, immune complex, chronic hepatitis, liver cirrhosis
Complement receptor type 1 (CR1, CD35, C3b/C4b receptor) is a transmembrane glycoprotein expressed on several circulating cells, including erythrocytes, neutrophils, monocytes/macrophages, B lymphocytes, and some T lymphocytes, as well as specific epithelial cells.1 CR1 on primate erythrocytes can bind C3b and C4b. This interaction allows primate erythrocytes to bind complement opsonised particles and immune complexes (IC), a phenomenon referred to as immune adherence. The physiological role of the erythrocyte CR1 (E-CR1) is primarily as an inert shuttle for circulating IC, safely directing IC to the organs of the monocyte-phagocytic system and reticuloendothelial system, including the liver and spleen, thus preventing indiscriminate IC deposition in vulnerable tissues and potentially regulating IC driven complement activation that might be associated with tissue injury.2,3
The number of E-CR1 is in part hereditarily determined in healthy individuals and has been shown to be genetically regulated by two codominant autosomal alleles.4,5 A genetic polymorphism of E-CR1 quantitative expression correlates with a Hind III restriction fragment length polymorphism of the CR1 gene.5 However, E-CR1 levels are reduced in patients with IC mediated diseases, such as systemic lupus erythematosus (SLE), autoimmune haemolytic anaemia, and acquired immunodeficiency syndrome (AIDS).3 In patients with SLE, the reduced number of CR1 on erythrocytes has been reported to be largely associated with disease activity and thus is an acquired and not a hereditarily determined parameter,6,7 although a genetic component for low E-CR1 levels cannot be ruled out.4 Acquired loss of E-CR1 has been considered to result from transport of IC to the liver,8,9 although the precise mechanism remains to be elucidated.
In chronic viral liver diseases, especially type C chronic liver diseases, increased serum levels of IC containing viral particles have been reported.10–12 Increased levels of IC in type C chronic liver diseases are thought to be associated with various extrahepatic manifestations, including arthritis, dermatitis, membranoproliferative glomerulonephritis, and cryoglobulinaemia.13 However, detailed mechanisms of IC regulation in chronic viral liver diseases have not yet been thoroughly analysed.
In order to investigate the role of E-CR1 in the clearance of IC from the circulation of patients with chronic viral liver diseases, we analysed IC, E-CR1, and a quantitative polymorphism of the CR1 gene in these patients and normal subjects.
MATERIALS AND METHODS
Blood samples from normal subjects and patients
The mean number of CR1 per erythrocyte (CR1/E) and levels of IC were measured in erythrocytes and serum samples from 149 patients with liver diseases; including 36 with chronic hepatitis B (CH-B) (mean age 30.3 (SD 10.2) years; men/women 27/9), 78 with chronic hepatitis C (CH-C) (49.8 (13.2) years, 47/31), 13 with liver cirrhosis B (LC-B) (52.1 (15.2) years, 11/2), and 22 with liver cirrhosis C (LC-C) (55.9 (12.0) years, 15/7). Those from 64 normal blood donors (controls) (48.1 (14.6), 43/21) were also analysed. Diagnosis was established in all patients by liver biopsy using peritoneoscopy. Erythrocytes and serum samples were stored at 4°C and −80°C, respectively, until measurement and the former were used for assay within three days of isolation.
Preparation of erythrocytes
Erythrocytes were isolated from 1.5 ml of human blood by centrifugation at 1000 g. After packing, plasma and buffy coat were removed and stored at −80°C for the analysis described below. Erythrocytes were washed three times in eight volumes of cold (4°C) phosphate buffered saline (PBS), with the buffy coat removed each time. Leucocyte contamination following this procedure was less than 0.01%, as measured in a cell counter (Coulter Electronics, Hialeah, Florida, USA). Packed erythrocytes were measured in a cell counter and finally resuspended at a concentration of 1×109/ml.
