Abstract
Background: Damage to the gastrointestinal mucosa results in the acute up-regulation of the trefoil factor family peptides TFF1, TFF2, and TFF3. They possess protective, healing, and tumour suppressive functions. Little is known about the regulation of TFF gene expression. The promoters of all three TFF genes contain binding sites (E box) for upstream stimulating factor (USF) and Myc/Max/Mad network proteins.
Aims: To determine the nature and function of transcription factors that bind to these E boxes and to understand their role for TFF gene expression.
Methods: TFF promoter activities were determined by reporter gene assays. DNA binding was monitored by electromobility shift assays and by chromatin immunoprecipitation analyses. Expression of endogenous TFF was determined by multiplex RT-PCR.
Results: It was observed that the TFF2 promoter is specifically and efficiently activated by USF transcription factors but not by c-Myc. USF displayed comparable binding to a high affinity Myc/Max binding site compared with the three TFF E boxes, while c-Myc exhibited lower affinity to the TFF E boxes. In contrast, pronounced binding differences were observed in cells with a strong preference for USF to interact specifically with the TFF2 E box, while Myc was not above background. Exogenous expression of USF was sufficient to activate the chromosomal TFF2 and to a lesser extent, the TFF1 gene.
Conclusion: These findings define USF factors as regulators of the TFF2 gene and suggest that promoter specific effects are important for a pronounced gene activation of this cytoprotective peptide.
Keywords: trefoil factor family, trefoil peptide, c-Myc, Max, upstream stimulating factor, chromatin crosslinking and immunoprecipitation
The three members of the trefoil factor gene family (TFF1, TFF2, and TFF3) encode secreted peptides (formerly designated pS2, SP, and ITF) characterised by three loop trefoil domains and expressed in specific epithelial cells of the gastrointestinal tract.1 In vitro and in vivo studies showed that TFFs protect epithelia against experimentally induced mucosal damage.2 In addition, TFFs stimulate cellular motility, promote mucosal defence and wound healing, and inhibit tumour cell proliferation.1,3 TFF2 is expressed early in response to experimentally induced ulceration in animals and has been proposed to be the principal cytoprotective trefoil peptide.4,5 The findings summarised above are supported by the analysis of the corresponding knock-out animals. Tff1-/- mice show aberrant gastric mucosa and develop gastric carcinoma6 whereas Tff3-/- mice exhibit impaired intestinal defence.7 Recently Tff2-/- mice were demonstrated to possess a decreased thickness and proliferation rate of the gastric mucosa and an increased sensitivity to indomethacin induced ulceration.8
Human TFFs are up-regulated near sites of damaged mucosa in ulcerative conditions and in a variety of gut cancers.4,9 Interestingly the expression of TFF1 and TTF2 is down-regulated in intestinal metaplasia and many gastric carcinoma.10 A role in anti-proliferation of TFF1 and TFF3 has also been supported by genetic analyses.3,6 In addition the three human TFF genes are clustered in a region on chromosome 21q22.3 that is frequently deleted in gastric cancer.9,11 This suggests that TFFs may have tumour suppressor activity.
The regulation of TFF gene expression is of considerable interest in respect to the above described functions of TFF peptides. We recently demonstrated activation of the three human TFF genes by the transcription factor HNF-3 and GATA-6,12,13 conserved factors known to potentiate the competence of gastrointestinal differentiation and development.14 In addition we noticed E box elements in all three TFF promoters9 (fig 1 ▶). These are recognised by several different transcriptional regulators, including members of the Myc/Max/Mad network and upstream stimulating factor (USF) that belong to the family of basic region/helix-loop-helix/leucine zipper factors.15 A considerable amount of data links the Myc/Max/Mad network to the control of cell behaviour.16 In particular Myc proteins are strongly associated with proliferation and with tumour development both in humans and in animal model systems.17 The ubiquitously expressed USF1 and USF2 have also been implicated in the control of cellular proliferation.18,19 However unlike Myc, USF proteins have been linked to growth inhibition. In particular transformation of primary cells by c-Myc and an activated Ha-Ras is repressed by both USF1 and USF2.20 Furthermore partial or complete loss of USF transcriptional activity is a common event in breast cancer cell lines.21 Thus Myc/Max and USF/USF complexes may antagonise each other at least in part by competing for the same DNA binding elements.
