Abstract
Cdc42 is a member of the Rho GTPase family that regulates multiple cellular activities, including actin polymerization, kinase-signaling activation, and cell polarization. MSE55 is a nonkinase CRIB (Cdc42/Rac interactive-binding) domain-containing molecule of unknown function. Using glutathione S-transferase-capture experiments, we show that MSE55 binds to Cdc42 in a GTP-dependent manner. MSE55 binding to Cdc42 required an intact CRIB domain, because a MSE55 CRIB domain mutant no longer interacted with Cdc42. To study the function of MSE55 we transfected either wild-type MSE55 or a MSE55 CRIB mutant into mammalian cells. In Cos-7 cells, wild-type MSE55 localized at membrane ruffles and increased membrane actin polymerization, whereas expression of the MSE55 CRIB mutant showed fewer membrane ruffles. In contrast to these results, MSE55 induced the formation of long, actin-based protrusions in NIH 3T3 cells as detected by immunofluorescence and live-cell video microscopy. MSE55-induced protrusion formation was blocked by expression of dominant-negative N17Cdc42, but not by expression of dominant-negative N17Rac. These findings indicate that MSE55 is a Cdc42 effector protein that mediates actin cytoskeleton reorganization at the plasma membrane.
During a variety of eukaryotic cellular processes such as shape changes, migration, and cytokinesis, the actin cytoskeleton must undergo dynamic alterations (1). These changes in the cytoskeleton are attributed to differences in assembly of polymerized actin, a process catalyzed by members of the Rho GTPase family. In Swiss 3T3 cells, Cdc42 induces filopodia (2, 3), whereas Rac and Rho induce membrane ruffling (4) and stress fiber formation (5), respectively. The ability of Cdc42 to activate Rac, which, in turn, activates Rho, has implicated this GTPase cascade in the coordination of cell shape changes associated with cell migration (3, 4). In addition to their pronounced effects on the actin cytoskeleton, Cdc42, Rac, and Rho also regulate a number of other cellular processes including kinase-signaling pathways (6, 7) and cell growth (8).
The diverse processes regulated by Cdc42, Rac, and Rho are mediated through numerous downstream Rho GTPase effectors (9). A common motif found in the majority of Cdc42 and/or Rac effector proteins is the conserved 16-aa CRIB (Cdc42/Rac interactive-binding) domain, which serves as a binding site for Cdc42 and/or Rac (10). One particular subgroup of CRIB proteins that is composed of effectors for both Rac and Cdc42 is the serine/threonine PAK kinase subgroup. PAK kinases, the mammalian homologues of Saccharomyces cerevisiae STE 20, bind both Cdc42 and Rac, but not Rho, in a GTP-dependent manner, with their catalytic activity being stimulated markedly when associated with the GTP-bound form of Cdc42 or Rac (11). Despite PAK binding to both Cdc42 and Rac GTPases, the role of PAK effectors in cytoskeletal reorganization remains controversial (12–17). It is clear, however, that PAK kinases function to activate the p38 (18–20) and JNK/SAPK (15–18, 21) pathways. Other CRIB-containing kinase effectors mediate cytoskeletal reorganization, including the Cdc42 effectors MRCK-α (22) and GEK (23). MRCK-α-mediated cytoskeletal reorganization results from phosphorylation of nonmuscle myosin light chain leading to actin–myosin contractility (22). MRCK-α and GEK kinase share homology with Rho kinase effectors, which function in actin stress fiber formation (22–24), suggesting that effectors of different Rho GTPases may use common mechanisms of actin cytoskeletal reorganization.
A number of Cdc42 CRIB effectors appear not to be protein kinases (10). In yeast, two structurally related proteins, Gic1p and Gic2p, bind Cdc42 and regulate both actin polymerization and cytoskeletal polarization (25). The mechanism by which Gic1p and Gic2p function in cytoskeletal reorganization is unclear, because there are no obvious catalytic or actin-binding domains within these proteins. In mammalian cells, WASP (26) and N-WASP (27, 28) function as Cdc42 effectors involved in actin polymerization. Although both WASP and N-WASP contain cofilin-like actin-binding domains, the effects of WASP and N-WASP on actin reorganization differ significantly (26, 28). WASP induces perinuclear aggregates of polymerized actin (26), whereas N-WASP induces actin-based filopodia similar to what are observed by expression of constitutively active Cdc42 (27, 28).
