Abstract
Background: Germline mutations in the mismatch repair (MMR) genes hMLH1 and hMSH2 can cause hereditary non-polyposis colorectal cancer (HNPCC). However, the functional in vitro analysis of hMLH1 and hMSH2 mutations remains difficult.
Aims: To establish an in vitro method for the functional characterisation of hMLH1 and hMSH2 mutations.
Methods: hMLH1 and hMSH2 wild type (wt) genes and several mutated subclones were transiently transfected in mismatch repair deficient cell lines (HCT-116 and LOVO). Apoptosis, proliferation, and regulation of mRNA expression and protein expression of interacting proteins were analysed by Hoechst staining, AlamarBlue staining, real time polymerase chain reaction, and western blotting, respectively.
Results: The protein expression of hMLH1 and hMSH2 mutants was significantly decreased after transfection compared with wild type transfections. The hMLH1 and hMSH2 interacting proteins hPMS2 and hMSH6 became detectable only after transfection of the respective wild type genes. In parallel, hMSH6 mRNA levels were increased in hMSH2 wt transfected cells. However, hPMS2 mRNA levels were independent of the mutation status of its interacting partner hMLH1, indicating a post-transcriptional regulating pathway. In the hMLH1 deficient HCT-116 cell line apoptosis was not affected by transfection of any mismatch repair gene, whereas complementation of hMSH2 deficency in LOVO cells increased apoptosis. Conversely, proliferative activity of HCT-116 was decreased by complementation with hMLH1wt and unaffected in hMSH2 deficient LOVO cells.
Conclusion: These data show that the cellular role of the MMR genes and its mutations are assessable in a simple transient transfection system and show the influence of MMR gene regulation on major cell growth regulating mechanisms. This method is applicable for the functional definition of mutations in hMLH1 and hMSH2 genes observed in patients with suspected HNPCC.
Keywords: mismatch repair genes, hMLH1, hMSH2, colorectal cancer, hereditary non-polyposis colorectal cancer
Mismatch repair (MMR) is an important cellular pathway that facilitates genome stability by excising mismatched nucleotides of the DNA. Germline mutations in two human MMR genes, hMSH2 and hMLH1, account for approximately 98% of hereditary non-polyposis colorectal cancer (HNPCC) cases.1 hMSH2, a homologue of Escherichia coli MutS protein, is required for mismatch recognition.2 Heterodimers formed between hMSH2 and substrate specificity modifying MutS homologues (hMSH3 and hMSH6) are essential for the following repair of mismatches. hMutSα, the heterodimer of hMSH2 and hMSH6 is known to recognise base-base mispairs, or single insertion/deletion loops, while hMutSβ, the heterodimer of hMSH2 and hMSH3 is primarily involved in the correction of larger DNA insertion/deletion loops.3,4 For the DNA repair process, activated hMutSα or hMutSβ interact with hMLH1, the human homologue of the E coli MMR gene MutL.5,6 In addition to hMLH1, other human MutL homologues have been identified (hMLH3, hPMS1, and hPMS2). While interactions between hMLH1, hMLH3, and hPMS1 have been reported,7–9 only the hMLH1/hPMS2 heterodimer has been shown to participate in mismatch repair.10
Recently, specific mutational inactivation of hMLH1 and hMSH2 leading to post-translational downregulation of the heterodimerising partners was proposed.11 Furthermore, MMR proteins were suggested to be involved in the regulation of apoptosis and proliferation.12–14 In these studies, however, different experimental models such as transfection of whole chromosomes and co-microinjection of expression plasmids and GFP vectors were used. The interpretation of these data remain difficult because effects caused by multiple gene expression and accumulation of multiple defects in different tumour cells cannot be excluded. No data have yet been reported determining the different cellular roles of hMLH1 and hMSH2 in one comprehensive system. Moreover, the clinical role of specific MMR mutations in the progression of disease and for the efficacy of chemotherapy is widely unclear. Thus, the functional analysis of MMR mutations may help to define the clinical significance of mutations.
In this study we expressed hMLH1 wild type (hMLH1 wt), hMSH2 wild type (hMSH2 wt), and several mutants in a well defined transient transfection system to investigate the interaction of hMSH2/hMSH6 and hMLH1/hPMS2 as well as the functional role of hMLH1 and hMSH2 in apoptosis and proliferation.
METHODS
Cell lines and cultures
The cell lines used in this study were purchased from the American Type Culture Collection (ATCC). HCT-116 and LOVO cells were cultured in a humidified atmosphere containing 5% carbon dioxide in McCoy’s 5A (Gibco, Karlsruhe, Germany) and RPMI 1640 medium (Gibco), respectively. All media contained 10% fetal calf serum (Gibco).
Construction of hMLH1 and hMSH2 mutants
The complete wild type cDNA for hMSH2 was subcloned from laboratory isolates into the pcDNA3.1+ vector (Invitrogen, Groningen, Netherlands), placing the cDNA under the control of the CMV promoter. Inserts for hMSH2 wt cDNA were amplified using primers bearing BamHI (sense) or XhoI (antisense) restriction sites and ligated in the appropriate sites of the vector. The pcDNA3.1+ vector containing the cDNA of hMLH1 was kindly provided by Dr Hong Zhang (University of Utah, Salt Lake City, Utah). The expression constructs for the mutant proteins were designed using the corresponding pcDNA3.1+ wild type vectors (hMLH1 or hMSH2) and site directed mutagenesis (Promega, Madison, UK). The following cDNA-mutant constructs were generated: hMLH1 Thr117Met, hMLH1 Lys618Thr, hMSH2 DEL782FS, and hMSH2 Cys697Arg, representing four mutations of patients with HNPCC identified in our clinic. Expression vectors containing the genes of interest were confirmed by sequencing and transcription/translation (Promega). Proteins were detected by appropriate monoclonal antibodies (anti-hMLH1, clone G168–728, PharMingen, Stuttgart, Germany; anti-hMSH2, Ab2, Calbiochem, La Jolla, CA).