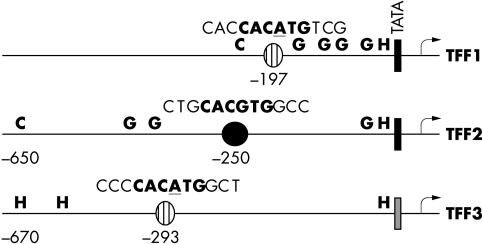
Figure 1.
Schematic representation of the promoters of the three TFF genes. The locations and sequences (deviation from the consensus are underlined) of E box elements in relation to the core promoters are given. Binding sites for C/EBP factors (C), HNF-3 (H), and GATA-6 (G) are indicated. The TATA box in TFF3 is CATAAA.
Our aim was to determine whether the expression of TFF genes is regulated through their E box DNA elements by Myc or USF proteins. We observed that USF, but not c-Myc, activated the TFF2 gene in gastrointestinal cells and bound to the TFF2 promoter in cells.
METHODS
Cell lines and reporter plasmids
The gastric adenocarcinoma cell line MKN45, the colon adenocarcinoma cell lines LS174T and HT-29, and COS-7 cells were cultivated as described.22,23 Reporter gene constructs were generated by cloning the promoter regions of human TFF1 (position −1100 to +38), TFF2 (position −821 to +61), and TFF3 (position −867 to +63) in front of luciferase genes.22 Site directed mutagenesis eliminated the TFF2-luc13 E box (from 5’-TGCACGTGGC to 5’-TGCA-G-GGC) resulting in mE-TFF2-luc. The M4-mintk-luc reporter gene has been described previously.24 A renilla luciferase reporter gene (pRL-CMV, Promega) was used to standardise for transfection efficiency.
Transient transfection assays
Transient transfections were performed as described before.22 For cotransfection experiments, 320 ng of reporter plasmid and the indicated amounts of the following expression plasmids were used: pUHD-USF1 and pUHD-USF1mutbr 25; psvUSF2 was kindly provided by M Sawadogo.20 Expression plasmids for c-Myc (pCMV-hu-c-myc), Mad1 (pCMV-mad1) and Maxp22 (pSP-maxp22) have been described previously.23 For the analysis of endogenous TFF expression, cells were seeded on 24 well plates, transfected with 1 μg of plasmids encoding USF1 and USF2 and total RNA was prepared 48 hours later. Multiplex PCR with TFF1, TFF2, TFF3, and GAPDH primer pools were performed as described.13,26 Transient transfection of COS-7 cells were performed as described.23
Chromatin crosslinking and immunoprecipitation (ChIP)
ChIP assays were performed as described previously.27,28 The following antibodies were used: anti-c-Myc (N-262), anti-Max (C-19), anti-c-Myb (H141), anti-USF-1 (C-20), anti-USF-2 (C-20) (all from Santa Cruz), and polyclonal sera specific for USF1 or USF2 obtained from M Sawadogo.
The following PCR primers were used:
hTFF1-f: 5’-GGCCTCTCAGATATGAGTAG,
hTFF1-r: 5’-TCCTCTGAGACAATAATCTCC,
hTFF2-f: 5’-TGTGGTCCCTGCCCACTC,
hTFF2-r: 5’-TCTCCCTGCTCGGTGATAC,
hTFF3-f: 5’-GGCTCTCTTGTCATGGGAC,
hTFF3-r: 5’-AAGCGGTAAGGGCGGATTC,
hADA-1: 5’-CCCTCCTCCTTTTTGTCTTCCTG,
hADA-2: 5’-GAAACTCAGTCTCCTTTGTTCCCC,
ODC-h1: 5’-GAGCAGAGCGCACCGGGATCA,
ODC-h2: 5’-CAGTACCTCGTGCCCGAGAGC,
eIF2α-h1: 5’-ttctcggaggacccagactctatg,
eIF2α-h2: 5’-tcacagagaccagacttgcttccc.