Here we report the functional characterization of the CRIB-containing protein MSE55 (10). MSE55 is a nonkinase effector protein that specifically binds GTP-bound Cdc42, but does not contain any recognizable signaling motifs with the exception of several proline-rich repeats. Expression of MSE55 in Cos-7 cells induced membrane ruffling, whereas expression in NIH 3T3 fibroblasts induced the formation of one to two long actin-based protrusions. Expression of an MSE55 CRIB mutant or coexpression of wild-type MSE55 with dominant-negative Cdc42 decreased the formation of cell protrusions. The localization of MSE55 to membrane ruffles in Cos-7 cells and its ability to induce membrane extensions in NIH 3T3 fibroblasts suggest that MSE55 may mediate actin cytoskeleton reorganization required for changes in cell polarity.
MATERIALS AND METHODS
MSE55 Mammalian Expression Vectors.
A cDNA encoding MSE55 (29) was placed downstream of Myc and FLAG epitope-tagged expression vectors. The N-terminal FLAG epitope-tagged vector was constructed in a modified cytomegalovirus promoter-driven cassette vector (pCAF1), and the Myc epitope-tagged MSE55 expression vector was constructed in a pcDNA-3 vector (Invitrogen). Site-directed mutagenesis of MSE55 was performed with sequence-specific oligonucleotides by using the QuickChange mutagenesis kit (Stratagene) per the manufacturer’s protocol. All constructs were sequenced to confirm their integrity.
Glutathione S-Transferase Capture Experiments.
Cos-1 cells were maintained in DMEM supplemented with 10% FCS/1 mM l-glutamine/100 units/ml penicillin/100 μg/ml streptomycin. Cos-1 cells were transfected by electroporation (400 V, 125 μF) with 10 μg of DNA/100-mm2 plate. Forty-eight hours after transfection, cells expressing Flag-tagged MSE55 were lysed at 4°C with 800 μl of lysis buffer A (50 mM Tris, pH 8.0/150 mM NaCl/1 mM EDTA/1% Triton X-100) containing phosphatase and protease inhibitors (0.5 mM PMSF/50 mM NaF/2 mM sodium orthovanadate/25 μg/ml aprotinin/50 μg/ml pepstatin/25 μg/ml leupeptin). The supernatant was obtained by centrifugation and precleared by incubation with Sepharose beads. Cytosol extracts containing Flag epitope-tagged MSE55 were incubated with glutathione S-transferase fusion proteins of L63RhoA, L61Rac1, or V12Cdc42 proteins bound to glutathione-Sepharose (3–5).
Recombinant L63RhoA, L61Rac1, or V12Cdc42 proteins (5 μg) immobilized to glutathione-Sepharose were loaded with GDP or guanosine 5′-[γ-thio]triphosphate (10) for 10 min at 30°C in 30 μl of 50 mM Tris, pH 7.5/5 mM EDTA/0.5 mg/ml BSA. Nucleotide exchange was stopped on ice by adding MgCl2 to 10 mM. The proteins were incubated for 30 min with equal amounts of cell supernatant from Flag-tagged MSE55-transfected cells. After four washes, bound proteins were eluted by boiling with SDS/PAGE loading buffer and analyzed on a 9% SDS/PAGE gel. After electrophoresis, Western blotting was performed with anti-FLAG M2 mAb (Sigma) to detect bound Flag-epitope MSE55 protein. After washing, goat anti-mouse horseradish peroxidase and enhanced chemiluminescence reagents (Pierce) were used to detect Flag-tagged MSE55.
Yeast Two-Hybrid Assay.
The yeast two-hybrid assay was performed in the PJ69–4A yeast strain (30) by using the pGBU GAL4-DNA-binding domain yeast vector (30) and pACT-II GAL4-activation domain yeast vectors (31) as described (30). MSE55 (amino acids 9–391) was placed downstream of the GAL4-DNA-binding domain of pGBU-C1. Wild-type and activated mutants of Cdc42, Rac, and Rho were placed downstream of the GAL4-activation domain in pACT-II. Interactions were assayed by using a β-galactosidase filter assay as described (32).
Transfection and Immunofluorescence.
Cos-7 cells were maintained as described for Cos-1 cells. Cos-7 cells were transfected by electroporation (400 V, 125 μF) and replated on glass coverslips coated with poly l-lysine (10 μg/ml). Cos-7 cells were transfected with 10 μg of expression vector for either FLAG-tagged MSE55 or FLAG-tagged MSE55 containing alanine mutations introduced at D36, P41, and H47 (MSE55-D36A, P41A, and H47A). Sixteen to twenty-four hours after transfection, cells were fixed in 3.7% formaldehyde (vol/vol) in PBS, pH 7.4, for 10 min and permeabilized with 0.05% Triton X-100 for 10 min. MSE55-transfected cells were identified by staining with anti-FLAG M2 mAb and then with a secondary goat anti-mouse IgG coupled to FITC (Molecular Probes). F-actin staining was visualized with Texas Red phalloidin (Molecular Probes).