Transcription/translation of plasmids
Protein production of the used plasmids was verified by the in vitro TNT Quick Coupled Transcription/Translation system (Promega).
Preparation of cells for transfection
HCT-116 and LOVO cells were grown to 50%–80% confluenty and subsequently transfected with 2 μg Genomed purified plasmid DNA (Genomed, Bad Oeynhausen, Germany) mixed with 6 μl FuGENE 6 (Boehringer Mannheim, Mannheim, Germany) and with 2 μg plasmid DNA mixed with 7.9 μl Tfx-20 reagent (Promega), respectively. pcDNA3.1+ vector without insert was transfected in both cell lines as mock control.
Western blotting and immunoprecipitation
Proteins were separated on denaturing 7.5% SDS-polyacrylamide gels, blotted onto PVDF membranes, and detected with the ECL-Kit (Amersham Pharmacia Biotech, Buckinghamshire, England). The purified mouse antihuman antibodies used for western blotting were the monoclonal anti-hMLH1 (Clone: G168–728, Pharmingen), the monoclonal anti-hPMS2 (Clone: A16–4, Pharmingen), the monoclonal anti-hMSH2 (Clone 27, Transduction Laboratories), and the monoclonal anti-GTBP (hMSH6, Clone: 44, Transduction Laboratories). Immunoprecipitation was carried out using a cellular labelling and immunoprecipitation kit (Boehringer Mannheim). The resulting precipitate was subsequently analysed by SDS-PAGE (7.5%).
Immunofluorescence
Cells were washed with phosphate buffered saline (PBS) 12, 24, 36, 48, 72, 96 hours, and seven days after transfection and fixed in 100% prechilled aceton overnight at −20°C. Fluorescence microscopy (Axiovert 135, Zeiss, Jena, Germany) was performed after incubation of the cells for five minutes with blocking buffer (PBS, 1% BSA, 0.2% Tween 20, 5 mM TRIS/HCl, pH 7.6), with the indicated primary antibody in blocking buffer for one hour at 37°C, and the corresponding Cy3 conjugated secondary antibody (antimouse IgH/L, Jackson Laboratories, UK) at room temperature for 30 minutes in the dark.
Quantitative PCR analysis of hPMS2 and hMSH6 mRNA
Total RNA was extracted from transfected HCT-116 and LOVO cells using Tristar (AGS GmbH, Heidelberg, Germany). First strand cDNA was prepared from total cellular RNA using random hexadeoxynucleotide primers and reverse transcriptase (SuperScriptII, Gibco). PCR was performed in a volume of 50 μl containing 50 mM KCl, 10 mM TRIS-HCl, 0.01 mM EDTA and 60 nM ROX (6-carboxy-x-rhodamine), 6 mM MgCl2, 400 μM of dATP, dCTP, dGTP and dTTP each, 100 nM of primers each (table 1▶), 100 nM of probe and 1.25 U of AmpliTaq Gold DNA polymerase. Thermal cycling involved preincubation at 95°C for 12 minutes, followed by 50 cycles at 95°C for 20 seconds and 55°C for 20 seconds. Fluorescence was measured real time during PCR (Taq-Man-PCR, Applied Biosystems, Weiterstadt, Germany). Emission ranges of both fluorescent dyes were detected independently by wavelength in the Perkin-Elmer 7700 Sequence Detection System (Applied Biosystems, Weiterstadt, Germany). The relative hPMS2 and hMSH6 RNA quantification values were obtained from the threshold cycle number from which the increase in signal associated with an exponential growth of PCR product became detectable (Sequence detection system software version 1.6, Applied Biosystems). The resulting ‡‡CT-factor enables relative quantification of the RNA of interest (Sequence Detector User Bulletin, Applied Biosystems). For quantification normalised to an endogenous control, standard curves were prepared for both the target (hPMS2 and hMSH6) and the endogenous reference (GAPDH). For calibration hPMS2 RNA and hMSH6 RNA were prepared from mismatch repair proficient colorectal tumour cells (SW 480). Quantitative PCR was performed in duplicate for each primer set and the mean of the two experiments was used as relative quantification value. Contamination precautions according to Kwok et al were strictly complied.15
Table 1 .
Primer for real time PCR. Probes for detection were labelled with FAM (6-carboxy-fluorescein), TAMRA (6-caboxy-N,N,N-,N-tetramethylrhodamin) or VIC (fluorescein derivat)
| Primer | Sequence (5′ → 3′) |
|---|---|
| hMSH6-probe | FAM-CAGGAGCTTTTATCAATGGCTA-TAMRA |
| hMSH6-Taq-S | TCTAGGTGGTTGTGTCTTCTACCTCA |
| hMSH6-Taq-AS | TAGTGCTGACTGTGTCAGAATCCA |
| hPMS2-probe | FAM-ACTGCTCTTAACACAAGCGAGATGAAGAA-TAMRA |
| hPMS2-Taq-S | CGGAAGTCGGTGATGATTGG |
| hPMS2-Taq-AS | TTGGCGATGTGTCTCATGGTT |
| GAPDH-probe | VIC-CAAGCTTTCCCGTTCTCAGCC-TAMRA |
| GAPDH-S | GAAGGTGAAGGTCGGAGTC |
| GAPDH-AS | GAAGATGGTGATGGGATTTC |
S, sense; AS, antisense.
Apoptosis
Air dried cells were fixed 12, 36, 48, and 72 hours after transfection with a freshly prepared paraformaldehyde solution (4% in PBS, pH 7.4) for 30 minutes at room temperature, washed with PBS and incubated with permeabilisation solution (1% Triton X-100, 0.1% sodium citrate in PBS) for two minutes at 4°C. Fluorescence microscopy (Axiovert 135, Zeiss) was performed after washing with PBS and staining with the DNA specific dye Hoechst 33342 (Sigma, Deisenhofen, Germany) in PBS (10 μg/ml).16 The intercalation of Hoechst 33342 in the DNA allows to distinguish compressed apoptotic from intact DNA.