Electrophoretic mobility shift assay (EMSA)
EMSA were performed as described23 using the following double stranded oligonucleotides:
CMD: 5’-TCAGACCACGTGGTCGGG,
TFF1: 5’-GATGACCTCACCACATGTCGTCTC,
TFF2: 5’-CAGACCTGCACGTGGCCGGTTTTC,
TFF3: 5’-CTGCCACCCCACATGGCTCCTGCAC.
RESULTS
Activation of TFF reporter genes by USF
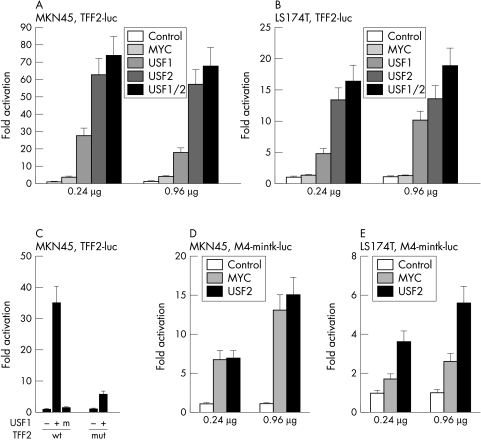
In gastric MKN45 cells, in colonic LS174T cells, and in several other gastrointestinal cell lines a strong activation of a TFF2 reporter gene construct by USF was observed whereas c-Myc showed little activity (fig 2A and B ▶ and data not shown). In both cell lines USF1 activated less efficiently than USF2 while coexpression of both USF proteins resulted in higher activation than either factor alone. This activation was specific as mutation of the E box strongly decreased activation (fig 2C ▶). Furthermore, a mutant of USF1 that cannot bind DNA did not activate the TFF2 reporter (fig 2C ▶).
Figure 2.
USF factors activate a TFF2-luc reporter gene construct. (A) The gastrointestinal cell line MKN45 was cotransfected with the TFF2-luc reporter gene construct and indicated amounts of expression plasmids encoding USF1, USF2, USF1/USF2, or c-Myc. (B) As in (A) but the cell line LS174T was used. (C) MKN45 cells were cotransfected with wild type (wt) or E box mutated (mut) TFF2-luc, with (+) or without (-) expression plasmids encoding USF1 or a mutant USF protein that cannot bind DNA (m). (D) As in (A) but the M4-mintk-luc reporter gene construct with four high affinity Myc/Max binding sites was used. (E) As in B but with the M4-mintk-luc reporter gene.
To determine whether the difference between the activation of TFF2 by USF and by c-Myc was promoter specific or cell line dependent, we measured the activity of these factors on M4-mintk-luc, a reporter gene with 4 high affinity Myc/Max binding sites (equivalent to the CMD oligonucleotide, see below). In MKN45 cells activation by USF and c-Myc was comparable while in LS174T cells USF was more active than c-Myc (fig 2D and E ▶). This suggested that c-Myc is capable of activating transcription to an extent that is comparable to other cell systems (see, for example, Sommer et al24) and that strong activation by USF appears specific for the TFF2 promoter.