Measurement of CR1/E
Mean numbers of CR1 antigenic sites on erythrocytes were measured using a direct RIA, as described previously by Wilson and colleagues4 with a minor modification using anti-CR1 monoclonal antibody 31R (isotype; IgG1 and kappa; Seikagaku-Kogyo Inc., Osaka, Japan),14 instead of using rabbit anti-CR1 polyclonal antibody. Radiolabelling of the monoclonal antibody was performed with 125I (Amersham Pharmacia Biotech KK, Tokyo) using Iodogen (Pierce, Rockford, Illinois, USA) to a specific activity of 1–2 μCi/μg. Assays were performed in duplicate in each patient and control subject, and data were expressed as the means of the assays. Each assay included duplicate twofold dilution series of the radiolabelled monoclonal antibody. Results were analysed by Scatchard plot, as described previously,15 and expressed as mean number of antigenic sites per erythrocyte (CR1/E). Day to day variability of CR1/E values was assessed by analysing three time points in five controls and the estimated coefficient of variation was 8%.
Measurement of IC
IC were measured in serum with an enzyme linked immunosorbent assay using mouse antihuman C3d monoclonal antibody (Quidel, San Diego, California, USA) and peroxidase conjugated antihuman immunoglobulins (Dako, Glostrup, Denmark), according to the method described previously.16 IC levels were determined according to the standard curve obtained from heat aggregated human immunoglobulin.17 The assays were performed in duplicate.
Genomic determination of CR1 quantitative polymorphism
Genomic DNA was extracted from the buffy coat stored at −80°C. CR1 quantitative polymorphism on erythrocytes was assayed using polymerase chain reaction (PCR) and Hind III restriction endonuclease digestion, as described previously.18
Analysis of IC binding to erythrocytes
Erythrocytes (5×104 in 100 μl of PBS) from healthy donors with CR1 gene polymorphism of HH, HL, and LL, prepared as described above, were incubated in vitro in triplicate at 37°C for 15 minutes with an equal volume of sera of CH-B or CH-C, which contain a high titre of IC. The incubated erythrocytes were washed extensively with PBS and analysed for viral genomes. DNA and RNA bound to erythrocytes were extracted by QIAamp DNA Blood Mini Kit or QIAamp Viral RNA Mini Kit (Qiagen, Tokyo, Japan), respectively, according to the instructions provided by the manufacturer. Hepatitis C virus (HCV) RNA was reverse transcribed to cDNA with M-MLV reverse transcriptase (Gibco BRL, Gaithersburg, Maryland, USA) and random hexamer. Hepatitis B virus (HBV) DNA and cDNA of HCV were amplified with primer sets, as described previously,19,20 and detected using ABI GeneAmp 5700 sequence detector (Applied Biosystems, Foster City, California, USA) using SYBR Green chemistry (Applied Biosystems) according to the instructions provided by the manufacturer. The assays were performed in triplicate.
Statistical methods
Data are expressed as median (range) values. Distributions of continuous variables were analysed by the Mann-Whitney U test or Kruskal-Wallis test, as indicated. The relationship between different variables was tested by linear regression analysis; p values <0.05 were considered statistically significant.
RESULTS
E-CR1 in controls and various liver diseases
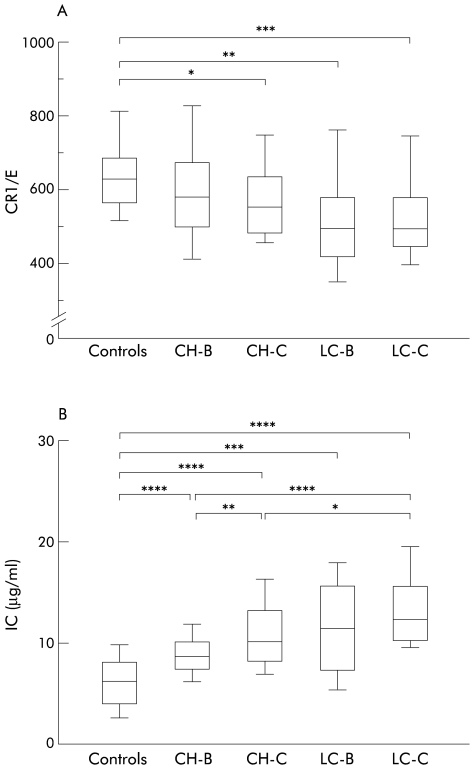
As the anti-CR1 monoclonal antibody 31R recognises a single epitope on the CR1 molecule,14 CR1/E estimated with this monoclonal antibody should be equal to the number of CR1 molecules on an erythrocyte. Namely, CR1/E in controls ranged from 349 to 1000, as reported previously.4,6 E-CR1 in chronic viral liver diseases and controls were compared (fig 1A ▶). CR1/E was significantly lower in CH-C (median 565 (25/75 percentiles 490–645); p=0.046), LC-B (507 (429–591); p=0.008), and LC-C (505 (455–592); p=0.007) than in controls (634 (570–691)). In contrast, there was no significant difference in CR1/E between CH-B (592 (509–682)) and controls (p=0.163).
