Abstract
Background and aims: Oesophageal cell lines derived from malignancies have numerous genetic abnormalities and therefore are of limited value for studying the early events in carcinogenesis. Reported attempts to establish normal human oesophageal cell lines either have failed to achieve immortalisation or have achieved it by disrupting important cell functions. We have used telomerase technology to establish normal human oesophageal cell lines.
Methods: Endoscopic biopsy specimens of normal oesophageal squamous epithelium were trypsinised, dispersed into single cell suspensions, and cocultivated with ATCC Swiss 3T3 cells. Oesophageal cells were infected with the catalytic subunit of human telomerase (hTERT) using a defective retroviral vector. The integrity of cell cycle checkpoints was tested by measuring p53 response to UV irradiation, and p16 response to infection with H-RasGV12. Expression of a differentiation marker was tested by measuring involucrin response to calcium exposure.
Results: Cultures of uninfected oesophageal cells had weak telomerase activity at baseline but exhibited loss of telomerase activity and progressive telomere shortening before undergoing senescence between population doublings (PD) 40–45. In contrast, hTERT infected cells exhibited sustained telomerase activity and stabilisation of telomere length. These cells have reached PD 100 with no diminution in growth rate, while cell cycle checkpoint integrity and involucrin response to calcium exposure have remained intact.
Conclusions: By introducing telomerase into normal human oesophageal squamous cells cocultivated with feeder layers, we have established a cell line that retains normal cell cycle checkpoints and normal differentiation markers. This cell line may be useful for studying the early events in oesophageal carcinogenesis.
Keywords: telomerase, oesophagus, squamous cells
Oesophageal squamous cell carcinoma develops from the stepwise accumulation of genetic alterations in oesophageal squamous epithelial cells. Although consumption of alcohol, tobacco smoking, and exposure to N-nitroso compounds have been linked to the development of oesophageal squamous cell carcinoma, the downstream molecular effects of these agents at the cellular level have not been well defined. Abnormalities of p53, cyclin D, epidermal growth factor, and p16 have been described in oesophageal carcinomas1–6 and several oesophageal cancer cell lines have been developed for study. However, frankly malignant oesophageal cells are of limited utility for investigating the early genetic changes of carcinogenesis. The development of a normal human oesophageal epithelial cell line would be of considerable value for the in vitro study of molecular mechanisms involved in neoplastic transformation. These reagents would be especially well suited for studies on the molecular effects of environmental toxins. Recent advances in understanding the role of telomeres in the regulation of cellular proliferative capacity have provided novel in vitro approaches for immortalising normal human cells.
The proliferative capacity of most normal human somatic cells in culture is a function of telomere length.7–9 Telomeres, located at the ends of each chromosome, contain many kilobases of non-coding TTAGGG repeats.10 Due to the inability of DNA polymerases to completely replicate the extreme 3′ chromosome ends, telomeric repeats are lost with each round of cell division.11,12 Eventually, one or more short telomeres triggers entry into a permanent growth arrest state known as cellular senescence.8,11 Telomerase is a ribonucleoprotein that utilises its RNA as a template for the de novo addition of telomeric DNA repeats to chromosome ends.13 Although most normal somatic cells lack telomerase activity, 85–90% of human cancers express high levels of the enzyme,14 suggesting that telomerase reactivation may be a major mechanism by which most human cancers become immortal. Consistent with this hypothesis, several groups have shown that the introduction of the catalytic subunit of human telomerase (hTERT) into cultures of normal human fibroblasts, retinal pigmented epithelial cells, myometrial cells, mammary epithelial cells, endothelial cells, and skin keratinocytes results in stabilisation of telomere length and immortalisation.15–19 We have recently demonstrated that normal human epidermal keratinocytes immortalised by ectopic hTERT expression maintain properties characteristic of normal keratinocytes, including appropriate p53 and p16 cell cycle checkpoint responses and the ability to undergo differentiation.18 Taken together, these results suggest that telomere maintenance may be an excellent strategy for immortalising normal human epithelial cells without affecting the integrity of their important phenotypic characteristics.
Several groups have attempted to establish normal human oesophageal cell lines with limited success.20–27 Compton et al established primary cultures of normal human oesophageal keratinocytes by cocultivation with 3T3 fibroblast feeder cells.23 However, the epithelial cells growth arrested after undergoing only four passages (approximately 20 population doublings) in culture. Inokuchi et al used a recombinant SV40 large T antigen expressing adenoviral vector to immortalise human oesophageal epithelial cells.24 The immortalised cells lost some of their original properties, such as the ability to induce expression of involucrin in response to calcium. Such effects may have been due to the abrogation of key cell cycle checkpoint proteins such as p53 and pRb (or other factors) by the introduced SV40 T antigen. We now report the generation of a line of normal human oesophageal squamous cells obtained from endoscopic biopsy specimens by the stable expression of hTERT. Telomerase expressing oesophageal squamous cells have undergone at least a threefold extension of lifespan compared with controls while maintaining intact p53 and Rb/p16 cell cycle checkpoints as well as differentiation functions.