Proliferation
Proliferation of transiently transfected HCT-116 and LOVO cells was quantified using the AlamarBlue Assay (Serotec, Oxford, Kidlington, UK). The assay works analogous to the MTT (3-(4,5)-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide) assay (De Fries et al17; Goegan et al18). The native, oxidised form of the AlamarBlue reagent is readily taken up by the cells and reduced intracellularly by oxidoreductases and the mitochondrial electron transport chain, with a corresponding shift in its absorbance.17,18 HCT-116 or LOVO cells were plated in 96 wells and transiently transfected with wild type proteins, mutant proteins or pcDNA3.1+ vector without insert as control. At 24, 48, and 96 hours after transfection cells were incubated with the alamarBlue dye (10% alamarBlue in growth medium) for three hours at 37°C. Absorption was monitored at 570/600 nm using an ELISA reader (SLT Rainbow, TECAN, Germany). Baseline readings were assessed from medium and alamarBlue without cells.
Data presentation and statistical analysis
Data are shown either as representative experiments or as mean (SD). Differences in apoptosis and proliferation data were analysed by Fisher’s exact test (BiAS Software version 7.04, Epsilon Verlag, Darmstadt, Germany). All p values are two tailed, and p<0.05 was considered significant.
RESULTS
Expression of mismatch repair proteins in deficient cell lines
hMLH1 expression was performed in the hMLH1 mismatch repair deficient human colon carcinoma HCT-116 cell line containing a homozygous non-sense mutation at codon 252 (stop codon) in exon 9. hMSH2 proteins were expressed in the hMSH2 deficient human colon carcinoma LOVO cell line that has a homozygous deletion in the hMSH2 gene from exon 3 to exon 8.19 The investigated mutants comprise mutations found in four patients with HNPCC: a well characterised hMLH1 mutation (hMLH1 Thr117Met) affecting the essential ATP binding domain in the aminoterminal portions,20 a hMLH1 mutation affecting the hPMS2 interaction domain (hMLH1 Lys618Thr)21 and two hMHS2 mutations, a 1-bp deletion-mutation leading to a truncated hMSH2 protein (hMSH2 DEL782FS), and a hMSH2 missense mutation (hMSH2 Cys697Arg). All expression plasmids were tested to produce protein using a coupled transcription/translation assay.
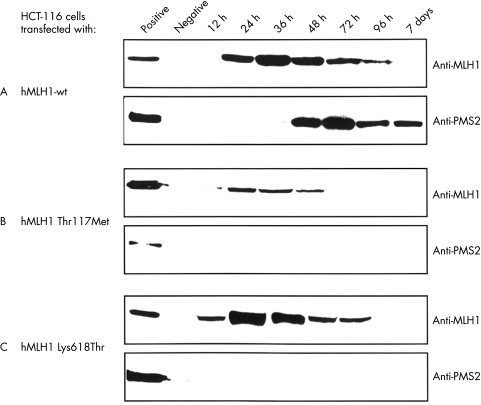
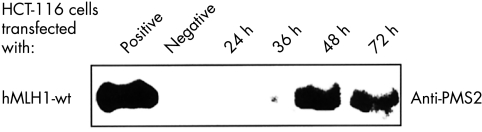
The time course of transient expression of wild type and mutant hMLH1 and hMSH2 was investigated by immunoblot (fig 1 and 2▶▶). Expression started 12 hours after transfection of mutant hMLH1 (fig 1B, C▶), hMSH2 wt, and mutant hMSH2 (fig 2A, B, C▶) and 24 hours after transfection of hMLH1 wt (fig 1A▶). Maximum expression was achieved after 24 hours (hMLH1 Thr117Met and hMLH1 Lys618Thr), 36 hours (hMLH1 wt, hMSH2 Cys697Arg and hMSH2 DEL782FS), and 48 hours (hMSH2 wt). Four to seven days after transfection, wild type and mutant MMR proteins declined to pre-transfection levels. In general, expression of mutant proteins decreased earlier than the expression of wild type proteins (fig 1 and 2▶▶). The expression of hMLH1 Thr117Met and hMSH2 DEL782FS was approximately 20% and 80% lower compared with the expression of hMLH1 wt and hMSH2 wt, respectively (fig 1A, B▶; fig 2A, C▶). However, similar expression levels were observed for hMLH1 wt and hMLH1 Lys618Thr as well as hMSH2 wt and hMSH2 Cys697Arg (fig 1A, C▶; fig 2A, B▶).
Figure 1 .
Expression of hMLH1 in human colorectal cancer cell line HCT-116. HCT-116 cells were transiently transfected either with pcDNA3.1+ vectors expressing hMLH1 wt (A), hMLH1 Thr117Met (B), hMLH1 Lys618Thr (C), or pcDNA3.1+ vector without insert (negative control). Total extracts of transiently transfected HCT-116 cells (20 μg/lane) were separated on a 7.5% SDS-PAGE and blotted onto PVDF membranes. The transferred proteins were visualised with either anti-hMLH1 antibody (upper lane) or anti-hPMS2 antibody (lower lane). hMLH1 proficient LOVO cells were used as positive control. Data shown are representative of four independent experiments.
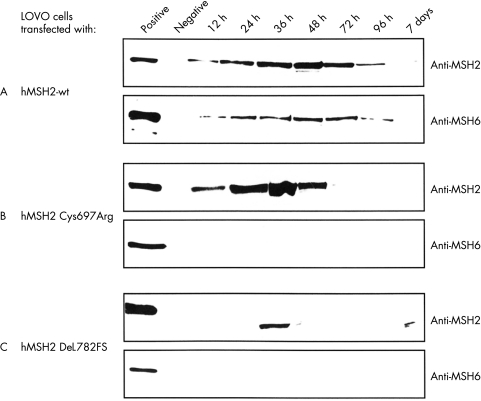
Figure 2 .