Figure 1.
Comparison of mean number of complement receptors type 1 per erythrocyte (CR1/E) (A) and immune complex (IC) levels (B) in controls and in chronic hepatitis B (CH-B), chronic hepatitis C (CH-C), liver cirrhosis B (LC-B), and liver cirrhosis C (LC-C). Box plots of CR1/E and IC levels are shown. The upper and lower limits of the boxes and the middle line across the boxes indicate the 75th and 25th percentiles and median, respectively. The upper and lower horizontal bars indicate the 90th and 10th percentiles, respectively. Comparison was performed by Kruskal-Wallis test. (A) *p=0.046, **p=0.008, ***p=0.007. (B) *p=0.013, **p=0.005, ***p=0.003, ****p<0.001.
Comparison of IC levels in controls and in various liver diseases
IC levels were significantly higher in CH-B (median 8.7 μg/ml (25/75 percentiles 7.5–10.2)), CH-C (10.3 (8.4–13.4)), LC-B (11.8 (7.6–16.0)), and LC-C (12.4 (10.5–15.7)) than in controls (6.3 (4.0–8.1)) (p<0.001 in all pairwise comparisons) (fig 1B ▶). In CH-B, IC levels were significantly lower than in CH-C and LC-C (p=0.005 and p<0.001, respectively). The level of IC in LC-C was significantly higher than in CH-C (p=0.013).
Correlation between IC and E-CR1
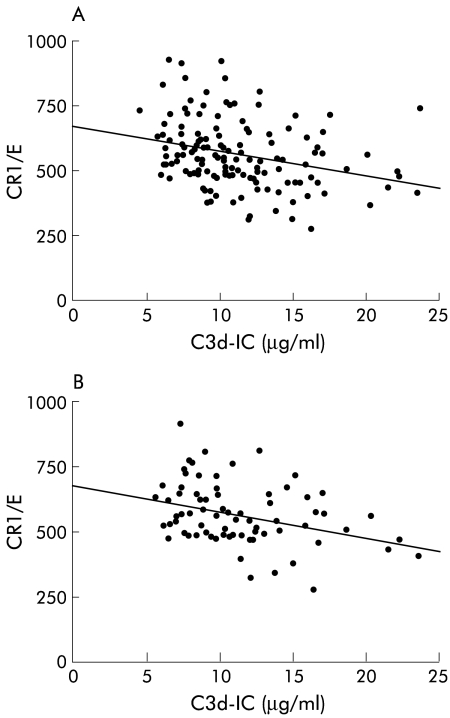
Linear regression analysis showed an inverse correlation between IC levels and CR1/E when data from the entire population sample were analysed (patients and controls), data from all patients with liver diseases, and data from those with chronic hepatitis only (CH-B and CH-C) (r=−0.327, p<0.001; r=−0.280, p<0.001; and r=−0.284, p<0.003, respectively) (fig 2A ▶ and data not shown). There was also an inverse correlation between the two variables in CH-C (r=−0.325, p=0.004) but not in CH-B, LC-B, LC-C, or controls (r=−0.141 p=0.429; r=0.503, p=0.097; r=0.159, p=0.521; and r=−0.101, p=0.548, respectively) (fig 2B ▶ and data not shown).
Figure 2.
Correlation between mean number of complement receptor type 1 per erythrocyte (CR1/E) and immune complexes (IC) in chronic liver diseases (A) and chronic hepatitis C (CH-C) (B). An inverse correlation was found in each group. The regression equation was y=−9.3x+669, r=−0.280 with p<0.001 for chronic liver diseases (A) and y=−9.4x+674, r=−0.325 with p=0.004 for CH-C (B).
Analysis of quantitative polymorphism of the CR1 gene
Three patterns of digested fragments were obtained by PCR-restriction fragment length polymorphism (RFLP) and were expressed as follows: HH, individuals homozygous for the CR1 high density allele; HL, individuals heterozygous for the CR1 gene polymorphism; and LL, individuals homozygous for the CR1 low density allele.18The overall allelic frequencies observed were 0.76 and 0.24 for the H and L alleles, respectively, which were also similar to those reported previously for Caucasians and Japanese (table 1 ▶).5,18,21 The gene frequencies of the H and L alleles and distribution of HH, HL, and LL genotypes were similar in controls and patients with liver diseases (table 1 ▶).
