Abstract
Objective: A significant proportion of individuals with chronic hepatitis C virus (HCV) infection have persistently normal alanine aminotransferase (ALT) levels. Although data are controversial, such patients usually have weaker histological damage and a lower progression rate of fibrosis. The aims of this study were: (1) to compare demographic, virological, and histological parameters of HCV patients with normal ALT values with those of HCV patients with elevated ALT levels; and (2) to determine whether HLA class II alleles contribute to the persistence of normal ALT levels in HCV patients.
Patients and methods: Eighty three patients with chronic HCV infection and persistently normal ALT values (group 1) and 233 patients with chronic HCV infection and elevated ALT levels (group 2) were studied. Histological features were expressed using Knodell and Metavir scores. HLA DRB1* and DQB1* genotyping was performed using hybridisation with sequence specific oligonucleotides after genomic amplification. The κ2 and Fisher’s exact tests were used to compare discrete variables and phenotype frequencies between the two groups, and Wilcoxon’s test was used for continuous variables. A multivariate logistic regression model was used to determine which variables predicted normal ALT values.
Results: ALT levels were correlated with the severity of liver damage. In group 1, 93% of patients had an F0 or F1 Metavir index of fibrosis compared with 47% of patients in group 2 (p<0.001). A longer duration of infection (p<0.001) and increased DRB1*11 phenotype frequency (pc=0.03) were observed among patients with normal ALT. The two groups did not differ with regard to the mode of contamination or viral genotype. After logistic regression, young age (p=0.0008), female sex (p=0.01), long duration of infection (p=0.0001), and HLA DRB1*11 (p=0.050) were more strongly associated with persistence of normal ALT.
Conclusions: Our study confirms that patients with chronic hepatitis C and normal ALT levels have less severe liver disease than those with elevated ALT levels. This particular biochemical outcome may be explained, at least in part, by host immunogenetic factors such as the presence of HLA-DRB1*11.
Keywords: chronic hepatitis, hepatitis C virus, major histocompatibility complex class II antigens, host response, asymptomatic carrier
Elevated alanine aminotransferase (ALT) levels characterise chronic hepatitis C virus (HCV RNA positive) but a substantial proportion of chronically infected patients have normal ALT levels.1 Subjects with active HCV replication and repeatedly normal ALT values represent approximately 25% of blood donors who are found to be anti-HCV positive on routine testing.2,3
The natural course of HCV infection in patients with persistent normal ALT levels is not well understood. Data concerning the histological characteristics of these patients are controversial. While some studies did not find any relationship between ALT levels and histological features,4,5 recent reports indicate that patients with normal ALT values usually have milder liver disease that progresses more slowly than patients with elevated ALT.6,7 Whether a specific mechanism of liver damage induced by HCV is responsible for this unusual disease profile is unclear. Viral factors such as viral load,4,6,8 HCV genotype,5–8 or quasispecies variability8,9 do not seem to be associated with ALT levels. Alternatively, the host immune response directed to HCV infected cells may be involved in the pathogenesis of HCV infection with normal ALT.
Major histocompatibility complex (MHC) alleles have been shown to influence the outcome of HCV infection. Self limiting HCV infection and persistent HCV infection are commonly associated with different HLA class II alleles which confer susceptibility or resistance to viral clearance or persistence.10–15 Few studies however have examined the relationship between HLA class II genotypes and ALT levels in chronically HCV infected patients.15–18
The aims of the present study were: (1) to evaluate demographic, virological, and histological features of chronically HCV infected patients with normal ALT; (2) to compare these parameters with those of chronically HCV infected patients with elevated ALT levels; and (3) to compare the distribution of HLA class II alleles in these two patient groups in order to establish whether the host immunogenetic factors influence this particular biochemical course of chronic HCV infection.
PATIENTS AND METHODS
Patients
A total of 316 HCV positive patients were studied. They were divided into two groups. Group 1 consisted of 83 consecutive chronically HCV infected patients with persistently normal ALT levels (36 men and 47 women; mean age 44.5 years (range 22–79)). All were Caucasians living in France and were prospectively recruited from hepatogastroenterology and internal medicine departments of 17 French hospitals between January 1999 and June 2000. All 83 patients had detectable serum HCV RNA by polymerase chain reaction (PCR). ALT normality was tested on at least five consecutive samples over a period of six consecutive months. In addition, because fluctuating ALT levels characterise chronic hepatitis C, the presence of an abnormal ALT value before this evaluation led to exclusion of the patient. Patients with hepatitis B surface antigen positivity, human immunodeficiency virus infection, alcohol consumption greater than 40 g/day for men and 20 g/day for women, other causes of chronic liver disease, or previous interferon and/or ribavirin therapy were excluded from the study. After enrolment in the study, patients underwent a physical examination, ultrasound liver evaluation, and percutaneous liver biopsy.