Expression of hMSH2 in human colorectal cancer cell line LOVO. LOVO cells were transiently transfected either with pcDNA3+ vectors expressing hMSH2 wt (A), hMSH2 Cys697Arg (B), hMSH2 DEL782FS (C), or pcDNA3+ vector without insert (negative control). Total extracts of transiently transfected LOVO cells (50 μg/lane) were separated on a 7.5% SDS-PAGE and blotted onto PVDF membranes. The transferred proteins were visualised with either anti-hMSH2 antibody (upper lane) or anti-hMSH6 antibody (lower lane). hMSH2 proficient HCT-116 cell were used as positive control. Data shown are representative of four independent experiments.
Stabilisation of interacting mismatch repair proteins
Expression of interacting proteins was analysed to determine functional consequences of transfected proteins. In HCT-116 cells hPMS2 became detectable by western blot analysis approximately 48 hours after transfection of hMLH1 wt gene (fig 1A▶), while hPMS2 remained undectable in cells expressing mutant hMLH1 proteins (fig 1B and 1C▶). The presence of hPMS2 protein was examined in LOVO cells transiently transfected with hMSH2 wt or hMSH2 mutants to obtain further evidence for the specific hMLH1 dependent expression of hPMS2 protein. The transfected plasmids, however, did not affect hPMS2 expression levels (data not shown).
Similarly, hMSH6 was only detectable in LOVO cells expressing hMSH2-wt protein (fig 2▶). The time course of hMSH6 expression was parallel to the expression of hMSH2 protein (fig 2A▶). In contrast, hMSH6 protein was not detectable in LOVO cells expressing mutant hMSH2 proteins (hMSH2 Cys697Arg and hMSH2 DEL782FS) (fig 2B and 2C▶). hMSH6 expression was not affected in HCT-116 cells transiently transfected with hMLH1 wt or hMLH1 mutant constructs, indicating specific hMSH2 dependent expression of hMSH6.
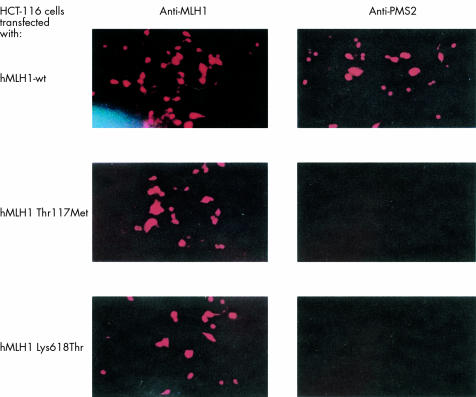
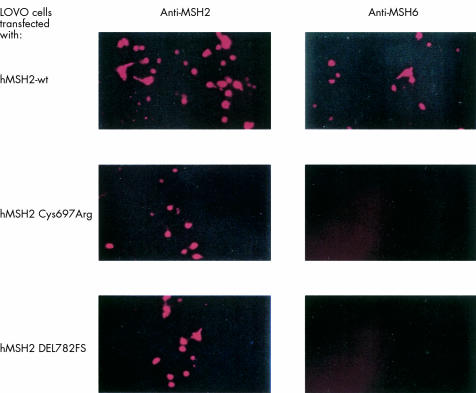
Both, hMLH1 wt and mutant proteins as well as hMSH2 wt and mutant proteins were detected by immunofluorescence in transfected HCT-116 and LOVO cells, respectively, with a similar time course as shown by western blot analysis. hPMS2 protein was detectable by immunofluorescence during expression of hMLH1 wt but not mutant protein in HCT-116 cells (fig 3▶). Similarly, hMSH6 protein was present in LOVO cells complemented with hMSH2 wt, but not in cells transfected with mutant hMSH2 constructs (fig 4▶). None of the respective proteins were detectable in HCT-116 and LOVO cells transfected with an empty pcDNA3.1+ vector (data not shown).
Figure 3 .
Immunofluorescence of hMLH1 deficient tumour cell line HCT-116 after transfection with hMLH1 wt, hMLH1 Thr117Met, or hMLH1 Lys618Thr. Protein expression after 48 hours was detected using anti-hMLH1 (left) or anti-hPMS2 antibody (right). Data shown are representative of four independent experiments.
Figure 4 .
Immunofluorescence of hMSH2 deficient tumour cell line LOVO after transfection with hMSH2 wt, hMSH2 Cys697Arg, or hMSH2 DEL782FS. Protein expression after 48 hours was detected using anti-hMSH2 (left) or anti-hMSH6 antibody (right). Data shown are representative of four independent experiments.
Coimmunoprecipitation of hMLH1-hPMS2 and hMSH2-hMSH6
hMLH1 was immunoprecipitated from HCT-116 cells transfected with hMLH1 wt, hMLH1 Thr117Met, and hMLH1 Lys618Thr using monoclonal anti-hMLH1 antibody. Analysis of the precipitates by western blotting showed the presence of hPMS2 only in HCT-116 cells expressing hMLH1 wt (fig 5▶), while hPMS2 protein was not detectable in precipitates from HCT-116 cells expressing hMLH1 Thr117Met or hMLH1 Lys618Thr and in HCT-116 cells transfected with empty pcDNA3.1+ plasmid (data not shown). These results indicate that hMLH1 wt protein interacts in vivo with hPMS2 to form the heterodimer hMutLα and that mutant hMLH1 loose function to stabilise hPMS2 protein. Experiments using LOVO cell extracts resulted in an increased background signal as a result of polyclonal anti-hMSH2 antibody binding. Using several antibodies, coimmunoprecipitiation was not sufficiently sensitive to detect hMSH2 expression and interaction with hMSH6.