Table 1.
Frequency of complement receptor type 1 (CR1) gene polymorphisms in various liver diseases and controls
| CR1 gene polymorphism | Allelic frequency | |||||
| Diagnosis | n | HH | HL | LL | H | L |
| CH-B | 36 | 20 (60.6)* | 10 (30.3) | 3 (9.1) | 0.76 | 0.24 |
| CH-C | 78 | 37 (56.0) | 25 (37.9) | 4 (6.1) | 0.75 | 0.25 |
| LC-B | 13 | 7 (63.6) | 2 (18.2) | 2 (18.2) | 0.73 | 0.27 |
| LC-C | 22 | 12 (54.5) | 8 (36.4) | 2 (9.1) | 0.73 | 0.27 |
| Controls | 64 | 37 (57.8) | 25 (39.1) | 2 (3.1) | 0.77 | 0.23 |
| Total | 213 | 113 (57.7) | 70 (35.7) | 13 (6.6) | 0.76 | 0.24 |
*Numbers in parentheses represent frequency (%) of each CR1 gene polymorphism.
CH-B, chronic hepatitis B; CH-C, chronic hepatitis C; LC-B, liver cirrhosis B; LC-C, liver cirrhosis.
Genotypes: HH, individuals homozygous for the CR1 high density allele; HL, individuals heterozygous for the CR1 gene polymorphism; LL, individuals homozygous for the CR1 low density allele.
Comparison of erythrocyte CR1 expression levels among quantitative polymorphisms of the CR1 gene
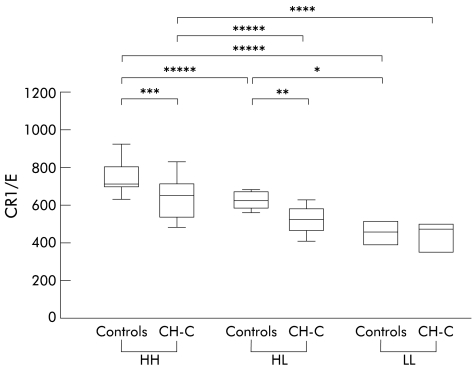
E-CR1 was highest in the genotype HH and lowest in the genotype LL in controls (fig 3 ▶), as reported previously.5 Similar results were observed in CH-C. Significant differences in E-CR1 levels between CH-C and controls were observed in genotypes HH and HL but not in genotype LL (p=0.013, 0.023, and 0.643, respectively) (fig 3 ▶).
Figure 3.
Comparison of mean number of complement receptors type 1 per erythrocyte (CR1/E) between CR1 gene polymorphisms in controls and chronic hepatitis C (CH-C). Box plots of CR1/E levels for each polymorphism are shown. The boxes and bars are as in fig 1 ▶. Genotypes are indicated at the bottom of the figure: HH, individuals homozygous for the CR1 high density allele; HL, individuals heterozygous for the CR1 gene polymorphism; LL, individuals homozygous for the CR1 low density allele. CR1 gene polymorphism and disease associated differences in CR1/E levels were noted. *p=0.025, **p=0.023, ***p=0.013, ****p=0.010, *****p<0.001.