Group 2 consisted of 233 HCV infected Caucasian patients (127 men and 106 women; mean age 47.3 years (range 18–78)) from the same geographic area who presented with elevated ALT levels, positive HCV RNA, and biopsy proven hepatitis.
Duration of HCV infection was estimated as the delay between the date of the first exposure to a risk factor (blood transfusion or first year of exposure to intravenous drug) and the date of inclusion in this study. The date of the earliest event was retained for patients with the two risk factors of infection. These data were determined in 50 of the 83 patients in group 1 (60.2%) and in 147 of the 233 patients in group 2 (63.0%).
The study was conducted in accordance with the guidelines of the 1975 Declaration of Helsinki. Each patient signed an informed consent form before enrolment.
Viral testing
Diagnosis of HCV infection was based on detection of anti-HCV antibodies by third generation enzyme linked immunosorbent assay (ELISA, Ortho Diagnosis Systems, Paris, France). Serum HCV-RNA was detected by standardised reverse transcription-PCR using the Amplicor HCV test (Roche Diagnostic Systems, Meylan, France). HCV genotype was determined in 262 patients by reverse hybridisation assay (InnoLiPA HCV; Innogenetics, Ghent, Belgium) containing 15 probe lines for the identification of HCV types 1–6 and subtypes 1a, 1b, 2a, 2b, 3a, 4a, 5a and, 6a. HCV RNA viral load was not analysed because there was a delay between the date of liver biopsy and the time of viral load measurement in a large number of patients with normal ALT levels.
HLA class II typing
For all patients, genomic DNA was isolated from peripheral blood cells using a conventional salting out procedure. HLA class II DNA typing was performed by means of hybridisation with sequence specific oligonucleotide probes, after amplification of the second exon of the DRB1 (all patients) and DQB1 (all patients in group 1 and 147/233 patients in group 2) genes using the InnoLipa HLA genotyping test (Abbott, France and Innogenetics, Ghent, Belgium).
Liver biopsy
The histological status of the liver specimens was scored for all patients according to the Metavir histological index19 and Knodell method20 by an independent observer who was blinded to the biochemical data.
Statistical analysis
HLA-DRB1 and DQB1 allele and phenotype frequencies were determined in the two patient groups by direct counting. The κ2 or Fisher’s exact tests were used to compare discrete variables and allele frequencies between the two groups, and Wilcoxon’s rank sum test was used for continuous variables. The level of significance was set at 0.05, and the Bonferonni correction for multiple tests based on Fisher’s exact test was applied by multiplying p by the number of alleles compared (n=25, pc). Data are expressed as mean (SD). A multivariate logistic regression model was used to determine which variables predicted normal ALT values. Any factors usually known to be correlated with normal ALT values or with a p level <0.25 in the univariate analysis were subsequently tested in the logistic regression model (stepwise selection). Continuous variables were entered into the model as covariables. Adjusted odds ratios (OR) and 95% confidence interval were derived from the coefficient of the final multivariate logistic model. Statistical analysis was performed using Statistical Analysis System (SAS 6.12).
RESULTS
Demographic and clinical features
The demographic and clinical characteristics of the two patient groups are shown in table 1 ▶. Patients with normal ALT levels were more likely to be female and were younger than those with elevated ALT values, although the differences were not statistically significant. The mode of contamination (blood transfusion, history of drug usage) was not significantly different between the two groups. The estimated duration of HCV infection was significantly higher in group 1 than in group 2 (p<0.001).
Table 1.
Demographic and clinical characteristics of 316 hepatitis C virus (HCV) chronically infected patients according to alanine aminotransferase (ALT) values
| Normal ALT | Abnormal ALT | ||
| Group 1 (n=83) | Group 2 (n=233) | p Value | |
| Females (F/M ratio) | 47 (0.57) | 106 (0.45) | 0.08 |
| Age at time of study (y) (mean (SD)) | 44.5 (11.9) | 47.3 (14.4) | 0.20 |
| Estimated duration of infection (y) (mean (SD))* | 19.5 (7.5) | 12.6 (6.6) | <0.001 |
| Fortuitous diagnosis (blood donation) (%) | 5 (6) | 9 (4) | 0.53 |
| Mode of contamination | |||
| Drug usage (%) | 25 (30) | 61 (26) | |
| Transfusion (%) | 25 (30) | 86 (37) | 0.61† |
| Others (%) | 33 (40) | 86 (37) |
*Number of patients with a known blood risk factor for HCV contamination: group 1=50/83 (60.2%); group 2=147/233 (63%).