NIH 3T3 cells were maintained in DMEM supplemented with 10% heat-inactivated calf serum/1 mM l-glutamine/100 units/ml penicillin/100 μg/ml streptomycin. NIH 3T3 cells were seeded in 12-well tissue culture dishes containing glass coverslips coated with poly-l-lysine (10 μg/ml). Eighteen hours after plating, cells were transfected with 1.0 μg of FLAG-tagged MSE55 by using Superfect Reagent (Qiagen). For quantitation of cellular extensions (>10 μm), 100 cells were counted from three independent experiments.
For cotransfection experiments, cells were transfected with 0.6 μg of MSE55 and 0.6 μg of either dominant-negative, Myc-tagged N17Cdc42 or Myc-tagged N17Rac. Cells were fixed 24 hr after transfection and stained with the anti-FLAG mouse mAb and rabbit anti-Myc polyclonal antibody (Santa Cruz Biotechnology) as described for Cos-7 immunostaining. After washing, the coverslips were incubated with goat anti-mouse IgG-Alexa 350 and goat anti-rabbit IgG-FITC (both from Molecular Probes) and Texas Red-phalloidin (Molecular Probes). Cells coexpressing both MSE55 (blue) and either Cdc42 or Rac (green) were identified and examined for actin staining (red). Slides were viewed and photographed on a Zeiss Photomicroscope III with a PH3, ×63 objective (Zeiss).
Microinjection and Time-Lapse Videography.
NIH 3T3 cells were seeded 24 hr before microinjection on poly l-lysine-coated CELLocate gridded coverslips (Eppendorf) in NIH 3T3 complete medium. Approximately 100 cells were microinjected on a heated stage by using the Eppendorf Micromanipulator 5171 and Transjector 5246. Cells were coinjected at 150 hPa for 0.2 sec with 0.300 μg/μl plasmid DNA containing equal amounts of MSE55 or MSE55 CRIB mutant expression vector along with the human enhanced green fluorescent protein (EGFP) expression vector pEGFP-C1 (CLONTECH). Cells then were incubated at 37°C with 5% CO2, and 3.5 hr later EGFP protein expression was detected on live cells by using a Zeiss Photomicroscope III equipped with a PH2 plan-Neofluor ×25 water-immersion lens (Zeiss). Cells then were imaged at 2-min intervals for 1 hr, returned to the incubator, and then, 6 hr later, imaged again for 1 hr. Imaging was done by time-lapse video microscopy by using an MTI CCD72S camera (Dage–MTI, Michigan City, IN). Digitized images were captured by using optimas 5.2 software (Optimas, Bothell, WA). MSE55 expression was confirmed at all time points by indirect immunofluorescence.
RESULTS
MSE55 Binds to Cdc42 in a GTP-Dependent Fashion.
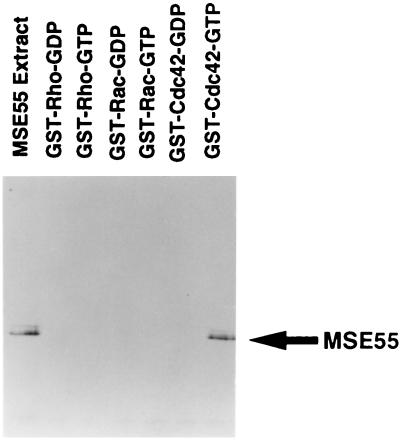
Previously, we demonstrated by using a filter-binding assay that the N terminus of MSE55 contained a functional CRIB domain (10). We utilized glutathione S-transferase capture experiments with fusion proteins of Rho, Rac, or Cdc42 GTPase and FLAG epitope-tagged full-length MSE55 produced in Cos1 cells to evaluate the interaction of MSE55 with these GTPases and to determine nucleotide dependence. We observed that MSE55 was highly expressed as a 55-kDa species upon transfection in these cells (Fig. 1). No interaction of MSE55 was detected with Rac or Rho by using either GDP- or GTP-loaded GTPases (Fig. 1), but MSE55 was found to associate with Cdc42 (Fig. 1). The interaction of MSE55 with Cdc42 required that Cdc42 be in the GTP-bound form, because GDP-bound Cdc42 did not interact with MSE55 (Fig. 1). These results confirm that MSE55 binds to Cdc42 in a GTP-dependent fashion and not to Rac or Rho and, therefore, is a likely effector for Cdc42.
Figure 1.