MATERIALS AND METHODS
Isolation of oesophageal squamous cells
The protocol was approved by the institutional review board on human studies at the Dallas VA Medical Center. Patients previously scheduled to undergo upper endoscopy for an appropriate clinical indication were invited to participate in the study. Patients with Barrett’s oesophagus, oesophagitis, or any other endoscopically obvious oesophageal mucosal abnormalities were excluded from the study. Twelve random biopsies of endoscopically normal squamous oesophagus were obtained from each patient during endoscopy using jumbo forceps, placed into cold Hank’s salt solution containing 1% antibiotic-antimycotic (both from Invitrogen, Carlsbad, California, USA), and processed within two hours of collection. Ten biopsies were placed into a fresh conical tube containing trypsin (Invitrogen) at 37°C for 30 minutes. Two biopsies were snap frozen in liquid nitrogen and stored at −80°C for analysis of telomere length and telomerase activity (see below). Following trypsinisation, biopsies were dispersed into a single cell suspension and cocultivated with ATCC Swiss 3T3 cells (∼1–3×106 cells/100 mm dish) treated with mitomycin C (10 μg/ml for two hours), as previously described.18,28,29
Culture media and methods
Oesophageal squamous cells were grown in a 3:1 mixture of Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F12 medium (Invitrogen) supplemented with 5% fetal bovine serum (Atlanta Biologicals), 0.4 μg/ml hydrocortisone, 5 μg/ml transferrin, 2×10−11 M 3, 3, 5-′triiodo-L-thyronine, 5 μg/ml insulin, 20 ng/ml recombinant epidermal growth factor, 180 μM adenine (all from Sigma), 10−10 M cholera toxin (Calbiochem, San Diego, California, USA), 100 units/ml penicillin, and 0.01 mg/ml streptomycin (Invitrogen). After removing feeder cells from dishes with EDTA (0.02% in 1×PBS, Sigma), 80% confluent squamous cells were trypsinised and resuspended in fresh medium containing 5% trypsin inhibitor (Sigma, St Louis, Missouri, USA). Cells were reseeded at a 1:8 split ratio and grown at 37°C in 5% CO2 with fresh media changes three times weekly. The number of population doublings (PD) per passage was calculated as log (number of cells harvested/number of cells plated)/log 2. On average, oesophageal cells underwent three PD per passage and had a doubling time of 48 hours.
Retroviral vectors and transductions
Oesophageal squamous cells were infected with hTERT using the Cre-lox recombination system, as previously described.30 Retroviral vector particles were generated by transfecting 30 μg of vector DNA into PhoenixE cells using Fugene (Roche Diagnostics, Indianapolis, Indiana, USA) according to the manufacturer’s instructions. Supernatants collected 48 hours after transfection were used to infect the amphotrophic packaging cell line PA317 (ATCC, CRL-9078). After a seven day selection with puromycin (3 μg/ml), supernatants containing the retroviral particles were harvested for infections and filtered through a 0.45 μm filter. Oesophageal cells at approximately 30–50% confluency growing in 100 mm plates in the presence of feeder layers were infected in the presence of 4 μg/ml of Polybrene (Sigma) for 10–12 hours. Seventy two hours later cells were selected with puromycin.
Telomerase activity and telomere length analysis
Telomerase activity was measured in the original biopsies and in cultured cells before and after introduction of hTERT using the TRAP-eze Telomerase Detection kit (Intergen, Burlington, Massachusetts, USA) as previously described.31 A telomerase expressing oesophageal adenocarcinoma cell line and lysis buffer only served as positive and negative controls, respectively. Telomerase activity was quantitated relative to a 36 base pair internal standard by determining the ratio of the signal from the entire telomerase ladder to the internal standard (ITAS), and the result was expressed as a per cent of levels in the positive control samples. The presence of TAQ polymerase inhibitors in any sample was excluded by amplification of the ITAS.
Mean telomere length was estimated by telomere restriction fragment analysis, a modification of Southern analysis, as previously described.30 Briefly, genomic DNA was digested to completion with a six-enzyme mixture comprising equal parts AluI, CfoI, HaeIII, HinfI, MspI, and RsaI (all from Roche). Digested samples (1–2 μg) were fractionated on a 0.7% agarose gel. The gel was denatured, dried, and hybridised with a 32 P-labelled telomeric (CCCTAA)4 probe. The gel was exposed on a PhosphorImager screen and telomere length was calculated as described previously.32
Protein extracts and immunoblotting
Cell pellets were lysed in 50 mM Tris-HCl (pH 7.0) containing 2% sodium dodecyl sulphate and 5% sucrose. Total protein concentration was determined using the BCA assay (Pierce Biotechnology, Rockford, Illinois, USA) according to the manufacturer’s instructions. Appropriate amounts of protein were electrophoresed on SDS polyacrylamide gels and transferred to nitrocellulose (Amersham Biosciences, Piscataway, New Jersey, USA). Non-specific binding sites were blocked by 5% non-fat Carnation dry milk and blots were incubated in primary antibody (p53 and p21 from Oncogene Research (San Diego, California, USA), p16 from PharMingen (San Diego, California, USA), involucrin from Novo Castra (Newcastle, UK)). Blots were incubated in an appropriate dilution of secondary antibody conjugated to horseradish peroxidase from the appropriate species (Amersham Pharmacia Biotech). Specific protein bands were developed using a chemiluminescent substrate (SuperSignal Substrate, Pierce) onto Hyperfilm MP (Amersham Pharmacia Biotech). Positive controls (MCF7, HeLa, H1299 cancer cells as appropriate) were included for each gel and Ponceau S staining (Sigma) was performed to verify equal loading in all lanes.
UV irradiation
Telomerase transfected and untransfected control cells were plated in triplicate at 106 cells per 100 mm culture dish containing growth medium and incubated overnight. After several rinses in phosphate buffered saline (PBS), cells were irradiated with 100 J/m2 UV radiation as previously described.33 Cells were counted and harvested for protein at 1, 2, 4, 8, 12, 24, and 30 days post irradiation. Non-irradiated cells served as controls for protein expression and growth arrest.