Figure 5 .
Immunoprecipitation of hMLH1. HCT-116 cells were transiently transfected with hMLH1 wt protein and harvested 24, 36, 48, and 72 hours after transfection. Immunoprecipitation was carried out with 5 μg monoclonal anti-hMLH1 antibody. Protein-A-agarose was resuspended in 40 μl SDS loading buffer and protein was resolved by 7.5% SDS-PAGE. hPMS2 was detected by monoclonal antibody. Data shown are mean (SD) of four individual experiments.
Quantitative PCR analysis
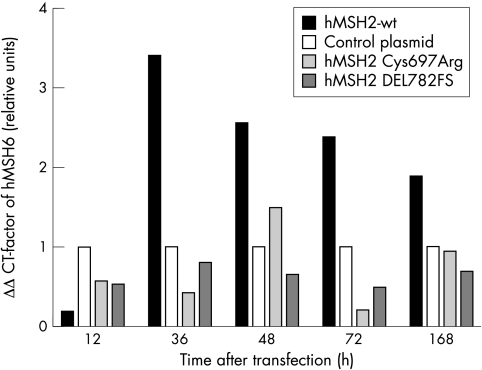
mRNA levels of hPMS2 and hMSH6 were quantified in transiently transfected HCT-116 and LOVO cells to differentiate between reduced transcription and protein stabilisation, respectively. Quantification of hPMS2 mRNA showed no difference at any time point between HCT-116 cells expressing hMLH1 wt or mutant proteins (data not shown). In contrast, two, three, and seven days after transfection, hMSH6 mRNA levels were increased in LOVO cells transfected with hMSH2 wt protein compared with LOVO cells transfected with hMSH2 Cys697Arg or hMSH2 DEL782FS (fig 6▶).
Figure 6 .
Quantitative PCR-analysis. Total RNA of transiently transfected LOVO cells expressing hMSH2 wt protein, hMSH2 Cys697Arg, hMSH2 DEL782FS, or control plasmid were extracted 12, 36, 48, 72 hours and seven days after transfection. hMSH6 mRNA levels were quantified using real time PCR. Results show the ‡‡CT-factor of hMSH6, representing the relative mRNA quantity of sample to control. ‡‡CT-factor was calculated for four independent experiments.
Expression of hMSH2 wt induces apoptosis
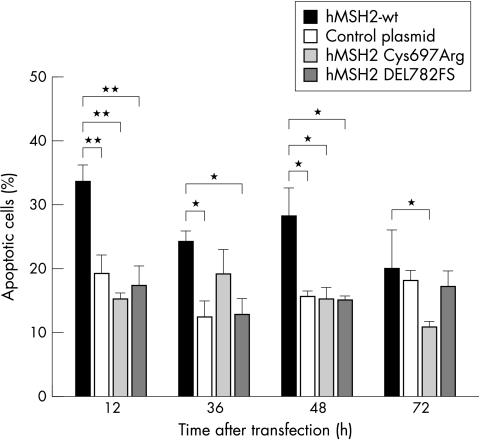
Apoptosis of HCT-116 and LOVO cells expressing hMLH1 wt, hMSH2 wt or mutant proteins was investigated to determine loss of function arising by mutant MMR proteins. Apoptosis of hMLH1 wt or hMLH1 mutant transfected HCT-116 cells was similar (data not shown). However, apoptosis of LOVO cells complemented with hMSH2 wt protein was increased compared with hMSH2 Cys697Arg or hMSH2 DEL782FS transfected LOVO cells (fig 7▶). Parallel to hMSH2 wt protein expression, apoptosis was observed 12 hours to 72 hours after transfection (fig 1A▶).
Figure 7 .
Measurement of apoptosis. LOVO cells were transiently transfected either with hMSH2 wt or mutant hMSH2. Cells were fixed 12, 36, 48, and 72 hours after transfection. Apoptosis was examined by fluorescence microscopy after membrane permeabilisation and staining of the nuclei with Hoechst 33342. Data shown are mean (SD) of four independent experiments. *p<0.05; **p <0.01.
Proliferation in transiently transfected HCT-116 and LOVO cells
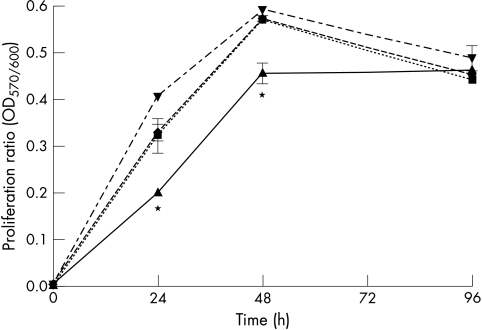
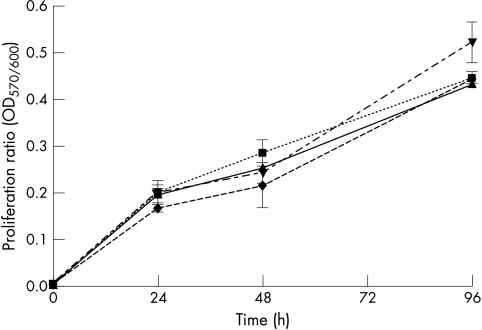
Conversion of AlamarBlue dye, an indicator of mitochondrial enzyme function, was measured to analyse the influence of hMLH1 and hMSH2 on cell proliferation. The assay has been previously validated as a substitute for the 3-(4,5)-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide assay.16,17 Proliferation of HCT-116 cells expressing hMLH1 wt was lower compared with HCT-116 cells transfected with an empty pcDNA3.1+ vector or cells expressing hMLH1 mutants (hMLH1 Thr117Met, hMLH1 Lys618Thr) (fig 8▶). Changes in proliferation of HCT-116 cells were similar to the time course of hMLH1 protein expression (fig 1A▶)—that is, HCT-116 proliferation normalised approximately four days after transfection as hMLH1 wt protein expression declined. Proliferation was not different in LOVO cells transiently transfected with hMSH2 wt, hMSH2 Cys697Arg or hMSH2 DEL782FS mutants (fig 9▶).