MSE55 interacts with Cdc42 in a GTP-dependent fashion. Cos-1 cells were transfected with FLAG epitope-tagged MSE55 eukaryotic expression vector. Cell lysates obtained from these cells were incubated with glutathione S-transferase-bound Rho, Rac, and Cdc42 that had been preloaded with either GDP or guanosine 5′-[γ-thio]triphosphate. After four washes, bound proteins were eluted and analyzed by Western blotting with M2 anti-FLAG mAb and goat anti-mouse-horseradish peroxidase and detected by enhanced chemiluminescence. An aliquot of cell lysate from FLAG-MSE55-transfected cells also is shown.
An MSE55-D36A, P41A, H47A CRIB Mutant Does Not Interact with Cdc42.
To further define the nature of the Cdc42-MSE55 interaction, we created several different point mutations within the CRIB domain of MSE55 by site-directed mutagenesis. Two mutants, MSE55-H47D and MSE55-P41A, contained single amino acid substitutions, whereas a third mutant MSE55-D36A, P41A, H47A contained a triple alanine substitution. We next tested these mutants in yeast two-hybrid assays with activated mutants of Cdc42, Rac, and Rho. Using an 5-bromo-4-chloro-3-indolyl β d-galactoside filter-lift assay, MSE55-H47D and MSE55-P41A mutants still bound L61Cdc42 and bound weakly to L61-Rac (Table 1). Similar results were found with wild-type Cdc42 and Rac (data not shown). These results are in contrast with work on an N-WASP-H208D CRIB mutant, which corresponds to our MSE55-H47D mutant, which was sufficient to prevent interaction with Cdc42 (28). A likely explanation for this is that various CRIB effector proteins interact differently with Cdc42 and that single mutations in the CRIB domain of MSE55 are not sufficient to block binding. The triple alanine mutant, MSE55-D36A, P41A, H47A, however, no longer interacted with L61Cdc42 or L61Rac (Table 1).
Table 1.
MSE55 interacts with Cdc42 in the yeast two-hybrid assay
| DNA-binding fusion domain | Activation domain fusion | Interaction |
|---|---|---|
| MSE55 | p-ACT | − |
| MSE55 | p-ACT-L61Cdc42 | +++ |
| MSE55 | p-ACT-L61Rac | + |
| MSE55 | p-ACT-L63Rho | − |
| MSE55-H47D | p-ACT-L61Cdc42 | +++ |
| MSE55-P41A | p-ACT-L61Cdc42 | +++ |
| MSE55-D36A, P41A, H47A | p-ACT-L61Cdc42 | − |
Summary of yeast two-hybrid interactions is shown between MSE55 with various baits. Interactions were tested for β-galactosidase activity as measured by the time taken for colonies to turn blue in 5-bromo-4-chloro-3-indolyl β-d-galactoside filter-lift assay (33). +++, <25 min; ++, 25–50 min; +, 50–100 min; and −, no activity.
MSE55 Localizes to Membrane Ruffles in Cos-7 Cells.
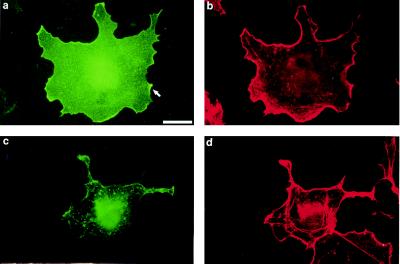
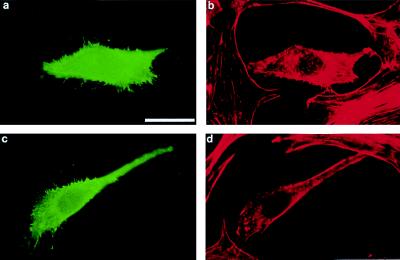
By indirect immunofluorescence, we examined the cellular distribution of transfected human MSE55 in Cos-7 cells by using a mAb against the N-terminal FLAG epitope tag. Expression of MSE55 in Cos-7 cells showed both diffuse cytoplasmic staining and strong ribbon-like membrane staining in transfected cells at 16–24 hr posttransfection (Fig. 2a). Staining of F-actin with Texas Red-phalloidin demonstrated that MSE55 colocalized with F-actin in lamellipodia (Fig. 2b). MSE55-expressing cells showed fewer stress fibers as compared with untransfected cells (Fig. 2b). Examination of MSE55-expressing Cos-7 cells 48 hr after transfection, when the level of endogenous Cos-7 cell ruffling had declined, revealed that MSE55-expressing cells showed enhanced F-actin staining in membrane ruffles and some small filopodia (data not shown). Based on these data, MSE55 may function to link Cdc42-regulated signaling to Rac signaling in Cos cells.
Figure 2.