Involucrin expression
hTERT expressing oesophageal epithelial cells on feeder layers were shifted to calcium free KGM-2 media (Clonetics, Walkersville, Maryland, USA) supplemented with 0.07 mM calcium for 72 hours. Parallel dishes were then transferred to 0.07 mM, 0.14 mM, or 1 mM calcium in KGM-2 for three and seven days. The treated cells were collected for determination of involucrin levels by immunoblotting.
Cytokeratin expression profile
To further characterise the telomerase expressing oesophageal squamous cells, we performed immunocytochemistry using a panel of cytokeratin antibodies as previously described.23,34–36 Oesophageal squamous cells were seeded at a density of 50 K cells per well onto two well glass chamber slides (Lab-Tek II; Nalge Nunc, Rochester, New York, USA) containing feeder cells and allowed to incubate for 48 hours. The slides were fixed for two minutes in methanol at room temperature and rinsed in PBS. Endogenous peroxidase was blocked by incubation with 0.3% hydrogen peroxide in PBS for 10 minutes. Primary antibodies were applied for two hours at room temperature in a humidified chamber as follows: cytokeratin (CK) 8/18 (1:500) and CK 13 (5 μg/ml). The appropriate secondary antibody and avidin-biotin complex were applied according to the manufacturer’s instructions (Vectastain; Vector Laboratories, Burlingame, California, USA). Slides were developed using diaminobenzadine (DAB; Sigma) as the chromogen and counterstained with haematoxylin. Additionally, we performed immunoblot assays using the same CK antibodies CK4 and CK 8/18. Immunoblotting was performed as described above using CK 4 (1:100) and CK 8/18 (1:500). The CK 4 antibody was from Novocastra, the CK 8/18 antibody was from Santa Cruz Biotechnology (Santa Cruz, California), and the CK13 antibody was from Research Diagnostics, Inc (Flanders, New Jersey, USA).
Soft agar assay
We sought to determine the ability of hTERT immortalised oesophageal cells to grow in soft agar. Growth in soft agar is a minimal requirement to demonstrate in vitro transformation of cells to their neoplastic counterparts. The soft agar assays were performed as previously described with minor modifications.37 Briefly, a 0.5% (w/v) bottom layer was prepared by melting 5% Noble agar (Sigma) and cooling it to 45°C. An agar mixture was then made by combining 1 ml 2×DMEM (Life Technologies, Incorporated, Gaithersburg, Maryland, USA), 1 ml serum (Invitrogen), 100 μl penicillin/streptomycin (Invitrogen), and 6.9 ml of 1×DMEM, and warming it to 37°C. The cooled agar (1 ml) was added to the mixture and after rapid stirring, 3 ml were plated per 35 mm dish. The agar was allowed to solidify and stored at 37°C until use. Cells were trypsinised and resuspended at dilutions of 0.5×104, 104, or 2×104 cells per ml of DMEM. In a 15 ml conical tube, 2 ml of the medium agar mixture were added to 1 ml of the cell suspension (final concentration 0.33% (w/v) agar). After rapid mixing, 1.5 ml of the cell solution was overlayed onto the solidified bottom layer and incubated in a humidified 37°C incubator with 5% CO2. Cells were fed weekly by addition of 200 μl of DMEM supplemented with 10% calf serum. After two weeks, colonies were scored by counting under a microscope. H1299 human lung adenocarcinoma cells were used as positive controls.
RESULTS
Isolation of normal oesophageal squamous (NOS) cells from endoscopic biopsies
Small outgrowths of epithelial cells were identified 72 hours after initial seeding. By day seven, 14–16 colonies were visualised. These were trypsinised, pooled, and reseeded at a 1:2 split ratio onto two dishes containing fresh feeder cells. Cells were grown in the presence of fibroblast feeder cells at all times and subcultured at 80% confluence (fig 1 ▶).
Figure 1.
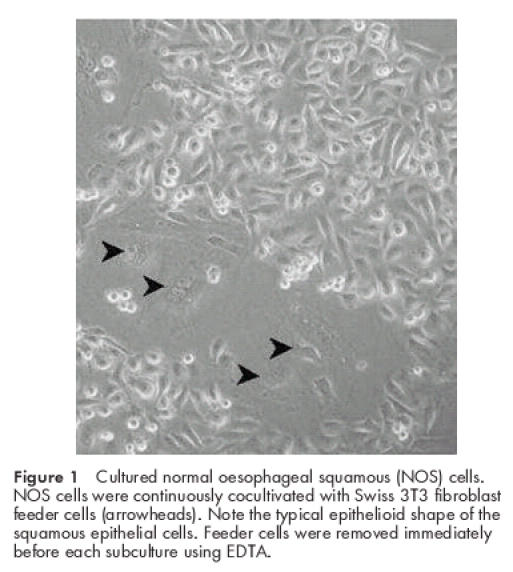
Cultured normal oesophageal squamous (NOS) cells. NOS cells were continuously cocultivated with Swiss 3T3 fibroblast feeder cells (arrowheads). Note the typical epithelioid shape of the squamous epithelial cells. Feeder cells were removed immediately before each subculture using EDTA.
Telomerase activity and telomere length in oesophageal squamous cells
Oesophageal cells exhibited progressive telomere shortening from 4.4 kb at PD 16 to 2.9 kb at senescence (PD 45, fig 2 ▶). Very weak telomerase activity (5% relative to positive control cells) was detected in the biopsy material of the normal oesophagus (fig 2 ▶) but this trace activity was lost by the third passage. Infection with hTERT at PD 20 resulted in an increase in telomere length to 7.7 kb, and this remained stable over time (fig 2 ▶), while cells expressed high telomerase levels (95% activity relative to positive control cells).