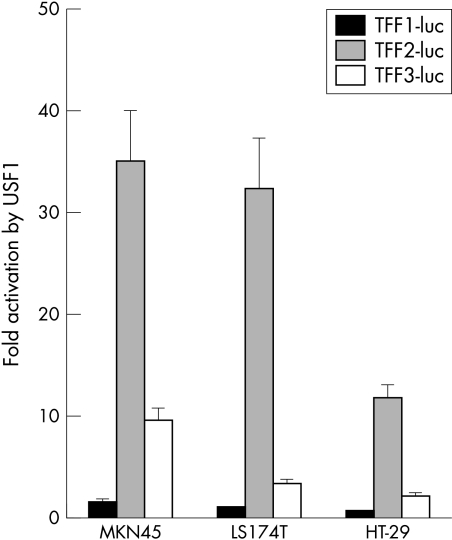
In addition to TFF2 we tested whether the two other TFF promoters were responsive to USF and Myc/Max/Mad proteins. Moderate activation of the TFF3 reporter gene was observed in three different cell lines (MKN45, LS174T, and HT-29) with USF1 while the TFF1 reporter gene was not activated (fig 3 ▶). c-Myc activated neither the TFF1 nor the TFF3 reporter gene (data not shown). Furthermore, other components of the Myc/Max/Mad network, including Max and Mad1, did not affect expression of any of the TFF reporter genes (data not shown). Together these findings identify USF proteins as potential activators of the TFF2 gene.
Figure 3.
Effects of USF1 on TFF1-luc and TFF3-luc. MKN45, LS174T, and HT-29 cells were cotransfected with the indicated TFF reporter gene constructs in the presence or absence of USF1. The fold induction by USF1 is shown.
Distinct reporter gene activation is not attributable to differences in DNA binding
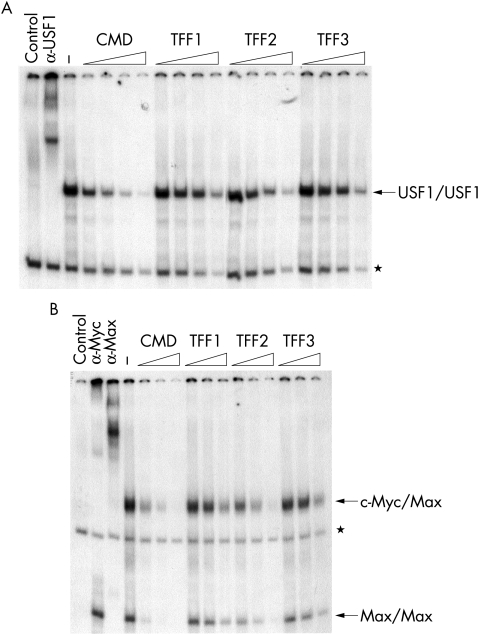
The difference in regulation of the three TFF reporters by USF and Myc/Max/Mad network members might be the result of distinct binding to the respective E boxes. Therefore binding of USF1/USF1, c-Myc/Max, and Max/Max complexes derived from COS-7 cells to CMD was compared in EMSA.23 Relative binding was determined in competition experiments that revealed comparable binding affinities of USF to the three TFF E box sequences whereas binding to CMD was slightly stronger (fig 4A ▶). Thus DNA binding is unlikely to be the reason for the differences in TFF reporter gene and M4-mintk-luc activation. c-Myc/Max and Max/Max complexes bound less efficiently to any of the three TFF derived E box elements than CMD (fig 4B ▶). Therefore weak binding of these complexes might explain in part the inability of c-Myc to activate the TFF reporter genes.
Figure 4.
Binding of USF, c-Myc/Max, and Max/Max complexes to TFF derived E box sequences. (A) Binding of COS-7 derived USF1/USF1 homodimers to radiolabelled CMD was competed with a 3-fold, 10-fold, 30-fold, or 100-fold excess of unlabelled CMD, or oligonucleotides spanning the E boxes of either TFF1, TFF2, or TFF3 as indicated. Supershift experiments were performed with an antibody specific for USF1 (α-USF1). Control: extract of mock transfected cells was used. * Non-specific complex. (B) Binding of COS-7 derived c-Myc/Max and Max/Max complexes were analysed as in (A) Supershift experiments were done with specific antibodies to c-Myc or Max (α-Myc or α-Max, respectively). The control is as in (A). * Non-specific complex.