MSE55 localizes to lamellipodia in Cos-7 cells. FLAG-tagged MSE55 was transfected into Cos-7 cells. Expression of wild-type MSE55 (a) or a CRIB mutant, MSE55-D36A, P41A, H47A (c), was detected with the M2 anti-FLAG mouse mAb and goat anti-mouse IgG-FITC. Staining of F-actin filaments was visualized with Texas Red-phalloidin (b and d). Fluorescent micrographs show MSE55 staining in membrane ruffles 18 hr after transfection (a and b). MSE55-D36A, P41A, H47A-expressing cells (c) showed less staining in the ruffles and more stress fibers (d). (Bar = 20 μm.)
In contrast to these results with wild-type MSE55, expression of the MSE55-D36A, P41A, H47A mutant (corresponding to the mutant that did not interact with Cdc42 in yeast two hybrid) no longer stained strongly at membrane ruffles (Fig. 2c). These data suggest that MSE55 localization at membrane ruffles depends on binding to Cdc42. Cells expressing this mutant showed increased stress fibers and contained intracellular vesicles not seen in untransfected cells or in MSE55-transfected cells (Fig. 2d). A similar phenomenon of increased stress fiber formation is seen in Cos-7 cells expressing a dominant-negative PAK kinase (14).
MSE55 Induces Long Cellular Extensions in NIH 3T3 Fibroblasts.
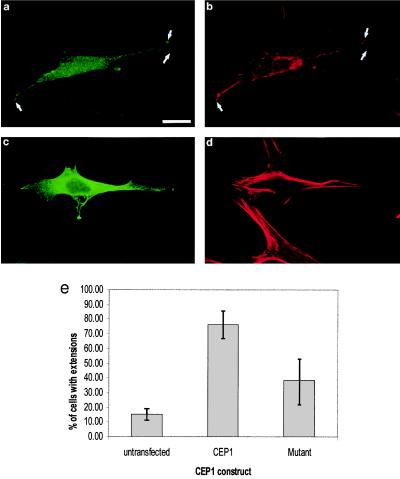
Much of the characterization of Rho GTPase function has been performed in fibroblasts. In these cells, expression of Cdc42 (2, 3) or TC10 (33), Rac (4), or Rho (5) induced filopodia, membrane ruffles, and stress fibers, respectively. NIH 3T3 cells expressing MSE55 produced one to two cellular processes that were greater than 10 μm in length (Fig. 3a). These processes were rich in F-actin as determined by staining with Texas Red-phalloidin (Fig. 3b). This phenotype is quite different from multiple filopodia seen in cells expressing the Cdc42 effector N-WASP (28, 34). NIH 3T3 cells expressing the MSE55-D36A, P41A, H47A CRIB mutant formed fewer cellular extensions and contained more stress fibers as compared with cells expressing wild-type MSE55 (Fig. 3 c and d). Extension formation was evaluated in MSE55- and MSE55-D36A, P41A, H47A-transfected cells by counting cells with extensions longer than 10 μm. Approximately 15% of untransfected cells formed long actin-based extensions whereas 80% of MSE55-expressing cells showed this phenotype (Fig. 3e). In contrast, only 40% of cells expressing the MSE55 CRIB mutant formed extensions (Fig. 3e). One explanation for why extension formation was higher in NIH 3T3 cells expressing the MSE55 mutant as compared with untransfected cells could be that high levels of mutant MSE55 protein may alter the actin cytoskeleton by alternative mechanisms independent of Cdc42 interactions.
Figure 3.
MSE55 induces long cellular extensions in NIH 3T3 cells. FLAG-tagged MSE55 was transfected into NIH 3T3 cells. Expression of wild-type MSE55 (a) or the MSE55-D36A, P41A, H47A CRIB mutant (c) was detected with M2 anti-FLAG mouse mAb and goat anti-mouse IgG-FITC. Staining of F-actin filaments also was visualized with Texas Red-phalloidin (b and d). Fluorescent micrographs show MSE55-expressing cells showing long cellular extensions (a) that colocalize with F-actin staining (b). Expression of MSE55-D36A, P41A, H47A did not show long cellular extensions (c and d). (Bar = 20 μm.) NIH 3T3 cells with extensions greater than 10 μm in length were counted in untransfected and both wild-type and mutant MSE55-transfected cells (e). Results are expressed as the mean percentage of cells (±SD) from three separate experiments.
Extensions in NIH 3T3 Cells Expressing MSE55 Are the Result of Membrane Protrusion.