Figure 2.
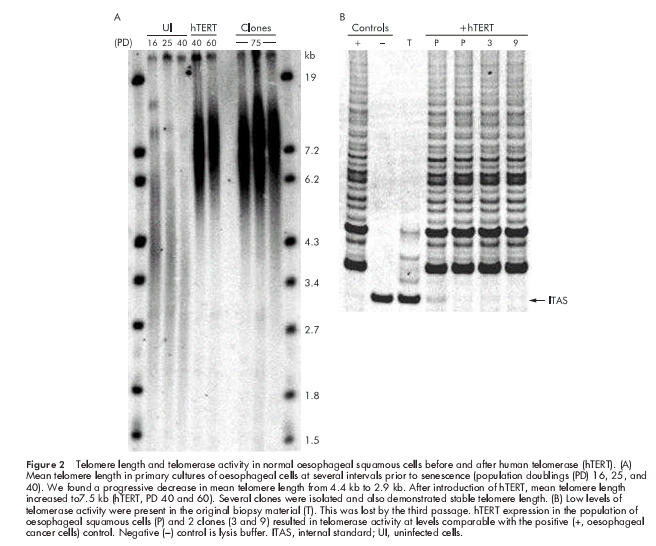
Telomere length and telomerase activity in normal oesophageal squamous cells before and after human telomerase (hTERT). (A) Mean telomere length in primary cultures of oesophageal cells at several intervals prior to senescence (population doublings (PD) 16, 25, and 40). We found a progressive decrease in mean telomere length from 4.4 kb to 2.9 kb. After introduction of hTERT, mean telomere length increased to7.5 kb (hTERT, PD 40 and 60). Several clones were isolated and also demonstrated stable telomere length. (B) Low levels of telomerase activity were present in the original biopsy material (T). This was lost by the third passage. hTERT expression in the population of oesophageal squamous cells (P) and 2 clones (3 and 9) resulted in telomerase activity at levels comparable with the positive (+, oesophageal cancer cells) control. Negative (−) control is lysis buffer. ITAS, internal standard; UI, uninfected cells.
Growth characteristics and lifespan of normal oesophageal keratinocytes
Parallel cultures of uninfected oesophageal cells in continuous culture underwent senescence between PD 40 and 45 (fig 3 ▶). In contrast, telomerase expressing oesophageal cells continued to divide, with a doubling time of 24 to 30 hours. The cell population has reached PD 100 with no diminution in growth rate. Ten clones were randomly selected, and all of these demonstrated growth rates comparable with the population. After 80 doublings, some of the cells were transferred to standard plastic culture dishes without fibroblast feeder cells and grown in keratinocyte serum free medium (Gibco/BRL Life Technologies). These cells growth arrested 9–10 doublings after transfer to plastic, suggesting cocultivation with feeder layers is required for continued proliferation of the oesophageal squamous cells (data not shown).
Figure 3.
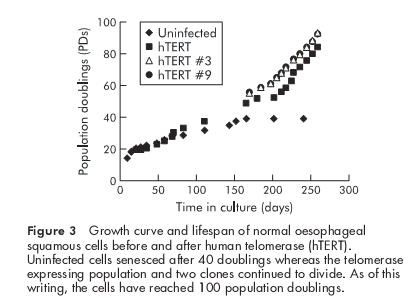
Growth curve and lifespan of normal oesophageal squamous cells before and after human telomerase (hTERT). Uninfected cells senesced after 40 doublings whereas the telomerase expressing population and two clones continued to divide. As of this writing, the cells have reached 100 population doublings.
Cell cycle checkpoints
The integrity of the p53 cell cycle checkpoint in the cultured oesophageal cells was tested by the response of p53 to DNA damage. Cells were harvested at various time intervals after exposure to 100 J/m2 UV. Protein levels of p53 increased within 24 hours after UV exposure and remained elevated for at least 96 hours. Levels of p21, a downstream effector of p53, increased 96 hours after UV exposure (fig 4 ▶). The increases in p53 and p21 were accompanied by a transient growth arrest as determined by cell count (data not shown). We investigated the integrity of the p16/Rb checkpoint by infecting cells growing on feeder layers with a retroviral vector encoding the Harvey Ras sarcoma viral oncogene homologue (H-RasG12V). Previous investigators have shown that exogenous Ras induces a premature growth arrest in normal human fibroblasts that is mediated at least in part by p16.38,39 Figure 5 ▶ shows a robust induction of p16 in the hTERT immortalised oesophageal cells following overexpression of Ras. Taken together, our results suggest that the combination of telomerase and cocultivation with feeder cells results in immortalisation of normal oesophageal epithelial cells without affecting the integrity of the p53 and p16/Rb cell cycle checkpoints.
Figure 4.
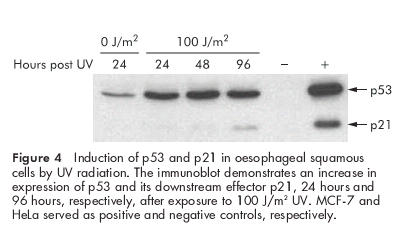
Induction of p53 and p21 in oesophageal squamous cells by UV radiation. The immunoblot demonstrates an increase in expression of p53 and its downstream effector p21, 24 hours and 96 hours, respectively, after exposure to 100 J/m2 UV. MCF-7 and HeLa served as positive and negative controls, respectively.