Comparison of IC levels among quantitative polymorphisms of the CR1 gene
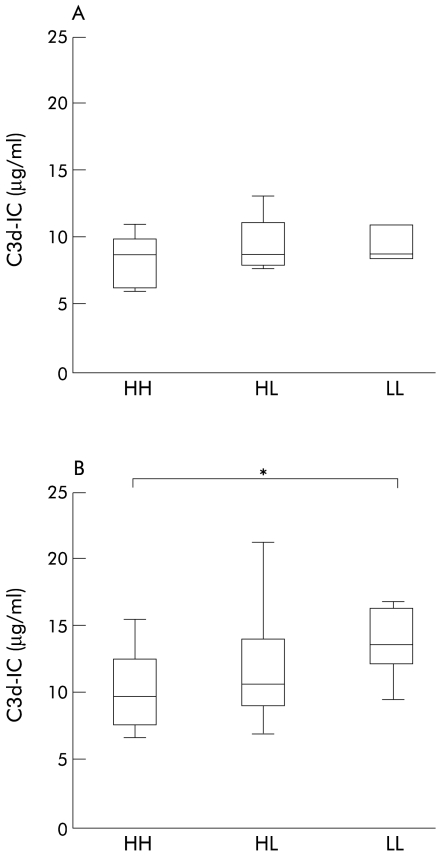
IC levels in CH-C were compared among CR1 quantitative polymorphisms (fig 4 ▶). Mean IC levels were highest to lowest in the order of LL, HL, and HH, respectively. IC levels for the genotype LL (13.9 (12.4–16.4) μg/ml) were significantly higher than those for the genotype HH (9.8 (7.6–12.5) μg/ml) (fig 4B ▶, p=0.011). There was no significant difference in IC levels between HL and HH (10.8 (9.3–14.1)) or LL (p=0.113 and p=0.411, respectively). There were also no significant differences in IC levels among CR1 genotypes in CH-B, LC-B, LC-C, and controls (fig 4A ▶ and data not shown).
Figure 4.
Comparison of immune complexes (IC) in complement receptor type 1 (CR1) gene polymorphisms in chronic hepatitis B (CH-B) (A) and chronic hepatitis C (CH-C) (B). Box plots of IC levels for each polymorphism are shown. Boxes and bars are as in fig 1 ▶. The genotypes are: HH, individuals homozygous for the CR1 high density allele; HL, individuals heterozygous for the CR1 gene polymorphism; LL, individuals homozygous for the CR1 low density allele. In CH-C, an additional three patients with genotype LL, whose data on CR1/E were not available, are included. (B) *p=0.011.
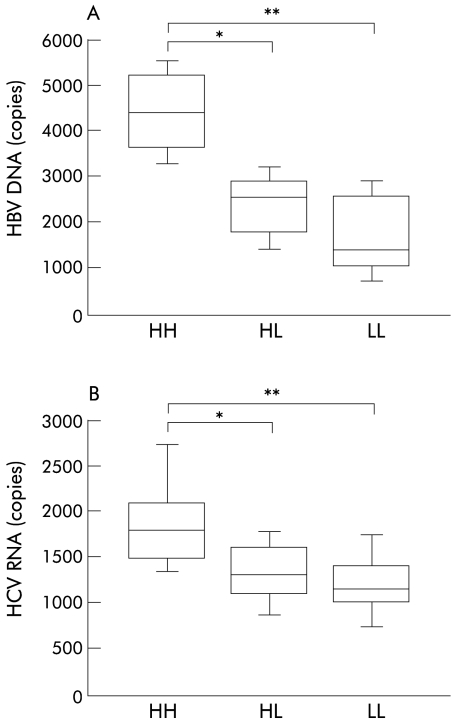
CR1 gene polymorphism dependent binding of IC to erythrocytes
Erythrocytes were incubated with IC and bound viral genomes were quantified and compared in terms of CR1 gene polymorphisms of the blood donors (fig 5 ▶). Significant differences in the amount of bound HBV DNA or HCV RNA were noted between HH and HL or LL (fig 5 ▶). These results indicate that the CR1 gene polymorphism is associated with the capacity of erythrocytes for binding to HBV or HCV containing IC.
Figure 5.
Complement receptor type 1 (CR1) gene polymorphism dependent binding of immune complexes (IC) to erythrocytes. Hepatitis B virus (HBV) DNA (A) and hepatitis C virus (HCV) RNA (B) bound to erythrocytes were quantified by real time detection polymerase chain reaction and box plots of the viral gene levels for each polymorphism are shown. Boxes and bars are as in fig 1 ▶. The genotypes are: HH, individuals homozygous for the CR1 high density allele; HL, individuals heterozygous for the CR1 gene polymorphism; LL, individuals homozygous for the CR1 low density allele. (A) *p=0.021, **p=0.007; (B) *p=0.008, **p=0.001.