The TFF2 promoter is occupied and regulated by USF but not c-Myc
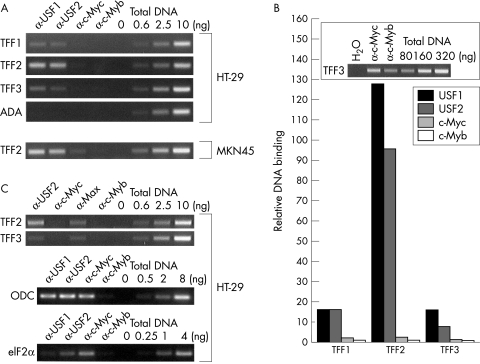
To determine whether endogenous USF is associated with the chromosomal TFF2 promoter, we performed chromatin immunoprecipitation (ChIP) experiments.28 We observed strong signals for USF1 and USF2 on the TFF2 promoter while binding to TFF1 and TFF3 was considerably weaker (fig 5A ▶). Rough quantification was obtained by comparing the signals from the ChIP with a dilution series of sonicated total input DNA. USF1 binding to TFF2 was about eightfold higher than binding to TFF1 and TFF3 whereas USF2 binding was about sixfold to 10-fold stronger (fig 5A and B ▶). No binding of USF factors was detectable to the locus control region of the ADA gene,29 which served as control (fig 5A ▶). The binding of c-Myc to either of the three TFF promoters was not significantly above the non-specific c-Myb control (fig 5B ▶) despite considerable expression of c-Myc in both cell lines (data not shown). In contrast, comparable binding of c-Myc and USF was observed to the E box of the ODC promoter and c-Myc binding was stronger than USF binding to the E box of the eIF2α promoter16 (fig 5C ▶). Interestingly Max was found on the TFF2 and TFF3 E boxes suggesting that other Myc/Max/Mad network complexes can bind to these promoters (fig 5C ▶). This defines specific interaction of USF factors preferentially with the TFF2 promoter.
Figure 5.
Binding of USF to the TFF2 promoter in cells. (A) ChIP analyses were performed on lysates of formaldehyde crosslinked HT-29 and MKN45 cells. One of 20 of the purified DNA from individual immunoprecipitations was subjected to PCR analysis (34 cycles). Fourfold dilution series of purified input DNA served as normalisation controls for the PCR reactions. (B) Quantification of ChIP reactions from HT-29 cells. Product intensities of samples and adequate twofold dilution series of purified input DNA were compared. Relative DNA binding defines the amount of PCR product obtained from immunoprecipitates compared with total input DNA with the amount of PCR product equivalent to 80 pg arbitrarily set as 1. The graph shows the mean value of two independent determinations. Insert: Lower amounts of input DNA were used to determine c-Myc and c-Myb binding equivalents (39 cycles). (C) ChIP analyses were performed as described under (A).
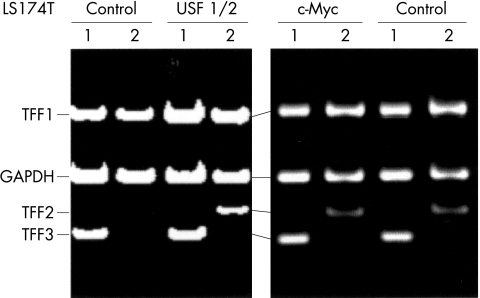
Finally, we determined whether USF factors could activate the endogenous TFF2 gene. USF1 and USF2 were expressed transiently and the expression of endogenous TFF genes monitored by multiplex RT-PCR. USF1/USF2 but not c-Myc were sufficient to activate the endogenous, chromosomal TFF2 gene in LS174T cells as well as other cell lines while little or no effect on TFF1 and TFF3 was observed (fig 6 ▶ and data not shown). Together our findings identify USF as a critical and specific modulator of TFF2 transcription.
Figure 6.
Exogenous USF stimulates the expression of the chromosomal copy of TFF2. LS174T cells were transiently transfected with empty vector or with plasmids encoding USF1 and USF2 or c-Myc as indicated. Endogenous TFF mRNA expression was analysed after 24 cycles of multiplex RT-PCR. Lanes 1: TFF1-PCR, GAPDH-PCR, TFF3-PCR reactions; lanes 2: TFF1-PCR, GAPDH-PCR, TFF2-PCR reactions.