Figure 8 .
Proliferative activity of transiently hMLH1 transfected HCT-116 cells. The hMLH1 deficient cell line HCT-116 was transfected with hMLH1 wt (triangles), hMLH1 Thr117Met (inverted triangles), hMLH1 Lys618Thr (diamonds), or control plasmid (squares). The mitochondrial activity as a parameter of proliferation was determined 24, 48, and 96 hours after transfection. Data shown are mean (SD) of four individual experiments. *p<0.05.
Figure 9 .
Proliferative activity of transiently hMSH2 transfected LOVO cells. The hMSH2 deficient LOVO cells were transfected with hMSH2 wt (triangles), hMSH2 Cys697Arg (inverted triangles), hMSH2 DEL782FS (diamonds) or control plasmid (squares). The mitochondrial activity as a parameter of proliferation was determined 24, 48, and 96 hours after transfection. Data shown are mean (SD) of four individual experiments. Differences observed did not achieve statistical significance.
DISCUSSION
The role of mismatch repair proteins was previously analysed in vitro by methods such as whole chromosome transfer, microinjections, and MMR assays, which are less suitable for large scale analyses of HNPCC mutations. Moreover, Umar et al22 indicated difficulties in chromosome complemented MMR deficient cell lines corresponding to unpredictable side effects of different amounts of transferred chromosomes. In contrast, transient transfection of mismatch repair deficient colorectal cancer cell lines with different hMLH1 and hMSH2 cDNA constructs permits direct functional study of mutations found in patients with HNPCC. Thus, in the present study wild type, hMLH1 (hMLH1 Thr117Met and hMLH1 Lys618Thr) and hMSH2 mutants (hMSH2 Cys697Arg and hMSH2 DEL782FS) were cloned and expressed in HCT-116 and LOVO cells, respectively.
MMR protein expression in transiently transfected cells was analysed by western blotting and immunofluorescense. Impaired expression of the hMLH1 Thr117Met mutant and shorter expression of hMLH1 Thr117Met and hMLH1 Lys618Thr mutants compared with hMLH1 wt protein was observed in HCT-116 cells. In LOVO cells, expression of hMSH2 DEL782FS mutant was impaired and expression of hMSH2 Cys697Arg and hMSH2 DEL782FS mutants was shorter compared with hMSH2 wt protein. These results indicate that mutant hMLH1 and hMSH2 proteins are either less stable than the hMLH1 and hMSH2 wt proteins, respectively, or less efficiently synthesised, for example, because of lower mutant MMR mRNA levels. These observations are in accordance with Curia et al who failed to extract and sequence mutant hMLH1 and hMSH2 mRNA but successfully extracted and sequenced hMLH1 wt and hMSH2 wt mRNA from patients with HNPCC.23 The data of this study may explain the inability to detect mutant MMR proteins in human tumour tissue of patients with HNPCC by immunohistochemistry.24,25
In addition, the expression of heterodimeric proteins of hMLH1 and hMSH2 were analysed. hPMS2 protein was restored in HCT-116 cells transfected with hMLH1 wt but not in cells transfected with hMLH1 mutants. hPMS2 mRNA levels as quantified by real time PCR showed no difference in hMLH1 transfected cells indicating that hMLH1 wt protein stabilises hPMS2 protein post-translationally. Moreover, the data shown by western blotting could be verified by coimmunoprecipitation. hPMS2 coimmunoprecipitated with hMLH1 wt but not with mutant hMLH1 proteins that failed to restore the expression of hPMS2. These results are in accordance with data discussed by Chang et al, Curia et al, and Guerrette et al.11,23,25
The interaction of hMSH6 with hMSH2 wt or mutant proteins was investigated in LOVO cells. hMSH6 protein was detectable after expression of hMSH2 wt, but not of hMSH2 mutants. Similiar data were previously reported by whole chromosome transfection experiments.27,28 Quantification of hMSH6 mRNA levels by real time PCR showed reduced hMSH6 mRNA levels in hMSH2 wt transfected LOVO cells indicating transcription associated regulation of hMSH2 and hMSH6. This may further account for the inability to detect hMSH6 in hMSH2 wt deficient cells, although it was previously attributed to post-translational stabilisation of hMSH6 by hMSH2 wt.11 However, hMSH2/hMSH6 interaction could not be demonstrated by coimmunoprecipitation in LOVO cells transfected with hMSH2. Because coimmunoprecipitation of hMSH6 with hMSH2 in MMR proficient cell lines was consistently successful, the efficiency of expression hMSH2 mutants in transfected LOVO cells may be insufficient for coimmunoprecipitation of hMSH6 and hMSH2 protein.
In this study the inability of hMLH1 mutants and hMSH2 mutants to complement deficiency of hMutLα or hMutSα in HCT-116 and LOVO cells, respectively, was demonstrated. It was previously shown in a yeast based hMLH1 functional assay that the investigated hMLH1 mutations alter mismatch repair.29 Moreover, all colorectal tumours of the patients with HNPCC from whom the hMLH1 and hMSH2 mutants were derived showed microsatellite instability.30 This provides evidence that the investigated mutations are associated with a defective DNA mismatch repair system. In this study we further focused on the possible functional consequences of wild type and mutant MMR proteins in the regulation of apoptosis and proliferation.