†p value relates to the difference between the three modes of contamination.
Genotype
HCV genotype was available for 262 patients (all patients in group 1 and 179 patients in group 2). Genotype 1 was the most common genotype in both groups (159 patients, 60.7%). The distribution of HCV genotypes did not differ significantly between the two patient groups (table 2 ▶).
Table 2.
Virological characteristics of 316 hepatitis C virus (HCV) chronically infected patients according to alanine aminotransferase (ALT) levels
| Genotype | Normal ALT Group 1 (n=83) | Elevated ALT Group 2 (n=233)* | p Value |
| 1 | 55 (67%) | 104 (58%) | 0.21 |
| 2 | 12 (14%) | 30 (17%) | 0.63 |
| 3 | 12 (14.5%) | 36 (20%) | 0.27 |
| 4 | 2 (2.5%) | 8 (4.5%) | 0.51 |
| 5 | 1 (1%) | 1 (0.5)% | 0.53 |
| 6 | 1 (1%) | 0 (0%) | 0.32 |
*HCV genotype was not available for 54/233 (23%) patients in group 2.
HLA-DR and DQ genotyping
The distribution of HLA-DRB1 and DQB1 phenotypes in the two patient groups is given in tables 3 and 4 ▶ ▶. The only significant difference concerned the DRB1*11 phenotype whose frequency was increased in patients with normal ALT levels compared with those with elevated ALT levels (43% v 24%; p<0.001, pc=0.03, OR 2.36 (1.39–4.0)). A negative association of DRB1*01 with normal ALT levels (6% v 19%; p=0.004) and a positive association with DRB1*08 (12% v 5%; p=0.02) were also observed although they did not reach significance after correction for multiple comparisons. The distribution of DQB1 alleles and phenotypes was similar in patients from groups 1 and 2. In particular, the frequency of DQB1*0301, in linkage disequilibrium with DRB1*11 in Caucasians, was not significantly different between the two groups (47% and 36%, respectively; p=0.13). Interestingly, several patients expressed DRB1*11 without DQB1*0301 but with DQB1*0501, *0602, *0604, or *0603.
Table 3.
Distribution of DRB1 phenotypes in hepatitis C virus (HCV) chronically infected patients with normal or elevated alanine aminotransferase (ALT) levels
| DRB1* | Normal ALT (n=83) (%) | Elevated ALT (n=233) (%) | p Value† (pc) | Odds ratio (95% CI) |
| 01 | 5 (6%) | 45 (19%) | 0.004 (0.07) | 0.26 (0.10 to 0.70) |
| 03 | 12 (14%) | 44 (19%) | ||
| 04 | 22 (27%) | 56 (24%) | ||
| 07 | 19 (23%) | 66 (28%) | ||
| 08 | 10 (12%) | 11 (5%) | 0.02 (0.58) | 2.76 (1.12 to 6.77) |
| 09 | 2 (2%) | 2 (1%) | ||
| 10 | 1 (1%) | 6 (3%) | ||
| 11 | 36 (43%) | 57 (24%) | <0.001 (0.03) | 2.36 (1.39 to 4.00) |
| 12 | 2 (2%) | 2 (1%) | ||
| 13 | 19 (23%) | 64 (27%) | ||
| 14 | 10 (12%) | 17 (7%) | ||
| 15 | 10 (12%) | 41 (18%) | ||
| 16 | 7 (8%) | 12 (5%) | ||
| 02‡ | 1 (1%) | 13 (6%) |
†Only significant p values are indicated.
‡DR2 subtyping (DR15, DR16) was not available for 14 individuals.
Table 4.
Distribution of DQB1 phenotypes in hepatitis C virus (HCV) chronically infected patients with normal or elevated alanine aminotransferase (ALT) levels
| DQB1* | Normal ALT (n=83) (%) | Elevated ALT (n=147)† (%) | p Value (pc) |
| 02 | 27 (32.5%) | 59 (40.1%) | |
| 0301/0304 | 39 (47%) | 53 (36.1%) | 0.13 |
| 0302 | 15 (18.1%) | 24 (16.3%) | |
| 0303 | 5 (6%) | 5 (3.4%) | |
| 0401/0402 | 11 (13.3%) | 5 (3.4%) | 0.01 (NS) |
| 0501 | 11 (13.3%) | 32 (21.8%) | |
| 0502 | 8 (9.6%) | 12 (8.2%) | |
| 0503 | 10 (12%) | 13 (8.8%) | |
| 0601 | 1 (1.2%) | 0 (0%) | |
| 0602 | 12 (14.5%) | 30 (20.4%) | |
| 0603 | 10 (12%) | 32 (21.8%) | |
| 0604/0609 | 7 (8.4%) | 9 (6.1%) |
†DQB1 typing data available for 147/233 patients with elevated ALT.