DISCUSSION
We have identified the TFF2 gene as a target for USF transcription factors. An E box DNA sequence element in the promoter of the TFF2 gene mediates this activation. Despite E box elements in TFF1 and TFF3 no significant stimulation by USF was observed. In addition, the Myc/Max/Mad network members c-Myc and Mad1 were unable to regulate any of the TFF genes. Thus it seems that the TFF2 promoter is unique among the TFF genes in that it supports strong activation by USF.
Our studies using EMSA suggest that this is most probably not attributable to differences in DNA binding affinity (fig 4 ▶). In contrast with these in vitro studies, DNA binding analyses in cells by ChIP showed striking differences. USF binding was predominantly found on TFF2 while binding to TFF1 and TFF3 was low (fig 5 ▶). Binding of c-Myc to any of these three genes was not significantly above background (fig 5 ▶). Thus DNA binding selectivity is considerably more pronounced in cells than in vitro offering a mechanistic basis for the observed specificity. This might reflect distinct accessibility to chromatin embedded binding sites or cooperative DNA binding effects resulting from interaction with other as yet unidentified transcriptional regulators.
Previous studies have shown that USF can activate a number of different promoters in reporter gene assays in many different cell types. In general this effect is rather modest (threefold to fivefold)24,30,32 while the activation of the TFF2 reporter is significantly higher (fig 2 ▶). Most probably this is attributable to cooperative effects with other transcriptional regulators. Indeed, cooperation of USF with other factors, including Egr-1, STAT1, and cAMP response element binding proteins, has been described.24,33,34 We postulate that these function at least in part in a tissue specific manner. This is indicated by the high tissue specific expression of TFF2, predominantly in the antral glands and the Brunner’s glands of the duodenum,4 whereas USF is expressed ubiquitously.
The endogenous levels of USF in LS174T as well as several other cell lines tested seem not to be sufficient to support high TFF2 expression22 despite the ability of USF to bind to the promoter of the TFF2 gene (fig 5 ▶ and data not shown). It is possible that the cooperative activation is inefficient in tumour cell lines because of insufficient factors available, which can be compensated by overexpressing USF (fig 6 ▶), or because of the lack of or the reduced sensitivity to activating signals. Indeed, the TFF2 gene is regulated in response to a number of different insults, to drugs such as 5-amino salicylic acid used for treatment of inflammatory bowel disease (EA and PG, unpublished observation) or aspirin and to circadian rhythm and food intake.35,36 TFF2 expression is predominantly controlled by transcriptional initiation,13,22,37 but it remains largely unclear how these stimuli affect the activity of the TFF2 promoter. USF might be involved in recognising stress signals38 or cooperate with factors targeted during signalling events, or both.24 As USF constitutes a major E box binding activity in most cell lysates, its interaction with the TFF2 promoter might be a prerequisite for signal stimulated activation. In addition regulatory elements that are not located within the reporter constructs could be important. This may be relevant for TFF1. In contrast with the reporter gene, the endogenous TFF1 gene was activated to a small extent (figs 3 and 6 ▶ ▶) suggesting the presence of activities that may cooperate with USF in regulating this gene.
USF is known to evoke anti-proliferative effects.18 In this respect it is noteworthy that several USF target genes, including BRCA2, p53, and transforming growth factor β2, are involved in inhibition of cell proliferation.18 The identification of USF as a potent regulator of TFF2 is in line with a more general role of this transcription factor in negative growth control. Together the data summarised above suggest that defining the control of TFF2 expression in more detail will potentially generate opportunities for therapeutic approaches of gastrointestinal diseases.
Acknowledgments
We thank J Lüscher-Firzlaff for the data in fig 5C and P ▶ Fegert and W Kornberger for excellent technical assistance. We are indebted to M Eilers, M Sawadogo, and P Pognonec for providing expression vectors and antisera and to R Paro for the ChIP protocol. This work was supported by the German Cancer Foundation (Deutsche Krebshilfe 10–1547Gö-2) to PG, the Interdisciplinary Centre of Clinical Research (IZKF) Tübingen to NB, and the DFG (Lu 466/1–4, GRK 139), and the Fonds der Chemischen Industrie to BL.