Apoptosis of LOVO cells was increased by hMSH2 wt but not by hMSH2 mutant expression, while no changes were observed after transfection of HCT-116 cells with hMLH1 wt or mutants. In contrast, Zhang et al showed that apoptosis was induced after microinjection of hMLH1 wt in HCT-116 cells.13 Because the GFP-plasmid was coinjected with the hMLH1 construct in that study, synergistic effects of overexpressed hMLH1 and GFP protein cannot be excluded. Indeed, we have obtained evidence that the GFP-plasmid transfection in colon carcinoma cell lines can induce apoptosis (unpublished data). The results suggesting that hMSH2 wt but not hMLH1 wt protein is required for induction of apoptosis, may have implications for the use of several apoptosis inducing chemotherapeutic agents.31–33
Furthermore, the proliferative activity of MMR transfected cells was investigated. HCT-116 cells complemented by hMLH1 wt protein showed reduced proliferative activity (24 hours and 48 hours after transfection) compared with cells transfected with the control plasmid or hMLH1 mutants. In accordance with Shin and coworkers,14 these data indicate that hMLH1 mutants fail to regulate proliferation. In contrast, no effect on proliferation was observed by complementation with hMLH2 wt or mutants, indicating that hMSH2 is not involved in the regulation of proliferation.
The analysis of microsatellite instability in colorectal tumours has substantially improved the management of patients with HNPCC.34,35 Nevertheless, the clinical role of specific MMR mutations in the progression of disease and for the efficacy of chemotherapy is widely unclear. Our experimental set up for the functional analysis of MMR mutations may be useful to evaluate their clinical significance. Chemotherapeutic agents act by induction of apoptosis or inhibition of proliferation.36 The data of this study show that mutations in the hMLH1 and hMSH2 genes lead to decreased apoptosis and increased proliferation, respectively. Chemotherapeutic agents may directly interact with the mismatch repair system,37–39 for example, hMLH1 but not hMSH2 seems to be involved in processing topoisomerase 1 poison induced damage.40 Our system may be suitable for further in vitro analysis of interaction of chemotherapeutic agents with the MMR system and may help to optimise and individualise treatment.
In conclusion, the data of this study demonstrate that hMLH1 wt protein, but not hMLH1 mutants increase hPMS2 protein levels and decrease proliferation. In contrast, hMSH2 wt protein but not hMSH2 mutants increase hMSH6 mRNA and protein levels as well as apoptosis. Thus, using these well defined transient transfection system the functional role of genetic variants (mutations/polymorphisms) of MMR genes found in HNPCC families can be easily studied.
Acknowledgments
The authors thank Dr Hong Zang (University of Utah, Salt Lake City, Utah) for providing the pcDNA3.1-hMLH1 wild type plasmid and Dr Josef Jiricny (University of Zurich, Zurich, Switzerland) for helpful discussion.
Abbreviations
HNPCC; hereditary non-polyposis colorectal cancer
MMR, mismatch repair
PCR, polymerase chain reaction
PBS, phosphate buffered saline
REFERENCES
- 1.Peltomaki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 1997;113:1146–58. [DOI] [PubMed] [Google Scholar]
- 2.Fishel R, Ewel A, Lescoe MK. Purified human MSH2 protein binds to DNA containing mismatched nucleotides. Cancer Res 1994;54:5539–42. [PubMed] [Google Scholar]
- 3.Palombo F, Galllinari P, Iaccarino I, et al. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science 1995;268:30. [DOI] [PubMed] [Google Scholar]
- 4.Alani E. The Saccharomyces cerevisiae hMSH2 and hMSH6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol Cell Biol 1996;10:5604–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen DJ, Makhov A, Grilley M, et al. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J 1997;16:4467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiricny J. Replication errors: cha(lle)nging the genome. EMBO J 1998;17:6427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raschle M, Marra G, Nystrom-Lahti M, et al. Identification of hMutLbeta, a heterodimer of hMLH1 and hPMS1. J Biol Chem 1999;274:32368–75. [DOI] [PubMed] [Google Scholar]
- 8.Leung WK, Kim JJ, Wu L, et al. Identification of a second MutL DNA mismatch repair complex (hPMS1 and hMLH1) in human epithelial cells. J Biol Chem 2000;275:15728–32. [DOI] [PubMed] [Google Scholar]
- 9.Lipkin SM, Wang V, Jacoby R, et al. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet 2000;24:27–35. [DOI] [PubMed] [Google Scholar]
- 10.Jiricny J. Mediating mismatch repair. Nat Genet 2000;24:6–8. [DOI] [PubMed] [Google Scholar]
- 11.Chang DK, Ricciardiello L, Goel A, et al. Steady-state regulation of the human DNA mismatch repair system. J Biol Chem 2000;275:18424–31. [DOI] [PubMed] [Google Scholar]
- 12.Toft NJ, Winton DJ, Kelly J, et al. MSH2 status modulates both apoptosis and mutation frequency in the murine small intestine. Proc Natl Acad Sci USA 1999;96:3911–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Richards B, Wilson T, et al. Apoptosis induced by overexpression of hMSH2 or hMLH1. Cancer Res 1999;59:3021–7. [PubMed] [Google Scholar]
