Abstract
Background and aims: Genetic variation in the chromosome 5q31 cytokine cluster (IBD5 risk haplotype) has been associated with Crohn’s disease (CD) in a Canadian population. We studied the IBD5 risk haplotype in both British and Japanese cohorts. Disease associations have also been reported for CARD15/NOD2 and TNF variants. Complex interactions between susceptibility loci have been shown in animal models, and we tested for potential gene-gene interactions between the three CD associated loci.
Methods: Family based association analyses were performed in 457 British families (252 ulcerative colitis, 282 CD trios) genotyped for the IBD5 haplotype, common CARD15, and TNF−857 variants. To test for possible epistatic interactions between variants, transmission disequilibrium test analyses were further stratified by genotype at other loci, and novel log linear analyses were performed using the haplotype relative risk model. Case control association analyses were performed in 178 Japanese CD patients and 156 healthy controls genotyped for the IBD5 haplotype.
Results: The IBD5 haplotype was associated with CD (p=0.007), but not with UC, in the British Caucasian population. The CARD15 variants and IBD5 haplotype showed additive main effects, and in particular no evidence for epistatic interactions was found. Variants from the IBD5 haplotype were extremely rare in the Japanese.
Conclusions: The IBD5 risk haplotype is associated with British CD. Genetic variants predisposing to CD show heterogeneity and population specific differences.
Keywords: Crohn’s disease, ulcerative colitis, inflammatory bowel disease, IBD locus
The inflammatory bowel diseases (IBD) Crohn’s disease (CD) and ulcerative colitis (UC) are chronic disorders of the gastrointestinal tract. Combined data from twin studies demonstrated a greater concordance for CD and UC in monozygotic twins than dizygotic twins—33% versus 4% and 13% versus 2%, respectively1–3—and the risk to siblings of affected individuals relative to the general population (λs) was 15–40 for CD and 8–10 for UC.4 These data show that there is a strong genetic component to disease susceptibility in IBD.
Genetic variants from three distinct loci have recently been shown to be associated with CD. Firstly, mutations in the monocyte CARD15/NOD2 gene have been shown to be associated with CD.5–7 The leucine rich repeat region of CARD15 is thought to have binding activity for bacterial components (including lipopolysaccharide), and activation of CARD15 stimulates the nuclear factor κB (NFκB) pathway and subsequent cytokine production.8 Secondly, we have described an association between IBD and the TNF−857C/T promoter polymorphism (located within the chromosome 6, IBD3 locus) in a British Caucasian population.9 We demonstrated higher stimulated tumour necrosis factor α (TNF) production in TNF−857C homozygotes, allele specific binding of the transcription factor OCT1, and an interaction between OCT1 and NFκB within the TNF promoter. Finally, genetic variation in a 250 kb cytokine gene cluster on chromosome 5q31 (IBD5 locus and risk haplotype) has been strongly associated with CD in a Canadian Caucasian population.10 Although the disease causing mutation has not been precisely characterised, genes contained within the risk haplotype include immunoregulatory cytokines.
Multiple genetic variants are thought to act to influence a complex disease like CD. It is possible for variants influencing phenotype expression (for example, whether or not an individual develops CD) to act independently of each other, in an additive manner, referred to as genetic heterogeneity. Alternatively, variants can interact with each other to affect phenotypic expression to a greater extent than would be expected by individual variants acting independently. Epistasis is defined as such non-additive effects due to complex interactions between multiple variants. In humans, epistatic interactions between genetic variants have been identified in Alzheimer’s disease, breast cancer, Hirschprung’s disease, and sickle cell anaemia.11–14 Furthermore, the interleukin 10 deficient mouse model of IBD demonstrates epistatic interactions between some susceptibility loci,15 and these findings suggested the investigation of possible epistatic interactions between the recently identified CD genetic variants.
We studied whether the IBD5 risk haplotype influenced British Caucasian CD, and assessed the association for the first time in UC and in a Japanese CD population. We performed further analyses to study the presence of genetic heterogeneity or epistasis between the different CD associated variants: CARD15 (chromosome 16), IBD5 (chromosome 5), and TNF (chromosome 6).
MATERIALS AND METHODS
Subjects
Northern European Caucasian families with one or more children affected with IBD were ascertained through the Oxford Gastroenterology Unit. Both parents were available from 457 families, of which 101 had two or more affected sibling pairs and 356 a single affected offspring. Data were obtained for a total of 282 CD trios, 252 UC trios, and 10 indeterminate colitis trios. Numbers of trios (fully genotyped for all markers) analysed after genetic stratification of the CD phenotype were: CD-CARD15-0: 184 trios; CD-CARD15–1+2: 95 trios; CD-IBD5-0: 77 trios; CD-IBD5-1+2: 200 trios; CD-TNF−857CC: 251 trios; and CD- TNF−857CT/TT: 27 trios.
Japanese samples were obtained from 178 CD patients who attended Tohoku University Hospital (Sendai, Japan). We recruited 156 Japanese volunteers living in the same area as healthy controls.
In both populations, the diagnosis of IBD was made on the basis of clinical symptoms and endoscopic, radiographic, and histological findings according to conventional criteria. Demographic and phenotype data of affected CD individuals in both populations are described in table 1 ▶. Ethics approval was obtained from both British and Japanese institutions.
Table 1.