Abbreviations
USF, upstream stimulating factor
TFF, trefoil factor family
ChIP, chromatin crosslinking and immunoprecipitation
EMSA, electrophoretic mobility shift assay
REFERENCES
- 1.Thim L. Trefoil peptides: from structure to function. Cell Mol Life Sci 1997;53:888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran CP, Cook GA, Yeomans ND, et al. Trefoil peptide TFF2 (spasmolytic polypeptide) potently accelerates healing and reduces inflammation in a rat model of colitis. Gut 1999;44:636–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uchino H, Kataoka H, Itoh H, et al. Overexpression of intestinal trefoil factor in human colon carcinoma cells reduces cellular growth in vitro and in vivo. Gastroenterology 2000;118:60–9. [DOI] [PubMed] [Google Scholar]
- 4.Wong WM, Poulsom R, Wright NA. Trefoil peptides. Gut 1999;44:890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Playford RJ, Marchbank T, Chinery R et al. Human spasmolytic polypeptide is a cytoprotective agent that stimulates cell migration. Gastroenterology 1995;108:108–16. [DOI] [PubMed] [Google Scholar]
- 6.Lefebvre O, Chenard MP, Masson R et al. Gastric mucosa abnormalities and tumorigenesis in mice lacking the pS2 trefoil protein. Science 1996;274:259–62. [DOI] [PubMed] [Google Scholar]
- 7.Mashimo H, Wu DC, Podolsky DK et al. Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science 1996;274:262–5. [DOI] [PubMed] [Google Scholar]
- 8.Farrell JJ, Taupin D, Koh TJ et al. TFF2/SP-deficient mice show decreased gastric proliferation, increased acid secretion, and increased susceptibility to NSAID injury. J Clin Invest 2002;109:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gött P, Beck S, Machado JC, et al. Human trefoil peptides: Genomic structure in 21q22.3 and coordinated expression. Eur J Hum Genet 1996;4:308–15. [DOI] [PubMed] [Google Scholar]
- 10.Nogueira AMMF, Machado JC, Carneiro F, et al. Patterns of expression of trefoil peptides and mucins in gastric polyps with and without malignant transformation. J Pathol 1999;187:541–8. [DOI] [PubMed] [Google Scholar]
- 11.Park WS, Oh RR, Park JY, et al. Mapping of a new target region of allelic loss at 21q22 in primary gastric cancers. Cancer Lett 2000;159:15–21. [DOI] [PubMed] [Google Scholar]
- 12.Beck S, Sommer P, Dos Santos Silva E, et al. Hepatocyte nuclear factor 3 (winged helix domain) activates trefoil factor gene TFF1 through a binding motif adjacent to the TATAA box. DNA Cell Biol 1999;18:157–64. [DOI] [PubMed] [Google Scholar]
- 13.Al-azzeh E, Fegert P, Blin N, et al. Transcription factor GATA-6 activates expression of gastroprotective trefoil genes TFF1 and TFF2. Biochim Biophys Acta 2000;1490:324–32. [DOI] [PubMed] [Google Scholar]
- 14.Zaret K. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev Biol 1999;209:1–10. [DOI] [PubMed] [Google Scholar]
- 15.Lüscher B, Larsson LG. The basic region/helix-loop-helix/leucine zipper domain of Myc proto-oncoproteins: function and regulation. Oncogene 1999;18:2955–66. [DOI] [PubMed] [Google Scholar]
- 16.Grandori C, Cowley SM, James LP, et al. The Myc/Max/Mad Network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 2000;16:653–99. [DOI] [PubMed] [Google Scholar]
- 17.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene 1999;18:3004–16. [DOI] [PubMed] [Google Scholar]