DISCUSSION
In this study, we investigated levels of E-CR1 and IC, and the prevalence of CR1 gene polymorphisms, in patients with chronic viral liver diseases. We observed a correlation between CR1/E and IC in all samples analysed, in liver disease, and in chronic hepatitis. Moreover, in CH-C, we found this correlation and significantly higher levels of IC in the CR1 genotypes HL and LL, which exhibited lower E-CR1 expression levels than the CR1 genotype HH, indicating an association between CR1 gene polymorphism and levels of IC in CH-C. These observations highlight the important role of E-CR1 in clearance of IC from the circulation in patients with liver diseases, especially of type C, although we cannot rule out the contribution of Fc receptors on monocytes and other immune cells for clearance of IC.
Previous studies indicated that increased IC is associated with extrahepatic manifestations12,13 in type C chronic liver diseases. These include membranous glomerulonephritis, dermatitis, arthritis, and cryoglobulinaemia.13 These extrahepatic manifestations in type C chronic liver diseases may be, at least in part, associated with the level of E-CR1, which regulates IC levels by removing IC from the circulation.3 If this is the case, the occurrence of extrahepatic manifestations in type C chronic liver diseases might be associated with polymorphism of the CR1 gene, which is known to be genetically associated with quantitative expression of CR1 on erythrocytes.5 As these extrahepatic manifestations in type C chronic liver diseases, that are obvious in clinical practice, are only occasionally observed in Japan,22 the relationship between these clinical features in patients with type C liver diseases and E-CR1 or the CR1 gene polymorphism is the subject of ongoing work in our department. On the other hand, CR1 allelic frequencies were similar between CH-C and LC-C, although a limited number of LC-C patients were examined. This indicates that CR1 polymorphism does not significantly influence liver disease progression of HCV infection itself.
We also demonstrated low levels of E-CR1 in patients with CH-C, LC-B, and LC-C but not in those with CH-B. The low level of E-CR1 was even significant between controls and CH-C in patients with the CR1 genotypes HH and HL. Reduced E-CR1 expression in CH-C, estimated by flow cytometry using monoclonal antibody E11 that recognises multiple epitopes on CR1 and thus could overestimate the number of E-CR1,23 has been reported.24 However, no quantitative data for CR1/E or data on the relationship between CR1 gene polymorphism in liver disease are available. Therefore, our data clearly indicate, for the first time, an obvious decrease in E-CR1 in CH-C, even after considering polymorphism of the CR1 gene, indicating that this has been acquired after the development of liver disease, as has been suggested for SLE and AIDS.3,6,7 For patients with genotype LL, the difference between CH-C and controls was not significant. This was probably due in part to the limited number of patients with this specific genotype enrolled in this study and the variable severity of disease activity or liver damage in these groups of CH-C, which might be associated with increased levels of IC.10,25 Alternatively, the relatively lower density of E-CR1 in LL compared with the HH and HL genotypes could be associated with the less pronounced decrease in E-CR1 in CH-C patients with genotype LL compared with genotypes HH and HL, as it has been shown that the decrease in E-CR1 was relatively greater in the subpopulation of erythrocytes with a higher density of CR1, namely those with genotype HH.9
On the other hand however there was substantial overlap between CH-C and controls, even in each CR1 polymorphism. This seems to be in part due to a modest and variable increase in levels of IC in CH-C (fig 1 ▶) relative to SLE in which extensive IC formation and a marked decrease in E-CR1 are often observed.6–8,26
Low E-CR1 numbers have been reported in patients with various diseases, including SLE,6 autoimmune haemolytic anaemia, and AIDS, in which an immunological disorder, particularly increased levels of IC, is involved in the pathogenesis.3 Various mechanisms have been proposed for low E-CR1. For example: (i) it has been suggested that the number of E-CR1 and erythrocyte size decrease when E-CR1 transports IC from the circulation to the liver,2,8,9 although the detailed mechanism is still unknown3; and (ii) masking of the E-CR1 epitope by IC or by autoantibodies against CR1 have been reported.27 The latter mechanism is not the case in our study however as binding of the anti-CR1 monoclonal antibody 31R to E-CR1 was not affected by binding of IC to erythrocytes in our assay system (data not shown), as has been described in other diseases.8,26 This finding is supported by the evidence that the monoclonal antibody 31R recognises a single epitope on the CR1 molecule that bears at least two C3b and C4b binding sites independent of structural allotypes.3,14 Moreover, autoantibodies against CR1 have been reported in a very limited number of cases of autoimmune disease, for example SLE,27 and incubation of erythrocytes with serum from patients with CH-C did not affect measurement of CR1/E with the particular monoclonal antibody 31R used in our study (data not shown). Therefore, in patients with CH-C and LC-C, the decrease in E-CR1 did not appear to be due to occupancy of E-CR1 by IC or autoantibodies against CR1, but rather to actual loss of the E-CR1 epitope. Whether loss of the E-CR1 epitope is associated with actual loss of the CR1 molecule itself on erythrocytes, or with conformational changes in E-CR1 structure during interaction with and release of IC, remains to be determined.3
We also examined and demonstrated the effect of the CR1 gene polymorphism on binding of erythrocytes to IC in patients with HBV and HCV. The results indicate that binding of a virus containing IC to erythrocytes depends on the CR1 gene polymorphism, suggesting an association between E-CR1 and efficiency of IC clearance from the circulation in CH-B and CH-C. However, not all viral particles form IC and not all IC in CH-B and CH-C contain viral particles or viral genomes. Therefore, further investigations are required with sensitive and quantitative methods to clarify this phenomenon.