Liver histology
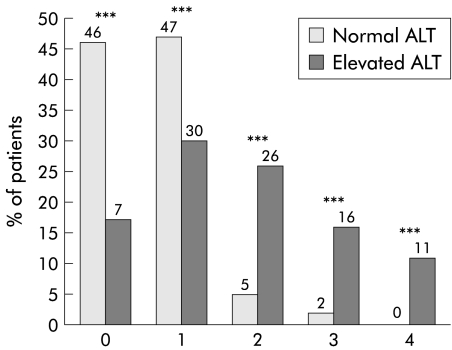
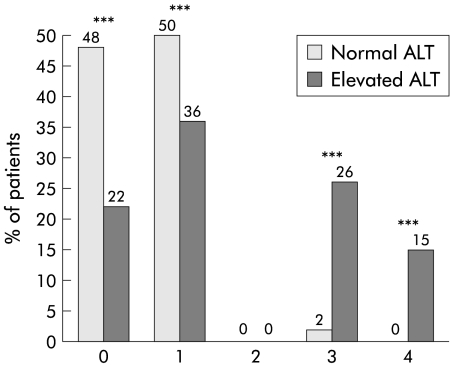
Five of the 83 patients in group 1 (6%) had normal liver histology (Metavir index and Knodell score). No patient in group 1 developed cirrhosis. ALT levels were correlated with the severity of liver damage. Index of activity was lower in group 1 than in group 2, as indicated by both a lower Metavir index (0.9 (0.7) and 1.6 (0.8), respectively) and a lower Knodell score (4.3 (2.8) and 6.8 (3.8), respectively). Index of liver fibrosis was also lower in group 1 than in group 2 (Metavir 0.6 (0.7) and 1.7 (1.2); Knodell 0.6 (0.6) and 1.8 (1.4), respectively). In group 1, 77 of 83 (93%) patients had an F0 or F1 Metavir index of fibrosis compared with 110 of 233 (47%) patients in group 2 (p<0.001) (fig 1 ▶). Four patients in group 1 had an F2 Metavir index and only two were scored as F3. Similar significant differences between the two patient groups were observed when comparing the Knodell index (fig 2 ▶). Although histological lesions were milder in patients with normal ALT values than in those with elevated ALT, the estimated duration of infection was significantly higher in the former group (19.5 v 12.6 years; p<0.001) (table 1 ▶).
Figure 1.
Distribution of fibrosis (index of Metavir (0–4)) for patients with normal and elevated levels of alanine aminotransferase (ALT). ***p<0.001 between the two groups (κ2 test).
Figure 2.
Distribution of fibrosis (Knodell’s index (0–4)) for patients with normal and elevated levels of alanine aminotransferase (ALT). ***p<0.001 between the two groups (κ2 test).
A significant association between the stage of liver fibrosis and DRB1 phenotype was observed (table 5 ▶). Using multivariate analysis, the DRB1* 11 phenotype was associated with no or only mild fibrosis (F0-F1 Metavir index) (p=0.03). Other factors independently associated with a low stage of fibrosis (F0-F1) were normal ALT value (p=0.0001), young age (p=0.006), and female sex (p=0.007).
Table 5.
Multivariate analysis of factors associated with absence of fibrosis or with portal fibrosis without septa (F0-F1 index of Metavir)
| Odds ratio (95% CI) | p Value | |
| Age (young) | 1.08 (1.03 to 1.13) | 0.006 |
| Female sex (v male) | 5.52 (1.61 to 18.89) | 0.007 |
| Normal ALT (v elevated ALT) | 14.27 (3.09 to 65.88) | 0.0001 |
| HLA-DRB1*11 (v other phenotypes) | 3.62 (1.03 to 12.71) | 0.03 |
| Duration of contamination (long) | 1.02 (1.1 to 0.95) | 0.46 |
ALT, alanine aminotransferase.
Factors associated with normal ALT
Age, sex, duration of infection, route of contamination, viral genotype, and HLA class II phenotype were analysed as predictors of normal ALT levels (table 6 ▶). After stepwise logistic regression, four variables were independently associated with normal ALT values: young age (p=0.0008), female sex (p=0.01), long duration of contamination (p=0.0001), and HLA DRB1*11 phenotype (p=0.050). There was no particular association between normal ALT values and viral genotype or mode of contamination.
Table 6.