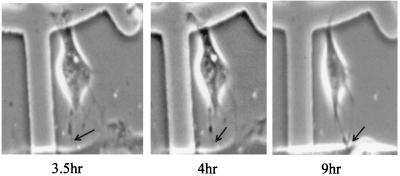
We used time-lapse video microscopy to confirm that the extensions observed in NIH 3T3 cells were a result of active actin polymerization leading to membrane protrusion rather than a result of membrane retraction. Specifically, we coinjected plasmids encoding both human EGFP (pEGFP-C1) and MSE55. After microinjection, cells were monitored for EGFP expression by live-cell fluorescent microscopy. By 3.5 hr after microinjection, pEGFP expression was detected and live-cell time-lapse videography was begun. Representative images were captured at 2-min intervals for 1 hr starting at 3.5 hr after microinjection and then again at 8.0 hr after microinjection. The cellular extension observed in NIH 3T3 cells expressing MSE55 was due to active membrane protrusion and not membrane retraction (Fig. 4). No retraction fibers were observed even up to 9 hr after microinjection (Fig. 4). Microinjection of pEGFP-C1 and the MSE55-D36A, P41A, H47A CRIB mutant did not cause extension formation at this early time point (data not shown). These results indicate that the extensions observed in NIH 3T3 cells expressing MSE55 were due to active membrane protrusions.
Figure 4.
MSE55 expression induces membrane protrusions. NIH 3T3 cells were seeded on gridded coverslips 24 hr before microinjection. Cells were comicroinjected with mammalian expression vectors for MSE55 and EGFP (used as a tracking agent). Upon detection of EGFP expression, time-lapse video microscopy was begun and digitized images were captured at 2-min intervals. Cell is shown at 3.5, 4, and 9 hr after microinjection.
Coexpression of MSE55 and N17Rac in NIH 3T3 Fibroblasts Produces Both Peripheral Filopodia and Cellular Extensions.
In addition to their independent roles in mediating actin cytoskeletal reorganization, Rho GTPases often work together in a cascade to modulate cell shape and motility (3). In this signaling cascade, Cdc42 activates Rac, leading to lamellipodia formation (3). Because of this hierarchical signaling cascade, we wanted to determine whether Rac activity contributed to extension formation in fibroblasts overexpressing MSE55. Thus, we tested the biological effect of MSE55 when cotransfected with a dominant-negative Rac inhibitor N17Rac (3) to block Rac signaling. NIH 3T3 cells expressing N17Rac alone formed many filopodia around the cell periphery (Fig. 5 a and b). Cells expressing both N17Rac and MSE55 formed peripherally localized filopodia and, additionally, showed the formation of one to two long actin-based extension(s) similar to those observed in NIH 3T3 cells expressing only MSE55 (compare Fig. 5 c and d with Fig. 3 a and b). These data indicate that in fibroblasts, MSE55-induced extensions do not require Rac-signaling activity.
Figure 5.
Extension formation does not require Rac Activity. NIH 3T3 cells were cotransfected with MSE55 and dominant-negative Rac. N17Rac transfected cells were detected with rabbit polyclonal anti-Myc antibody and goat anti-rabbit IgG-FITC (green). N17Rac-expressing cells produce multiple thin filopodia (a and b). N17Rac, MSE55, and F-actin were detected by using triple immunofluorescence. Cells coexpressing N17Rac and MSE55 form multiple thin filopodia and additionally form a single, long cellular extension (c and d). MSE55 was detected with M2 anti-FLAG mouse mAb and goat anti-mouse IgG-Alexa 350 (data not shown). Expression of dominant-negative N17Rac (green) was detected with a rabbit polyclonal antibody against c-Myc and with anti-rabbit IgG-FITC. F-actin was detected with Texas Red-phalloidin (red). (Bar = 20 μm.)
We addressed further the function of Cdc42 in MSE55-mediated extension formation by cotransfecting MSE55 with dominant-negative N17Cdc42 and MSE55. Cells expressing only N17Cdc42 were rounded and reduced in size (data not shown). Cells coexpressing N17Cdc42 and MSE55 lacked extension formation, and the majority of cells were rounded and reduced in size as was observed in cells expressing only N17Cdc42 (data not shown). Taken together, these results indicate that Cdc42, but not Rac, is required for MSE55-mediated extension formation.
DISCUSSION
It has become apparent that a fairly large number of effector proteins are responsible for transducing signals from the various Rho GTPases. Our work here indicates that MSE55 is a Cdc42 effector that induces the formation of long actin-based extensions in fibroblasts. MSE55 now can be considered a bona fide Cdc42 effector protein based on the following findings. First, glutathione S-transferase-capture experiments demonstrate that MSE55 interacts with Cdc42, but not with Rac or Rho, in a GTP-dependent fashion. Second, MSE55-induced extension formation in NIH 3T3 cells depended on Cdc42 activity, but not on Rac activity. Third, an MSE55 CRIB mutant markedly impaired extension formation. Finally, cells coexpressing MSE55 and dominant-negative Rac formed both filopodia and cellular extensions. Taken together, these results indicate that MSE55 is a Cdc42 effector protein that mediates actin cytoskeletal reorganization leading to membrane-extension formation in NIH 3T3 cells.