Figure 5.
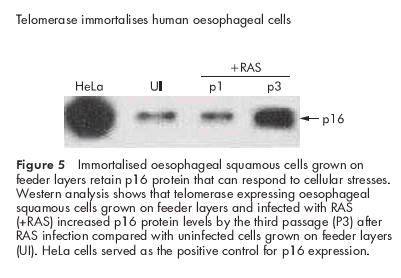
Immortalised oesophageal squamous cells grown on feeder layers retain p16 protein that can respond to cellular stresses. Western analysis shows that telomerase expressing oesophageal squamous cells grown on feeder layers and infected with RAS (+RAS) increased p16 protein levels by the third passage (P3) after RAS infection compared with uninfected cells grown on feeder layers (UI). HeLa cells served as the positive control for p16 expression.
Calcium and involucrin
We examined the ability of hTERT immortalised oesophageal epithelial cells to express differentiated markers. Previous reports have shown that normal oesophageal epithelial cells can be induced to express involucrin, a keratinocyte marker of terminal differentiation, by a variety of stimuli including high calcium concentrations and phorbol esters.24 We found an increase in involucrin expression in hTERT expressing oesophageal squamous cells after a seven day exposure to increasing concentrations of calcium (fig 6 ▶).
Figure 6.
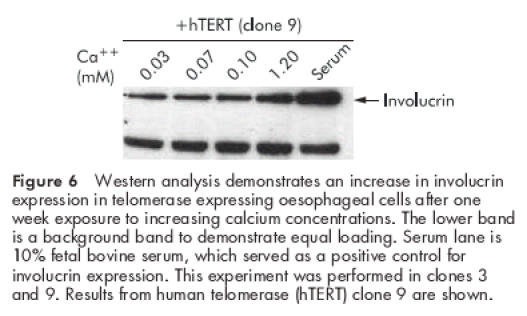
Western analysis demonstrates an increase in involucrin expression in telomerase expressing oesophageal cells after one week exposure to increasing calcium concentrations. The lower band is a background band to demonstrate equal loading. Serum lane is 10% fetal bovine serum, which served as a positive control for involucrin expression. This experiment was performed in clones 3 and 9. Results from human telomerase (hTERT) clone 9 are shown.
Cytokeratin expression
Using standard immunocytochemical techniques, several groups have previously demonstrated that tissue from the squamous oesophagus exhibits a characteristic pattern of CK staining.23,34–36 We sought to compare the pattern of staining in our cells to that reported for normal squamous oesophagus. Using immunoblot analysis, we detected positive expression for CKs 4 and 8/18 in the immortalised oesophageal squamous cells (fig 7A ▶). In contrast, fibroblast feeder cells were consistently negative for all of these cytokeratin markers. Using immunocytochemistry, we detected cytoplasmic staining for CKs 13 and 8/18.
Figure 7.
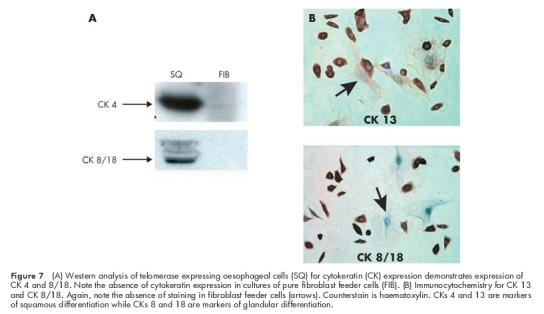
(A) Western analysis of telomerase expressing oesophageal cells (SQ) for cytokeratin (CK) expression demonstrates expression of CK 4 and 8/18. Note the absence of cytokeratin expression in cultures of pure fibroblast feeder cells (FIB). (B) Immunocytochemistry for CK 13 and CK 8/18. Again, note the absence of staining in fibroblast feeder cells (arrows). Counterstain is haematoxylin. CKs 4 and 13 are markers of squamous differentiation while CKs 8 and 18 are markers of glandular differentiation.
Soft agar assay
Telomerase expressing oesophageal cells failed to grow in soft agar (data not shown). Positive control cancer cells developed approximately 30–40 colonies per 2×104 cells.
DISCUSSION
Telomerase activity has been demonstrated in normal tissues from renewal organs such as skin, small intestine, and the lymphoproliferative system. Renewal organs must maintain the ability to proliferate in response to certain stimuli over the lifetime of an individual. Like skin, the oesophageal epithelium is normally in a state of slow renewal and has the capacity for accelerated cell proliferation in response to injury. Our finding that trace telomerase activity is present in biopsies of endoscopically normal oesophagus is consistent with the concept that the oesophagus is a renewal organ. We have previously demonstrated that expression of telomerase RNA (hTR) localises to the basal proliferative cells of the oesophageal squamous epithelium.40 Although cells from the oesophagus and other renewal organs normally express low levels of telomerase, studies in the small intestine and in CD34+ stem cells have demonstrated that such telomerase expression can only attenuate, but not prevent, telomere loss. Substantially higher levels of telomerase expression are needed for telomere maintenance and the immortalisation of normal oesophageal epithelial cells.