In conclusion, the results of this study emphasise the important role of E-CR1 in the regulation of circulating IC levels in patients with chronic viral liver diseases, in particular of type C. Low expression of E-CR1 in chronic viral liver diseases, especially of type C, appears to be an acquired phenomenon and may be associated with reduced clearance of IC from the circulation in these patients.
Acknowledgments
The authors thank Dr Douglas T Fearon for helpful discussions and critical review of this paper. This work was supported in part by grants from the Study Group for Intractable Hepatitis Research Committee, the Ministry of Health, Labour and Welfare, Japan (TT).
Abbreviations
AIDS, acquired immunodeficiency syndrome
CH-B, chronic hepatitis B
CH-C, chronic hepatitis C
controls, normal blood donors
CR1, complement receptor type 1
CR1/E, mean number of CR1 per erythrocyte
E-CR1, erythrocyte CR1
HBV, hepatitis B virus
HCV, hepatitis C virus
HH, individuals homozygous for the CR1 high density allele
HL, individuals heterozygous for the CR1 gene polymorphism
IC, immune complex
LC-B, liver cirrhosis B
LC-C, liver cirrhosis C
LL, individuals homozygous for the CR1 low density allele
PBS, phosphate buffered saline
PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism
SLE, systemic lupus erythematosus
REFERENCES
- 1.Fearon DT, Ahearn JM. Complement receptor type 1 (C3b/C4b receptor; CD35) and complement receptor type 2 (C3d/Epstein-Barr virus receptor; CD21). In: Lambris JD, ed. Current topics in microbiology and immunology. Berlin: Springer Verlag, 1989:83–98. [DOI] [PubMed]
- 2.Cornacoff JB, Hebert LA, Smead WL, et al. Primate erythrocyte-immune complex-clearing mechanism. J Clin Invest 1983;71:236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birmingham DJ. Erythrocyte complement receptors. Crit Rev Immunol 1995;15:133–54. [DOI] [PubMed] [Google Scholar]
- 4.Wilson JG, Wong WW, Schur PH, et al. Mode of inheritance of decreased C3b receptors on erythrocytes of patients with systemic lupus erythematosus. N Engl J Med 1982;307:981–6. [DOI] [PubMed] [Google Scholar]
- 5.Wilson JG, Murphy EE, Wong WW, et al. Identification of a restriction fragment length polymorphism by a CR1 cDNA that correlates with the number of CR1 on erythrocytes. J Exp Med 1986;164:50–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iida K, Mornaghi R, Nussenzweig V. Complement receptor (CR1) deficiency in erythrocytes from patients with systemic lupus erythematosus. J Exp Med 1982;155:1427–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holme E, Fyfe A, Zoma A, et al. Decreased C3b receptors (CR1) on erythrocytes from patients with systemic lupus erythematosus. Clin Exp Immunol 1986;63:41–8. [PMC free article] [PubMed] [Google Scholar]
- 8.Ross GD, Yount WJ, Walport MJ, et al. Disease-associated loss of erythrocyte complement receptors (CR1, C3b receptors) in patients with systemic lupus erythematosus and other diseases involving autoantibodies and/or complement activation. J Immunol 1985;135:2005–14. [PubMed] [Google Scholar]
- 9.Cosio FG, Shen XP, Birmingham DJ, et al. Evaluation of the mechanisms responsible for the reduction in erythrocyte complement receptors when immune complexes form in vivo in primates. J Immunol 1990;145:4198–206. [PubMed] [Google Scholar]
- 10.Araki K, Nagashima H, Tsuji T. Detection and characterization of circulating immune complexes during acute exacerbation of chronic viral hepatitis. Clin Exp Immunol 1982;47:520–6. [PMC free article] [PubMed] [Google Scholar]