Cdc42 is best characterized for its regulation of cytoskeletal reorganization leading to filopodia formation (2, 3). Fibroblast cells expressing either Cdc42 (2, 3), FGD1, a Cdc42-specific exchange factor (34, 35), or the Cdc42 effector N-WASP (28) all induce a similar phenotype, with filopodia forming around the cell periphery. The RalA GTPase also induces filopodia and acts downstream of Cdc42 to recruit the actin-bundling protein filamin (36). Here, we show that in NIH 3T3 cells, MSE55 induces membrane extensions distinct from filopodia consisting of one to two long actin-based protrusions. Unlike MSE55 expression in NIH 3T3 cells, expression of MSE55 in Cos-7 cells enhanced actin polymerization at membrane ruffles. These distinct phenotypes likely are due to the unique cytoskeletal architecture and/or signaling activities between Cos-7 and NIH 3T3 cells. These results suggest that MSE55 functions to stabilize actin polymerization at distinct sites on the plasma membrane in different ways depending on the cell type.
Several Cdc42-specific effectors induce cytoskeletal reorganization including MSE55, N-WASP, and WASP. Although MSE55, N-WASP, and WASP all affect actin polymerization, each does so in qualitatively different ways. MSE55 induces membrane protrusions, N-WASP causes the formation of multiple filopodia (27, 28), and WASP causes actin polymerization in the perinuclear region (26). In epithelial cells, MSE55 localizes to membrane ruffles, N-WASP localizes to filopodia (27, 28), and WASP shows a punctate staining pattern in the cytoplasm of endothelial cells (26). These data suggest that effector protein localization may explain the differences seen in actin cytoskeleton organization.
It is now clear that the Cdc42 effector protein N-WASP modulates actin polymerization through multiple mechanisms. Although N-WASP originally was shown to depolymerize actin through a cofilin-like sequence (28), it coordinates additional steps in actin polymerization. For example, Cdc42 regulates Arp2/3 complex-mediated actin nucleation (37) through N-WASP (38). In particular, the C terminus of N-WASP binds to the Arp 2/3 complex and stimulates Arp 2/3-mediated actin nucleation (38). N-WASP also mediates actin reorganization through its interactions with the actin-binding protein profilin (39). These results suggest that MSE55, like N-WASP, may regulate cellular extension formation by multiple mechanisms.
Although MSE55 does not contain a cofilin-like actin-binding sequence, there are some structural similarities between MSE55 and N-WASP. Both MSE55 and WASP family members contain stretches of positively charged amino acids N-terminal to the CRIB domain and a stretch of negatively charged amino acids at the C terminus of the protein. Miki et al. (28) have shown that Cdc42 binding to the CRIB domain of N-WASP disrupts electrostatic interaction between the positively charged N terminus and the negatively charged C terminus, thus exposing the cofilin-like actin-severing domain. Because MSE55 contains similar stretches of charged amino acids, MSE55 activity is likely to be regulated by Cdc42 binding. Like N-WASP and WASP, MSE55 contains proline-rich sequences (29). In particular, MSE55 contains eight repeats of the amino acid sequence PAANPPA that may interact with actin-binding proteins such as profilin or other SH3-containing signaling molecules (29). Thus, the simplest model of MSE55 function involves Cdc42 binding to MSE55, leading to the exposure of a unique domain (i.e., the polyproline repeats) that is involved in actin polymerization, leading to extension formation. Additional insight into the mechanism of MSE55 signaling may come from our identification of four additional, MSE55-like molecules that define a new family of Cdc42 effectors (D.M.S. and P.D.B., unpublished results). The potential role of each of these individual effectors, as well as the function of their conserved domains, is under current investigation.
Acknowledgments
We thank Dr. Chris Connelly for valuable technical assistance in electroporation and microscopy. We are also indebted to the Hall laboratory for various Rho, Rac, and Cdc42 constructs. We thank Dana M. Pirone for the critical reading of this manuscript. P.D.B. was supported by a Hitchings–Elion fellowship from the Burroughs Wellcome Fund and an R29 from the National Cancer Institute (R29-CA 77459-01). This work also was supported in part by National Institutes of Health Grant GM43880 to S.S.