Multivariate analysis of factors associated with normal alanine aminotransferase (ALT) levels
| Odds ratio (95% CI) | p Value | |
| Age (young) | 1.07 (1.03 to 1.12) | 0.0008 |
| Female gender (v male) | 2.83 (1.19 to 6.73) | 0.01 |
| Duration of contamination (long) | 1.19 (1.11 to 1.27) | 0.0001 |
| HLA-DRB1*11 (v other phenotypes) | 2.37 (0.99 to 5.66) | 0.050 |
DISCUSSION
The main findings of the present study were: (1) absence of significant differences in HCV genotype distribution between HCV chronically infected patients with normal or elevated ALT levels; (2) mild histological liver disease among patients with normal ALT values; and (3) positive association between normal ALT levels and the DRB1*11 phenotype.
Discrepancies still persist with regard to the epidemiological, virological, and histological characteristics of HCV patients with persistently normal ALT levels. Such controversies may be related to the small size of the patient groups studied, inclusion of patients with near normal ALT values,21 or inclusion of patients with heavy alcohol consumption. To our knowledge, our patient group represents the largest series of HCV chronically infected patients with normal ALT described to date. Alcohol abusers were excluded from the study. In addition, our definition of persistently normal ALT, which was at least five consecutive normal ALT values over a period of six months with no abnormal values before the date of inclusion, was particularly stringent. However, it is impossible to rule out fluctuations in ALT values prior to HCV detection or after the end of patient follow up. Accordingly, one could imagine that “normal ALT” is a snapshot of a dynamic process that includes abnormal ALT.22
Factors influencing this particular disease presentation are unclear. Our data indicated that HCV carriers with normal ALT levels were more likely to be female (57%), as previously reported.23 Conversely, most patients with raised ALT were men. Previous studies noted a female predominance in this setting among volunteer blood donors24 or among patients assessed during clinical practice.3,23 In our study, this observation cannot be attributed to alcohol consumption as alcohol abusers were excluded. Another possible explanation could be related to the lower values of basal ALT in females, as shown by Piton et al in a population of blood donors without chronic hepatitis.3
Patients with chronic hepatitis C and normal ALT values usually have asymptomatic liver disease.2,4 Because of the absence of clinical and biochemical abnormalities, the diagnosis of HCV infection in such individuals is generally fortuitous at the time of blood donation.23 Indeed, in our study, none of the 83 patients with normal ALT reported symptoms of liver disease, and only 6% underwent routine screening for HCV infection at the time of blood donation. This may explain why these patients had a longer delay to HCV diagnosis and consequently a longer duration of infection than patients with elevated ALT values. At variance with previous reports showing a decrease in ALT levels with age in HCV positive patients, we found that patients with normal ALT levels were significantly younger that those with elevated ALT.22,25 It is difficult to explain this finding, especially as the duration of contamination was higher in the former group of patients.
In most studies, virological factors, in particular HCV genotype, did not significantly differ between HCV patients with normal or abnormal ALT levels.5–7 Two Italian groups however reported a strong association between genotype 2a and normal ALT values.4,26 We did not find any significant difference in the distribution of viral genotype according to ALT levels, genotype 1 being predominant in both groups. Furthermore, HCV genotype distribution did not influence the severity of liver lesions in normal ALT patient groups (data not shown). These findings confirm that the severity of chronic hepatitis C is not strongly associated with HCV genotype.27
Our study showed that patients with normal ALT levels have less severe liver disease, both in terms of histological activity and fibrosis, compared with those with elevated ALT. The fact that alcohol abusers were excluded from our study may explain, at least in part, why none of our normal ALT patients presented with cirrhosis, and why only two showed a fibrosis score higher than 2. In other words, we observed that the majority of patients with normal ALT levels and without elevated alcohol consumption did not present with severe liver histological changes, despite a mean duration of infection of approximately 20 years. The mean histological grading and staging after this time suggests that their prognosis is good. Liver biopsies are standard care for those with elevated ALT levels but are controversial in those with normal ALT values due to incomplete knowledge of the natural history of chronic HCV infection in this setting.28 The absence of a rationale for the latter group is closely related to controversies regarding the severity of histological disease. Several authors have suggested that liver biopsy should routinely be offered to all viraemic patients regardless of ALT levels in order to evaluate the severity of the liver disease.4,5,29–31 The EASL International Consensus on Hepatitis C also recommended liver biopsy in this setting.4,32 In contrast, Mathurin et al and Jamal et al, who analysed fibrosis progression per year, concluded that patients with normal ALT values have a low risk of developing severe liver lesions.6,7 In clinical practice, these observations, together with our data, cast doubt on the usefulness of a regular follow up by liver biopsy in normal ALT patients, particularly in those with no alcohol abuse. However, frequent ALT determinations must be performed to rapidly detect a mild ALT elevation that requires appropriate investigation and treatment. Whether interferon should be a therapeutic option for patients with normal ALT levels is an open issue. Absence of clear cut evidence that there is a risk of progression to severe disease, combined with the poor effectiveness of interferon treatment, argue against an active therapeutic strategy.33–35 A wait and see attitude might be more appropriate. However, if a more effective and better tolerated therapy were to become available, such patients might be treated to eliminate the virus.