We found low levels of telomerase activity both in our original biopsy material and in our cultured oesophageal cells in early passages. Telomerase activity was lost by the third passage however. A similar loss of telomerase activity has been reported for squamous cells (keratinocytes) cultured from other organs, such as skin. Using specific markers for skin stem cells, other investigators have demonstrated that telomerase activity in early cultures of skin cells emanates from a small subpopulation of slowly dividing stem cells.41,42 After a few passages, rapidly proliferating telomerase negative keratinocytes overgrow the stem cells, resulting in loss of telomerase activity in the culture. These data suggest that loss of telomerase activity in our cultured oesophageal squamous cells may have resulted from similar events—that is, telomerase negative cells overgrowing a smaller population of telomerase positive stem cells. Unfortunately, no markers are available to distinguish stem cells from other proliferating cells in the squamous oesophagus and, consequently, the events underlying the loss of telomerase activity in our cultures are not clear.
Previous studies attempted to establish lines of normal oesophageal epithelial cells from biopsy or resection material with limited success. Compton et al used the feeder cell system to establish cultures of oesophageal squamous cells for extended periods.23 These investigators were able to achieve excellent cell growth for about 20 doublings before the population growth arrested. In our study, we achieved a lifespan greater than that of Compton et al in our control cells, and then demonstrated that the observed growth limitations were due to the loss of telomeric DNA. The ability to stabilise telomeres by the ectopic expression of telomerase thus represents a conceptually sound strategy for generating immortal oesophageal squamous cells.
In addition to induction of telomerase expression, we found that cocultivation of oesophageal squamous cells with fibroblast feeder layers is essential for their successful immortalisation. Unlike some normal fibroblasts, which can be immortalised with ectopically expressed telomerase alone, several investigators have suggested that additional genetic alterations are necessary to immortalise normal epithelial cells.43–45 For example, Dickson and colleagues and Brenner and colleagues44,45 reported that normal human skin keratinocytes and mammary epithelial cells require abrogation of the p16/Rb tumour suppressor pathway in order to bypass an initial telomere independent growth arrest. We have recently shown however that this premature growth arrest is induced by suboptimal cell culture conditions which lead in turn to an increase in p16 expression. When grown on fibroblasts feeder cells to improve cell culture conditions, p16 levels in skin keratinocytes remain below the threshold that induces growth arrest. Under these improved conditions, telomerase induction alone is sufficient to immortalise epithelial cells.18
Alternative approaches to immortalising normal oesophageal cells have been tried. Inokuchi and colleagues24 used a recombinant SV40 adenoviral vector to generate cultures of immortal human oesophageal epithelial cells. SV40 T antigen abrogates the p53 and Rb/p16 pathways, interrupting the normal signalling mechanism by which short telomeres lead to senescence.46–48 In the presence of SV40 T antigen, cells are forced to replicate despite critically short telomeres. These cells eventually enter a state known as “crisis”, which is characterised by a balance between cell division and massive cell death. As a rare event, a cell escapes this crisis period by reactivating its telomerase and stabilising its telomeres. Thus SV40 T antigen expressing cells become immortal by telomerase reactivation. Blockade of p53 and Rb/p16 cell cycle checkpoints prevents these cells from responding normally to DNA damage however, and predisposes them to genomic instability. By using the combination of introduced telomerase and fibroblast feeder layers, we have eliminated the need to abrogate the p53 and p16/Rb pathways. An important advantage of our approach is that the integrity of these important tumour suppressor pathways (and thus genomic stability) remains intact. The appropriate growth arrest of our telomerised oesophageal cells after UV radiation induced DNA damage indicates that the p53 pathway is intact, and that telomerase expression does not prevent a normal cellular response to noxious (and potentially oncogenic) stimuli. In addition, our demonstration that the introduction of oncogenic Ras readily induces p16 expression with premature growth arrest is further evidence that the p16/Rb cell cycle checkpoint axis is intact and functional. The ability to immortalise normal human oesophageal cells without disturbing their critical cell cycle checkpoints represents a novel approach to the generation of normal human epithelial cell lines.
Cytokeratins (CKs) are a family of at least 20 structural proteins found in the cytoskeletons of epithelial cells. Certain epithelia exhibit characteristic patterns of cytokeratin expression. Several investigators have reported that CKs 4 and 13, both markers of squamous differentiation, are expressed in the normal oesophagus.23,34–36 Markers of glandular differentiation (CKs 8, 18, 19) generally are not expressed in mature oesophageal squamous cells,35,36 but staining for CKs 8, 18, and 19 has been observed in cells from the basal layer of the squamous oesophagus.34,36 We used both immunocytochemistry and immunoblot assays to generate a cytokeratin expression profile for our telomerase expressing oesophageal squamous cells, and found positive staining for CKs 4, 8, 13, and 18. Thus the pattern of CK staining we observed in our immortalised squamous cells is consistent with published literature for squamous oesophagus.
Primary cultures of oesophageal epithelial cells respond to increasing calcium concentrations by inducing expression of the differentiation marker involucrin. Inokuchi et al found that SV40 T antigen immortalised cells were unable to respond appropriately to calcium treatment, suggesting that SV40 T expression interferes with the differentiation of oesophageal squamous cells.24 In contrast, our telomerase immortalised cells demonstrated an appropriate increase in involucrin expression in response to increasing calcium concentrations. Our results suggest that immortalisation by telomerase does not interfere with the differentiating ability of oesophageal cells.
Although immortality is a fundamental characteristic of most human cancers, the immortal phenotype alone is not sufficient to transform normal cells into their fully malignant counterparts. Generally, transformation of cells in culture is defined by the ability to grow in an anchorage independent fashion, the ability to grow in the absence of serum, the ability to grow in soft agar, and by the development of tumours in nude mice; of these four criteria, the latter two are, by far, the most stringent. Hahn et al reported that coinduction of oncogenic ras, the SV40 t antigen, and the SV40 T antigen were required in addition to telomerase for the successful transformation of cultured normal human fibroblasts.49,50 None of these transforming genetic alterations occurred spontaneously in telomerase expressing fibroblasts despite continuous cultivation for over 300 population doublings.51 The failure of our telomerase immortalised oesophageal cells to grow in soft agar provides strong evidence that our cells are not neoplastic. Taken together, the data suggest that telomerase expression alone should not predispose immortalised oesophageal squamous cells to cancer.