- 11.Hijikata M, Shimizu YK, Kato H, et al. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J Virol 1993;67:1953–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sansonno D, Iacobelli AR, Cornacchiulo V, et al. Immunochemical and biomolecular studies of circulating immune complexes isolated from patients with acute and chronic hepatitis C virus infection. Eur J Clin Invest 1996;26:465–75. [DOI] [PubMed] [Google Scholar]
- 13.Pawlotsky JM, Ben YM, Andre C, et al. Immunological disorders in C virus chronic active hepatitis: a prospective case-control study. Hepatology 1994;19:841–8. [PubMed] [Google Scholar]
- 14.Hara T, Kojima A, Fukuda H, et al. Levels of complement regulatory proteins, CD35 (CR1), CD46 (MCP) and CD55 (DAF) in human haematological malignancies. Br J Haematol 1992;82:368–73. [DOI] [PubMed] [Google Scholar]
- 15.Iwasaki Y, Takabatake H, Monestier M, et al. Idiotypic diversity and variable region gene usage by mouse anti-HLA-DQ3 mAb. Immunogenetics 1995;42:90–100. [DOI] [PubMed] [Google Scholar]
- 16.Jordan SC, Gautier E, Sakai R, et al. Quantitation of circulating immune complexes in human serum by the Raji cell and F(ab')2 anti-C3 micro enzyme immunoassays. J Immunol Methods 1985;83:363–70. [DOI] [PubMed] [Google Scholar]
- 17.Hay FC, Nineham LJ, Roitt IM. Routine assay for the detection of immune complexes of known immunoglobulin class using solid phase C1q. Clin Exp Immunol 1976;24:396–400. [PMC free article] [PubMed] [Google Scholar]
- 18.Cornillet P, Philbert F, Kazatchkine MD, et al. Genomic determination of the CR1 (CD35) density polymorphism on erythrocytes using polymerase chain reaction amplification and HindIII restriction enzyme digestion. J Immunol Methods 1991;136:193–7. [DOI] [PubMed] [Google Scholar]
- 19.Abe A, Inoue K, Tanaka T, et al. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J Clin Microbiol 1999;37:2899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeuchi T, Katsume A, Tanaka T, et al. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 1999;116:636–42. [DOI] [PubMed] [Google Scholar]
- 21.Satoh H, Yokota E, Tokiyama K, et al. Distribution of the HindIII restriction fragment length polymorphism among patients with systemic lupus erythematosus with different concentrations of CR1. Ann Rheum Dis 1991;50:765–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka K, Aiyama T, Imai J, et al. Serum cryoglobulin and chronic hepatitis C virus disease among Japanese patients. Am J Gastroenterol 1995;90:1847–12. [PubMed] [Google Scholar]
- 23.Quadri RA, Schifferli JA. Over-estimation of the number of complement receptor type 1 (CR1) on erythrocytes. Scand J Immunol 1992;36:125–30. [DOI] [PubMed] [Google Scholar]
- 24.Kanto T, Hayashi N, Takehara T, et al. Low expression of erythrocyte complement receptor type 1 in chronic hepatitis C patients. J Med Virol 1996;50:126–34. [DOI] [PubMed] [Google Scholar]
- 25.Tsai JF, Margolis HS, Jeng JE, et al. Circulating immune complexes in chronic hepatitis related to hepatitis C and B viruses infection. Clin Immunol Immunopathol 1995;75:39–44. [DOI] [PubMed] [Google Scholar]
- 26.Thomsen BS, Nielsen H, Bendixen G. Erythrocyte CR1 determination using monoclonal antibody in a microtiter plate ELISA; receptors are not masked by immune complexes. Allergy 1986;41:479–86. [DOI] [PubMed] [Google Scholar]
- 27.Wilson JG, Jack RM, Wong WW, et al. Autoantibody to the C3b/C4b receptor and absence of this receptor from erythrocytes of a patient with systemic lupus erythematosus. J Clin Invest 1985;76:182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]