Several studies have evidenced the participation of MHC class II alleles in susceptibility to HCV infection36 as well as in susceptibility or resistance to persistent HCV infection.10–15,37–39 Interestingly, HLA-DRB1*11 and/or DQB1*0301 alleles have been associated with spontaneous clearance of circulating HCV.10,13,14,39 Because DRB1*11 is in strong linkage disequilibrium with DQB1*0301, it has been difficult to determine which of these two alleles is responsible for the observed association. Other studies have demonstrated a close association between some HLA-DR alleles and histological changes in patients with chronic HCV infection, suggesting an influence of HLA class II in the course of liver disease.17,40–43 Few studies however have examined the association between HLA phenotype and ALT levels in patients with persistent infection. We showed that DRB1*11 was almost twice as frequent in HCV patients with normal ALT levels than in those with elevated ALT. DQB1*0301 frequency was not significantly different in the two groups. This may be related to the fact that some DRB1*11 patients presented with unusual DR/DQ combinations—that is, expressed the DQB1 allele different from DQB1*0301—and may suggest that DRB1*11 is dominant in determining the outcome of infection, as already proposed.17,44 Our data extend previous observations of an increased DRB1*11 frequency in smaller series of healthy HCV carriers.16,17 Therefore, it appears that a vigorous DRB1*11 restricted T cell response is associated with viral clearance at the time of infection, suggesting that DR11 may present HCV epitopes to CD4 cells more efficiently than other alleles. In addition, in subjects who do not clear the virus, DRB1*11 may also be critical in limiting the spread of the virus so that liver injury remains minimal. In contrast, in DRB1*11 negative patients, continuous liver damage mostly caused by non-specific bystander inflammatory processes may accelerate the development of fibrosis.
In conclusion, we have shown in a large series of well documented HCV patients that the natural history of HCV chronic infection in patients with persistently normal ALT is better than for those with elevated ALT, and is associated with a delay in the development of liver damage, particularly when there is no alcohol abuse. This good biochemical and histological outcome could be controlled, at least in part, by host immunogenetic factors as HLA DRB1*11 frequency is significantly higher in patients with normal ALT, a finding also observed in patients who spontaneously recover from HCV infection.
Acknowledgments
We are grateful to H Khiri and R Deydier for their excellent technical assistance (Alphabio, Marseille, France). We also thank E Sablon and T James for DQB genotyping and for language editing (Innogenetics, Ghent, Belgium). This work was supported by a grant from Produits Roche, Neuilly-sur-Seine, France.
Abbreviations
HCV, hepatitis C virus
ALT, alanine aminotransferase
MHC, major histocompatibility complex
HLA, human leucocyte antigen
PCR, polymerase chain reaction
REFERENCES
- 1.Zylberberg H, Pol S, Nalpas B, et al. Significance of repeatedly normal aminotransferase activities in hepatitis C virus-infected patients. Hepatology 1998;27:1169. [DOI] [PubMed] [Google Scholar]
- 2.Marcellin P, Lévy S, Erlinger S. Therapy of hepatitis C: patients with normal aminotransferase levels. The National Institute of Health Consensus Development Conference. Hepatology 1997;26(suppl 1):133–6S. [DOI] [PubMed] [Google Scholar]
- 3.Piton A, Poynard T, Imbert-Bismut F, et al. Factors associated with serum alanine transaminase activity in healthy subjects: consequences for the definition of normal values, for selection of blood donors, and for patients with chronic hepatitis C. MULTIVIRC Group. Hepatology 1998;27:1213–19. [DOI] [PubMed] [Google Scholar]
- 4.Puoti C, Magrini A, Stati T, et al. Clinical, histological, and virological features of hepatitis C virus carriers with persistently normal or abnormal alanine transaminase levels. Hepatology 1997;26:1393–8. [DOI] [PubMed] [Google Scholar]