In summary, we report the generation of a line of normal human oesophageal epithelial cells by the introduction of human telomerase. Our cell line retains normal cell cycle checkpoints and normal differentiation. Telomerase immortalised oesophageal cells should be valuable reagents for investigations of human oesophageal carcinogenesis.
Acknowledgments
This research was supported by Advanced Technology Program grant (0244–1999) from the Texas Higher Education Coordinating Board to CPM and JWS and a Merit Entry Grant from the VA Medical Center to CPM.
Abbreviations
hTERT, human telomerase
PBS, phosphate buffered saline
PD, population doublings
CK, cytokeratin
DMEM, Dulbecco’s modified Eagle’s media
REFERENCES
- 1.Lu S, Hsieh LL, Luo FC, et al. Amplification of EGF receptor and c-myc genes in human esophageal cancers. Int J Cancer 1988;42:502–5. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein MC, Metcalf RA, Welsh JA, et al. Frequent mutation of the p53 gene in human esophageal cancer. Proc Am Assoc Cancer Res 1990;87:9958–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boynton FR, Huang Y, Blount PL, et al. Frequent loss of heterozygosity at the retinoblastoma locus in human esophageal cancer. Cancer Res 1991;51:5766–9. [PubMed] [Google Scholar]
- 4.Tsuda T, Tahara E, Kajiyama G, et al. High incidence of coamplification of int-2 and hst-1 genes in human esophageal carcinomas. Cancer Res 1991;49:5505–8. [PubMed] [Google Scholar]
- 5.Jiang WJ, Kahn SM, Tomita N, et al. Amplification and expression of the human cyclin D gene in esophageal cancer. Cancer Res 1992;52:2980–3. [PubMed] [Google Scholar]
- 6.Wang DD, Hong JY, Oui SL, et al. Accumulation of p53 protein in human esophageal precancerous lesions: a possible early biomarker for carcinogenesis. Cancer Res 1993;53:1783–7. [PubMed] [Google Scholar]
- 7.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature 1990;345:458–60. [DOI] [PubMed] [Google Scholar]
- 8.Allsopp RC, Vaziri H, Patterson C, et al. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci U S A 1992;89:10114–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright WE, Brasiskyte D, Piatyszek MA, et al. Experimental elongation of telomeres extends the lifespan of immortal x normal cell hybrids. EMBO 1996;15:1734–41. [PMC free article] [PubMed] [Google Scholar]
- 10.Moyzis RK, Buckingham JM, Cram LS, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Pro Natl Acad Sci U S A 1988;85:6622–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allsopp RC, Chang E, Kashefi-Aazam M, et al. Telomere shortening is associated with cell division in vitro and in vivo. Exp Cell Res 1995;220:194–200. [DOI] [PubMed] [Google Scholar]
- 12.Bacchetti S. Telomere dynamics and telomerase activity in cell senescence and cancer. Semin Cell Dev Biol 1996;7:31–9. [Google Scholar]
- 13.Villenponteau B. The RNA components of human and mouse telomerase. Semin Cell Dev Biol 1996;7:15–21. [Google Scholar]
- 14.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer 1997;33:787–91. [DOI] [PubMed] [Google Scholar]
- 15.Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science 1998;279:349–52. [DOI] [PubMed] [Google Scholar]
- 16.Condon J, Yin S, Mayhew B, et al. Telomerase immortalisation of human myometrial cells. Biol Reprod 2002;67:506–14. [DOI] [PubMed] [Google Scholar]
- 17.Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol 1998;8:279–82. [DOI] [PubMed] [Google Scholar]
- 18.Ramirez RD, Morales CP, Herbert B-S, et al. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev 2001;15:398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X-R, Jimenez G, Chang E, et al. Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat Genet 1999;21:111–14. [DOI] [PubMed] [Google Scholar]
- 20.Resau JH, Phelps PC, Zhu S, et al. Long-term culture of human esophageal explants and cells. Cytotechnology 1990;3:61–73. [DOI] [PubMed] [Google Scholar]
- 21.Vocci MJ, Combs JW, Hillman EA, et al. The cell kinetics of the adaptation of human esophagus to tissue culture. In vitro 1983;19:881–91. [DOI] [PubMed] [Google Scholar]
- 22.Mothersill C, Cusack A, Seymour CB, et al. Optimization of media for the culture of normal human epithelial cells from esophageal mucosa. Cell Biol Int Rep 1989;13:625–33. [DOI] [PubMed] [Google Scholar]
- 23.Compton CC, Warland G, Nakagawa H, et al. Cellular characterization and successful transfection of serially subcultured normal human esophageal keratinocytes. J Cell Physiol 1998;177:274–81. [DOI] [PubMed] [Google Scholar]
- 24.Inokuchi S, Handa H, Imai T, et al. Immortalisation of human esophageal epithelial cells by a recombinant SV40 adenovirus vector. Br J Cancer 1995;71:819–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banks-Schlegel SP. Isolation, cultivation and characterization of normal human esophageal epithelial cells. J Tissue Cult Methods 1985;9:95–105. [Google Scholar]
- 26.Banks-Schlegel SP, Harris CC. Tissue specific expression of keratin proteins in human esophageal and epidermal epithelium and their cultured keratinocytes. Exp Cell Res 1983;146:271–80. [DOI] [PubMed] [Google Scholar]
- 27.Banks-Schlegel SP, Green H. Involucrin synthesis and tissue assembly by keratinocytes in natural and cultured human epithelia. J Cell Biol 1981;90:732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: The formation of keratinizing colonies from single cells. Cell 1975;6:331–43. [DOI] [PubMed] [Google Scholar]
- 29.Rheinwald JG. Methods for clonal growth and serial cultivation of normal human epidermal keratinocytes and mesothelial cells. In: Baserga R, ed. Cell growth and division: a practical approach. Oxford, England: IRL Press, 1989:81–94.