- 18.Qyang Y, Luo X, Lu T, et al. Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol Cell Biol 1999;19:1508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirito M, Lin Q, Deng JM, et al. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc Natl Acad Sci 1998;95:3758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo X, Sawadogo M. Functional domains of the transcription factor USF2: atypical nuclear localization signals and context-dependent transcriptional activation domains. Mol Cell Biol 1996;16:1367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ismail PM, Lu T, Sawadogo M. Loss of USF transcriptional activity in breast cancer cell lines. Oncogene 1999;18:5582–91. [DOI] [PubMed] [Google Scholar]
- 22.Beck S, Sommer P, Blin N, et al. 5’-flanking motifs control cell specific expression of trefoil factor genes (TFF). Int J Mol Med 1998;2:353–61. [DOI] [PubMed] [Google Scholar]
- 23.Sommer A, Bousset K, Kremmer E, et al. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J Biol Chem 1998;273:6632–42. [DOI] [PubMed] [Google Scholar]
- 24.Sommer A, Burkhardt H, Keyse SM, et al. Synergistic activation of the mkp-1 gene by protein kinase A signaling and USF, but not c-Myc. FEBS Lett 2000;474:146–50. [DOI] [PubMed] [Google Scholar]
- 25.Desbarats L, Gaubatz S, Eilers M. Discrimination between different E-box-binding proteins at an endogenous target gene of c-myc. Genes Dev 1996;10:447–60. [DOI] [PubMed] [Google Scholar]
- 26.Dos Santos Silva E, Ulrich M, Döring G, et al. Trefoil factor family-domain peptides in the human respiratory tract. J Pathol 2000;190:133–42. [DOI] [PubMed] [Google Scholar]
- 27.Strutt H, Paro R. Mapping DNA target sites of chromatin proteins in vivo by formaldehyde crosslinking. Methods Mol Biol 1999;119:455–67. [DOI] [PubMed] [Google Scholar]
- 28.Bouchard C, Dittrich O, Kiermaier A, et al. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev 2001;15:2042–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ess KC, Whitaker TL, Cost GJ, et al. A central role for a single c-Myb binding site in a thymic locus control region. Mol Cell Biol 1995;15:5707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis PL, Miron A, Andersen LM, et al. Isolation and initial characterization of the BRCA2 promoter. Oncogene 1999;18:6000–12. [DOI] [PubMed] [Google Scholar]
- 31.Outram SV, Owen MJ. The helix-loop-helix containing transcription factor USF activates the promoter of the CD2 gene. J Biol Chem 1994;269:26525–30. [PubMed] [Google Scholar]
- 32.Wang D, Sul HS. Upstream stimulatory factor binding to the E-box at -65 is required for insulin regulation of the fatty acid synthase promoter. J Biol Chem 1997;272:26367–74. [DOI] [PubMed] [Google Scholar]
- 33.Carrasco-Serrano C, Campos-Caro A, Viniegra S, et al. GC- and E-box motifs as regulatory elements in the proximal promoter region of the neuronal nicotinic receptor alpha7 subunit gene. J Biol Chem 1998;273:20021–8. [DOI] [PubMed] [Google Scholar]
- 34.Muhlethaler-Mottet A, Di Berardino W, Otten LA, et al. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity 1998;8:157–66. [DOI] [PubMed] [Google Scholar]
- 35.Azarschab P, Al-azzeh E, Kornberger W, et al. Aspirin promotes TFF2 gene activation in human gastric cell lines. FEBS Lett 2001;488:206–10. [DOI] [PubMed] [Google Scholar]
- 36.Semple JI, Newton JL, Westley BR, et al. Dramatic diurnal variation in the concentration of the human trefoil peptide TFF2 in gastric juice. Gut 2001;48:648–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lüdeking A, Fegert P, Blin N, et al. Osmotic changes and ethanol modify TFF gene expression in gastrointestinal cell lines. FEBS Lett 1998;439:180–4. [DOI] [PubMed] [Google Scholar]
- 38.Andrews GK. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem Pharmacol 2000;59:95–104. [DOI] [PubMed] [Google Scholar]