- 5.Stanley AJ, Haydon GH, Piris J, et al. Assessment of liver histology in patients with hepatits C and normal transaminase levels. Eur J Gastroenterol Hepatol 1996;8:869–72. [PubMed] [Google Scholar]
- 6.Mathurin P, Moussalli J, Cadranel JF, et al. Slow progression rate of fibrosis in hepatitis C virus patients with persistently normal alanine transaminase activity. Hepatology 1998;27:868–72. [DOI] [PubMed] [Google Scholar]
- 7.Jamal MM, Soni A, Quinn PG, et al. Clinical features of hepatitis C-infected patients with persistently normal alanine transaminase levels in the Southwestern United States. Hepatology 1999;30:1307–11. [DOI] [PubMed] [Google Scholar]
- 8.Shindo M, Arai K, Sokawa Y, et al. The virological and histological states of anti-hepatitis C virus-positive subjects with normal liver biochemical values. Hepatology 1995;22:418–25. [PubMed] [Google Scholar]
- 9.Naito M, Hayashi N, Moribe T, et al. Hepatits C viral quasispecies in hepatitis C virus carriers with normal liver enzymes and patients with type C chronic liver disease. Hepatology 1995;22:407–12. [PubMed] [Google Scholar]
- 10.Thursz M, Yallop R, Goldin R, et al. Influence of MHC class II genotype on outcome of infection with hepatitis C virus. The HENCORE group. Hepatitis C European Network for Cooperative Reseach. Lancet 1999;354:2119–24. [DOI] [PubMed] [Google Scholar]
- 11.Barrett S, Ryan E, Crowe J. Association of the HLA-DRB1*01 allele with spontaneous viral clairance in an Irish cohort infected with hepatitis C virus via contaminated anti-D immunoglobulin. J Hepatol 1999;30:979–83. [DOI] [PubMed] [Google Scholar]
- 12.Fanning LJ, Levis J, Kenny-Walsh E, et al. Viral clearance in hepatitis C (1b) infection: relationship with human leukocyte antigen class II in a homogeneous population. Hepatology 2000;31:1334–7. [DOI] [PubMed] [Google Scholar]
- 13.Alric L, Fort M, Izopet J, et al. Genes of the major histocompatibility complex class II influence the outcome of hepatitis C virus infection. Gastroenterology 1997;113:1675–81. [DOI] [PubMed] [Google Scholar]
- 14.Minton EJ, Smillie D, Neal KR, et al. Association between MHC class II alleles and clearance of circulating hepatitis C virus. J Infect Dis 1998;178:39–44. [DOI] [PubMed] [Google Scholar]
- 15.Mangia A, Gentile R, Cascavilla I, et al. HLA class II favors clearance of HCV infection and progression of the chronic liver damage. J Hepatol 1999;30:984–9. [DOI] [PubMed] [Google Scholar]
- 16.Kuzushita N, Hayashi N, Moribe T, et al. Influence of HLA haplotypes on the clinical courses of individuals infected with hepatitis C virus. Hepatology 1998;27:240–4. [DOI] [PubMed] [Google Scholar]
- 17.Asti M, Martinetti M, Zavaglia C, et al. Human leukocyte antigen class II and III alleles and severity of hepatitis C virus-related chronic liver disease. Hepatology 1999;29:1272–9. [DOI] [PubMed] [Google Scholar]
- 18.McKiernan SM, Hagan R, Curry M, et al. The MHC is a major determinant of viral status, but not fibrotic stage, in individuals infected with hepatitis C. Gastroenterology 2000;118:1124–30. [DOI] [PubMed] [Google Scholar]
- 19.The METAVIR cooperative group. Inter and intra-observer variation in the assessment of liver biopsy of chronic hepatitis C. Hepatology 1994;20:15–20.8020885 [Google Scholar]
- 20.Knodell RG, Ishak KG, Black WC, et al. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepalology 1981;1:431–5. [DOI] [PubMed] [Google Scholar]
- 21.Van Thiel DH, Caraceni P, Molloy PJ, et al. Chronic hepatitis C in patients with normal or near normal alanine aminotransferase levels: the role of interferon alpha 2b therapy. J Hepatol 1995;23:503–8. [DOI] [PubMed] [Google Scholar]
- 22.Inglesby TV, Rai R, Astemborski J, et al. A prospective, community-based evaluation of liver enzymes in individuals with hepatitis C after drug use. Hepatology 1999;29:590–6. [DOI] [PubMed] [Google Scholar]
- 23.Gholson CF, Morgan K, Catinis G, et al. Chronic hepatitis C with normal aminotranferase levels: a clinical histologic study. Am J Gastroenterol 1997;92:1788–92. [PubMed] [Google Scholar]
- 24.Yuki N, Hayashi N, Takehara T, et al. Serum hepatitis C virus RNA levels and liver injury in volunteer blood donors. Am J Gastroenterol 1994;89:1462–6. [PubMed] [Google Scholar]