ABBREVIATIONS
- CRIB
Cdc42/Rac interactive binding
- EGFP
enhanced green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Stossel T P. Science. 1993;260:1086–1094. doi: 10.1126/science.8493552. [DOI] [PubMed] [Google Scholar]
- 2.Kozma R, Ahmed S, Best A, Lim L. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nobes K, Hall A. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 4.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 5.Ridley A J, Hall A. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 6.Coso O A, Chiariello M, Yu J-C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 7.Minden A, Lin A, Claret F-X, Abo A, Karin M. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 8.Olson M F, Ashworth A, Hall A. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 9.Lim L, Manser E, Leung T, Hall C. Eur J Biochem. 1996;242:171–185. doi: 10.1111/j.1432-1033.1996.0171r.x. [DOI] [PubMed] [Google Scholar]
- 10.Burbelo P D, Drechsel D, Hall A. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 11.Manser E, Leung T, Salihuddin H, Zhao Z, Lim L. Nature (London) 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 12.Manser E, Huang H-Y, Loo Z-Q, Chen X-Q, Dong J-M, Leung T, Lim L. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sells M A, Knaus U G, Bagrodia S, Ambrose D M, Bokoch G M, Chernoff J. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Z-S, Manser E, Chen X-Q, Chong C, Leung T, Lim L. Mol Cell Biol. 1998;18:2153–2163. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamarche N, Tapon N, Stowers L, Burbelo P D, Aspenstrom P, Bridges T, Chant J, Hall A. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 16.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. Science. 1997;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 17.Westwick J K, Lambert Q T, Clarke G J, Symons M, Van Aelst L, Pestell R G, Der C J. Mol Cell Biol. 1997;17:1324–1335. doi: 10.1128/mcb.17.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagrodia S, Derijard B, Davis R J, Cerione R A. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 19.Polverino A, Frost J, Yang P, Hutchison M, Neiman A M, Cobb M H. J Biol Chem. 1995;270:26067–26070. doi: 10.1074/jbc.270.44.26067. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S J, Han J, Sells M A, Chernoff J, Knaus U G, Ulevitch R J, Bokoch G M. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 21.Brown J L, Stowers L, Baer M, Trejo J A, Coughlin S, Chant J. Curr Biol. 1996;6:598–605. doi: 10.1016/s0960-9822(02)00546-8. [DOI] [PubMed] [Google Scholar]
- 22.Leung T, Chen X Q, Tan I, Manser E, Lim L. Mol Cell Biol. 1998;18:130–140. doi: 10.1128/mcb.18.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo L, Lee T, Tsai L, Tang G, Jan L Y, Jan Y N. Proc Natl Acad Sci USA. 1997;94:12963–12968. doi: 10.1073/pnas.94.24.12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura K, Ito M, Amano M, Chihara K, Fuata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, et al. Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 25.Brown J L, Jaquenoud M, Gulli M-P, Chant J, Peter M. Genes Dev. 1997;11:2972–2982. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Symons M J, Derry M J, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 27.Miki H, Miura K, Takenawa T. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 28.Miki H, Sasaki T, Takai Y, Takenawa T. Nature (London) 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 29.Bahou W F, Cambell A D, Wichia M S. J Biol Chem. 1992;267:13986–13992. [PubMed] [Google Scholar]
- 30.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 32.Bartel P L, Chien C-T, Sternglanz R, Fields S. In: Cellular Interactions in Development: A Practical Approach. Hartley D, editor. Oxford: Oxford Univ. Press; 1993. pp. 153–179. [Google Scholar]
- 33.Neudauer C L, Joberty G, Tatsis N, Macara I G. Curr Biol. 1998;8:1151–1160. doi: 10.1016/s0960-9822(07)00486-1. [DOI] [PubMed] [Google Scholar]
- 34.Olson M F, Pasteris N G, Gorski J L, Hall A. Curr Biol. 1996;6:1628–1633. doi: 10.1016/s0960-9822(02)70786-0. [DOI] [PubMed] [Google Scholar]
- 35.Zheng Y, Fischer D J, Santos M F, Tigyi G, Pasteris N G, Gorski J L, Xu Y. J Biol Chem. 1996;271:33169–33172. doi: 10.1074/jbc.271.52.33169. [DOI] [PubMed] [Google Scholar]
- 36.Ohta Y, Suzuki N, Nakamura S, Hartwig J H, Stossel T P. Proc Natl Acad Sci USA. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma L, Rohatgi R, Kirschner M W. Proc Natl Acad Sci, USA. 1998;95:15362–15367. doi: 10.1073/pnas.95.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner M W. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 39.Suetsugu S, Miki H, Takenawa T. EMBO J. 1998;17:6516–6526. doi: 10.1093/emboj/17.22.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]