- 30.Steinert S, Shay JW, Wright WE. Transient expression of human telomerase extends the life span of normal human fibroblasts. Biochem Biophys Res Commun 2000;273:1095–8. [DOI] [PubMed] [Google Scholar]
- 31.Holt SE, Norton JC, Wright WE, et al. Comparison of the telomeric repeat amplification protocol (TRAP) to the new TRAP-eze telomerase detection kit. Methods Cell Sci 1996;18:237–48. [Google Scholar]
- 32.Ouellette M, Liao M, Herbert B-S, et al. Subsenescent telomere lengths in fibroblasts immortalised by limiting amounts of telomerase. J Biol Chem 2000;275:10072–6. [DOI] [PubMed] [Google Scholar]
- 33.Morales CP, Holt SE, Ouellette M, et al. Lack of cancer-associated changes in human fibroblasts after immortalisation with telomerase. Nat Genet 1999;21:115–18. [DOI] [PubMed] [Google Scholar]
- 34.Glickman JN, Chen Y-Y, Wang HH, et al. Phenotypic characteristics of a distinctive multilayered epithelium suggests that it is a precursor in the development of Barrett’s esophagus. Am J Pathol 2001;25:569–78. [DOI] [PubMed] [Google Scholar]
- 35.Boch JA, Shields HM, Antonioli DA, et al. Distribution of cytokeratin markers in Barrett’s specialized columnar epithelium. Gastroenterology 1997;112:760–5. [DOI] [PubMed] [Google Scholar]
- 36.Salo JA, Kivilaakso EO, Kiviluoto TA, et al. Cytokeratin profile suggests metaplastic epithelial transformation in Barrett’s Oesophagus. Ann Med 1996;28:305–9. [DOI] [PubMed] [Google Scholar]
- 37.Clark GJ, Cox AD, Graham SM, Balch WE, Der CJ, Hall A, editors. Methods in Enzymology. San Diego: Academic Press, 1995:395–412. [DOI] [PubMed]
- 38.Serrano M, Lin AW, McCurrach ME, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997;88:593–602. [DOI] [PubMed] [Google Scholar]
- 39.Robles S, Adami GR. Agents that cause DNA double strand breaks lead to p16INK4a enrichment and the premature senescence of normal fibroblasts. Oncogene 1998;16:1113–23. [DOI] [PubMed] [Google Scholar]
- 40.Morales CP, Lee EL, Shay JW. In situ hybridization for the detection of telomerase RNA in the progression from Barrett’s esophagus to esophageal adenocarcinoma. Cancer 1998;83:652–9. [PubMed] [Google Scholar]
- 41.Kunimura C, Kikuchi K, Ahmed N, et al. Telomerase activity in a specific cell subset co-expressing integrin â1/EGFR but not p75 NGFR/bcl2/integrin â4 in normal human epithelial cells. Oncogene 1998;17:187–97. [DOI] [PubMed] [Google Scholar]
- 42.Yasumoto S, Kunimura C, Kikuchi K, et al. Telomerase activity in normal human epithelial cells. Oncogene 1996;13:433–9. [PubMed] [Google Scholar]
- 43.Kiyono T, Foster SA, Koop JI, et al. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalise human epithelial cells. Nature 1998;396:84–8. [DOI] [PubMed] [Google Scholar]
- 44.Dickson MA, Hahn WC, Ino Y, et al. Human keratinocytes that express hTERT and also bypass a p16INK4a-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol 2000;20:1436–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brenner AJ, Stampfer MF, Aldaz CM. Increased p16 expression with first senescencearrest in human mammary epithelial cells and extended growth capacity with p16 inactivation. Oncogene 1998;17:199–205. [DOI] [PubMed] [Google Scholar]
- 46.Shay JW, Wright WE, Werbin H. Defining the molecular mechanisms of human cell immortalisation. Biochim Biophys Acta 1991;1072:1–7. [DOI] [PubMed] [Google Scholar]
- 47.Wright WE, Pereira-Smith OM, Shay JW. Reversible cellular senescence: A two-stage model for the immortalisation of normal human diploid fibroblasts. Mol Cell Biol 1989;9:3088–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shay JW, Pereira-Smith OM, Wright WE. A role for both Rb and p53 in the regulation of human cellular senescence. Exp Cell Res 1991;196:33–9. [DOI] [PubMed] [Google Scholar]
- 49.Hahn WC, Counter CM, Lundberg AS, et al. Creation of human tumour cells with defined genetic elements. Nature 1999;400:464–8. [DOI] [PubMed] [Google Scholar]
- 50.Hahn WC, Dessain SK, Brooks MW, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol 2002;22:2111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morales CP, Holt SE, Ouellette M, et al. Absence of cancer-associated changes in human fibroblasts immortalised with telomerase. Nat Genet 1999;21:115–18. [DOI] [PubMed] [Google Scholar]