- 25.Shakil AO, Conry-Cantilena C, Alter HJ, et al. Volunteer blood donors with antibody to hepatitis C virus: clinical, biochemical, virologic, and histologic features. The Hepatitis C Study Group. Ann Intern Med 1995;123:330–7. [DOI] [PubMed] [Google Scholar]
- 26.Silini E, Bono F, Cividini A, et al. Differential distribution of hepatitis C virus genotypes in patients with and without liver function abnormalities. Hepatology 1995;21:285–90. [PubMed] [Google Scholar]
- 27.Yamada M, Kakumu S, Yoshioka K, et al. Hepatitis C virus genotypes are not responsible for development of serious liver disease. Dig Dis Sci 1994;39:234–9. [DOI] [PubMed] [Google Scholar]
- 28.Everhart JE, Stolar M, Hoofnagle JH. Management of hepatitis C: a national survey of gastroenterologists and hepatologists. Hepatology 1997;26:78–82S. [DOI] [PubMed] [Google Scholar]
- 29.Prati D, Capelli C, Zanella A, et al. Influence of different hepatitis C genotypes on the course of asymptomatic hepatitis C virus infection. Gastroenterology 1996;110:178–83. [DOI] [PubMed] [Google Scholar]
- 30.Naito M, Hayashi N, Hagiwara N, et al. Serum hepatitis C virus RNA quantity and histological features of hepatitis C virus carriers with presistently normal ALT levels. Hepatology 1994;19:871–5. [PubMed] [Google Scholar]
- 31.Okanoue T, Yasui K, Sakamoto S, et al. Circulating HCV-RNA, HCV genotype, and liver histology in asymptomatic individuals reactive for anti-HCV antibody and their follow-up study. Liver 1996;16:241–7. [DOI] [PubMed] [Google Scholar]
- 32.EASL International Consensus Conference on Hepatitis C. Consensus statement. J Hepatol 1999;30:956–61. [PubMed] [Google Scholar]
- 33.Serfaty L, Chazouilléres O, Pawlotsky JM, et al. Interferon alfa therapy in patients with chronic hepatitis C and persistently normal aminotransferase activity. Gastroenterology 1996;110:291–5. [DOI] [PubMed] [Google Scholar]
- 34.Rossini A, Ravaggi A, Biasi L, et al. Virological response to interferon treatment in hepatitis C virus carriers with normal aminotransferase levels and chronic hepatitis. Hepatology 1997;26:1012–17. [DOI] [PubMed] [Google Scholar]
- 35.Sangiovanni A, Morales R, Spinzi G, et al. Interferon alfa treatment of HCV RNA carriers with persistently normal transaminase levels: a pilot randomized controlled study. Hepatology 1998;27:853–6. [DOI] [PubMed] [Google Scholar]
- 36.Congia M, Clemente MG, Dessi C, et al. HLA class II genes in chronic hepatitis C virus-infection and associated immunological disorders. Hepatology 1996;24:1338–41. [DOI] [PubMed] [Google Scholar]
- 37.Lechmann M, Schneider EM, Giers G, et al. Increased frequency of the HLA-DR15 (B1*15011) allele in German patients with self-limited hepatitis C virus infection. Eur J Clin Invest 1999;29:337–43. [DOI] [PubMed] [Google Scholar]
- 38.Höhler T, Gerken G, Notghi A, et al. MHC class II genes influence the susceptibility to chronic active hepatitis C. J Hepatol 1997;27:259–64. [DOI] [PubMed] [Google Scholar]
- 39.Cramp ME, Carucci P, Underhill J, et al. Association between HLA class II genotype and spontaneous clearance of hepatitis C viraemia. J Hepatol 1998;29:207–13. [DOI] [PubMed] [Google Scholar]
- 40.Peano G, Menardi G, Ponzetto A, et al. HLA-DR5 antigen. A genetic factor influencing the outcome of hepatitis C virus infection? Arch Intern Med 1994;154:2733–6. [DOI] [PubMed] [Google Scholar]
- 41.Tillmann HL, Chen DF, Trautwein C, et al. Low frequency of HLA-DRB1*11 in hepatitis C virus induced end stage liver disease. Gut 2001;48:714–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amoroso A, Berrino M, Canale L, et al. Are HLA class II and immunoglobulin constant region genes involved in the pathogenesis of mixed cryoglobulinemia type II after hepatitis C virus infection? J Hepatol 1998;29:36–44. [DOI] [PubMed] [Google Scholar]
- 43.Haruna Y, Miyamoto T, Yasunami R, et al. Human leukocyte antigen DRB1 1302 protects against bile duct damage and portal lymphocyte infiltration in patients with chronic hepatitis C. J Hepatol 2000;32:837–42. [DOI] [PubMed] [Google Scholar]
- 44.Thursz M. MHC and the viral hepatitides. Q J Med 2001;94:287–91. [DOI] [PubMed] [Google Scholar]