- 14.Shin KH, Han HJ, Park JG. Growth suppression mediated by transfection of wild-type hMLH1 in human cancer cells expressing endogenous truncated hMLH1 protein. Int J Oncol 1998;12:609–15. [DOI] [PubMed] [Google Scholar]
- 15.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature 1989;339:237–8. [DOI] [PubMed] [Google Scholar]
- 16.Weber GF, Daley J, Kraeft SK, et al. Measurement of apoptosis in heterogenous cell populations. Cytometry 1997;27:136–44 [DOI] [PubMed] [Google Scholar]
- 17.De Fries R, Mitsuhashi M. Quantification of mitogen induced human lymphocyte proliferation: comparison of alamarBlueTM assay to 3H-Thymidine incorporation assay. J Clin Lab Anal 1995;9:89–95. [DOI] [PubMed] [Google Scholar]
- 18.Goegan P, Johnson G, Vincent R. Effects of serum protein and colloid on the AlamarBlue assay in cell cultures. Toxic in vitro 1995;9:267–6. [DOI] [PubMed] [Google Scholar]
- 19.Umar A, Boyer JC, Thomas DC, et al. Defective mismatch repair in extracts of colorectal and endometrial cancer cell lines exhibiting microsatellite instability. J Biol Chem 1994;20:14367–70. [PubMed] [Google Scholar]
- 20.Trojan J, Zeuzem S, Randolph A, et al. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology 2002;122:211–19. [DOI] [PubMed] [Google Scholar]
- 21.Han HJ, Maruyama M, Baba S, et al. Genomic structure of human mismatch repair gene, hMLH1, and its mutation analysis in patients with hereditary non-polyposis colorectal cancer (HNPCC). Hum Mol Genet 1995;4:237–42. [DOI] [PubMed] [Google Scholar]
- 22.Umar A, Koi M, Risinger JI, et al. Correction of hypermutability, N-methyl-N’-nitro-N-nitroguanidine resistance, and defective DNA mismatch repair by introducing chromosome 2 into human tumor cells with mutations in MSH2 and MSH6. Cancer Res 1997;57:3949–55. [PubMed] [Google Scholar]
- 23.Curia MC, Palmirotta R, Aceto G, et al. Unbalanced germ-line expression of hMLH1 and hMSH2 alleles in hereditary nonpolyposis colorectal cancer. Cancer Res 1999;59:3570–5. [PubMed] [Google Scholar]
- 24.Lo Muzio L, Nocini P, Mignogna MD, et al. Immunocytochemical detection of hMSH2 and hMLH1 expression in oral melanoma. Anticancer Res 2000;20:741–8. [PubMed] [Google Scholar]
- 25.Leach FS, Velasco A, Hsieh JT, et al. The mismatch repair gene hMSH2 is mutated in the prostate cancer cell line LNCaP. J Urol 2000;164:1830–3. [PubMed] [Google Scholar]
- 26.Guerrette S, Acharya S, Fishel R. The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem 1999;274:6336–41. [DOI] [PubMed] [Google Scholar]
- 27.Glaab WE, Tindall KR. Mutation rate at the hprt locus in human cancer cell lines with specific mismatch repair-gene defects. Carcinogenesis 1997;18:1–8. [DOI] [PubMed] [Google Scholar]
- 28.Davis TW, Wilson-Van Patten C, Meyers M, et al. Defective expression of the DNA mismatch repair protein, hMLH1, alters G2-M cell cycle checkpoint arrest following ionizing radiation. Cancer Res 1998;15:767–78. [PubMed] [Google Scholar]
- 29.Shimodaira H, Filosi N, Shibata H, et al. Functional analysis of human MLH1 mutations in Saccharomyces cerevisiae. Nat Genet 1998;19:384–9. [DOI] [PubMed] [Google Scholar]
- 30.Raedle J, Brieger A, Trojan J, et al. Evaluation of rapid microsatellite analysis of paraffin-embedded specimens in screening for hereditary nonpolyposis colorectal cancer. Mod Pathol 1999;12:485–91. [PubMed] [Google Scholar]
- 31.de las Alas MM, Aebi S, Fink D, et al. Loss of DNA mismatch repair: effects on the rate of mutation to drug resistance. J Natl Cancer Inst 1997;89:1537–41. [DOI] [PubMed] [Google Scholar]
- 32.Simon JA, Szankasi P, Nguyen DK, et al. Differential toxicities of anticancer agents among DNA repair and checkpoint mutants of Saccharomyces cerevisiae. Cancer Res 2000;60:328–33. [PubMed] [Google Scholar]
- 33.Sobrero A, Kerr D, Glimelius B, et al. New directions in the treatment of colorectal cancer: a look to the future. Eur J Cancer 2000;36:559–66. [DOI] [PubMed] [Google Scholar]
- 34.Brueckl WM, Jung A, Wein A, et al. Microsatellite instability in colorectal adenomas: relevance and clinical importance. Int J Colorectal Dis 2000;15:189–96. [DOI] [PubMed] [Google Scholar]
- 35.Raedle J, Trojan J, Brieger A, et al. Bethesda guidelines: relation to microsatellite instability and MLH1 promoter methylation in patients with colorectal cancer. Ann Intern Med 2001;16:566–76. [DOI] [PubMed] [Google Scholar]
- 36.Easmon J, Puerstinger G, Roth T, et al. 2-benzoxazolyl and 2-benzimidazolyl hydrazones derived from 2-acetylpyridine: a novel class of antitumor agents. Int J Cancer 2001;94:89–96. [DOI] [PubMed] [Google Scholar]
- 37.Larson ED, Drummond JT. Human mismatch repair and G*T mismatch binding by hMutSalpha in vitro is inhibited by adriamycin, actinomycin D, and nogalamycin. J Biol Chem 2001;30:9775–83. [DOI] [PubMed] [Google Scholar]
- 38.Pepponi R, Graziani G, Falcinelli S, et al. hMSH3 overexpression and cellular response to cytotoxic anticancer agents. Carcinogenesis 2001;22:1131–37. [DOI] [PubMed] [Google Scholar]
- 39.Lin X, Ramamurthi K, Mishima M, et al. P53 modulates the effect of loss of DNA mismatch repair on the sensitivity of human colon cancer cells to the cytotoxic and mutagenic effects of cisplatin. Cancer Res 2001;61:1508–16. [PubMed] [Google Scholar]
- 40.Fedier A, Schwarz VA, Walt H, et al. Resistance to topoisomerase poisons due to loss of DNA mismatch repair. Int J Cancer 2001;93:571–76. [DOI] [PubMed] [Google Scholar]