Clinical characteristics of Crohn’s disease individuals
| British Caucasian affected offspring (n=282) | Japanese cases (n=178) | |
| Sex (M/F) | 35%/65% | 73%/27% |
| Age at diagnosis (y)* | 22.4 | 26.0 |
| Disease location† | ||
| Ileal | 72% | 76% |
| Colonic | 60% | 70% |
| Perianal | 21% | 10% |
†Disease location defined according to previously published criteria.23
*Mean years.
Genotyping
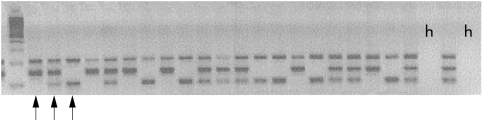
Eleven single nucleotide polymorphisms (SNPs) were previously reported that had alleles unique to the 250 kb IBD5 risk haplotype, and were essentially identical in their genetic information by virtue of being in nearly total linkage disequilibrium with one another and of similar allele frequencies.10 We genotyped two SNPs from the 5′ and 3′ ends of the IBD5 haplotype (http://www.genome.wi.mit.edu/humgen/IBD5) to confirm a similar extent of linkage disequilibrium in our populations, and enable subsequent accurate identification of the IBD5 risk haplotype. Genotyping for IGR2060a_1 and IGR3096a_1 was performed separately using multiplexed sequence specific polymerase chain reaction (PCR) amplification (Amplitaq Gold, Applied Biosystems standard protocol). For IGR2060a_1, we used primers flanking the polymorphism and specific to each allele at the most 3′ base (for G allele, 5′-CATTACATCCTTGCAACCCTG-3′; for C allele, 5′-AGCTCAGTCATTACCTTTGCG- 3′), with the outer primers (5′-AATGTGGGAGGGAAGTTGTG-3′ and 5′-TGTA AAATGGGGACAATTACAGTG-3′) to form two allele specific PCR products of different size (fig 1 ▶). For IGR3096a_1, allele specific primers were: T allele (5′-GAACCCAAACA TCCTGGAGAAT-3′), C allele (5′-CCTTGGTAGGTTCTCAGCTG-3′), and outer primers (5′-GGACAAAAATAGGGCCACAAG-3′ and 5′- TGCCACCTCCCATCTCTAAG-3′). For both SNPs, the two outermost primers also provided a larger positive control band (not allele specific). PCR products were electrophoresed on agarose gels and visualised with ethidium bromide staining (fig 1 ▶).
Figure 1.
Agarose gel image of IGR2060a_1 genotyping. First lane is 100 bp DNA size marker, followed by products of 24 individual polymerase chain reactions (PCR). PCR product sizes are control (212 bp), C allele (149 bp), and G allele (105 bp). Arrows mark, in order, CC homozygote, GC heterozygote, GG homo- zygote. Lanes marked “h” are negative controls (containing no DNA).
Genotyping for common CARD15 variants (Arg702Trp, Gly908Arg, and Leu1007fsinsC) and the TNF promoter −857C/T variant was performed by PCR restriction fragment length polymorphism assay, as described previously.9,16
Statistical analysis
Linkage disequilibrium (D′) between variants was calculated by the ldmax algorithm implemented in the GOLD software package.17 We carried out family based association analyses (transmission disequilibrium test (TDT))18 using the ASPEX program (ftp://lahmed.stanford.edu/pub/aspex). Simulations were done to calculate empirical probabilities for the TDT χ2 statistic by permuting parent alleles while fixing the identity by descent status of siblings within a family. Thus the p value for the TDT reflects an association independent of linkage when multiple siblings from the same family are tested, allowing the use of both simplex and multiply affected families in the TDT (simplex and multiply affected families were combined for all analyses). We also performed further analyses stratified on carriage of the associated disease associated mutations by recoding CD affection status as unaffected dependent on genotype. We stratified CD affected offspring by carriage or non- carriage of the IBD5, CARD15, and TNF−857 variants. TDT analyses for each variant were performed for CD overall, and with genotype stratified groups. We performed Kaplan-Meier analyses of the effect of the IBD5 haplotype on age at diagnosis, using all CD affected individuals (including parents) from the family cohort and the log rank test to assess significance (SPSS Inc., Chicago, Illinois, USA).
To formally test for genetic heterogeneity, we selected a single affected offspring per family. In the case of multiply affected families, a single offspring was chosen at random, and similar results were obtained with several different random seeds (data not shown). IBD5, CARD15, and TNF−857 alleles transmitted to affected offspring from each parent were determined using MERLIN.19 We then constructed a case control study from the family data (using the haplotype relative risk model of transmitted/untransmitted alleles to a single affected offspring, including those from homozygous parents20) and performed log linear analyses of the main effects and interactions using the CATMOD procedure implemented in the SAS analysis suite (SAS Institute Inc., Cary, North Carolina, USA).
RESULTS
Association studies of the IBD5 haplotype in the British Caucasian population.
Strong linkage disequilibrium between variants from the 5′ and 3′ extremes of the IBD5 haplotype (IGR2060a_1 and IGR3096a_1) was observed in 96 genotyped healthy Caucasian blood donors (D′=0.81, p<0.00001). These data confirmed a similar (minimum) extent of linkage disequilibrium in the British population as found in a Canadian population.10 Subsequent genotyping was therefore performed using IGR2060a_1 only.
TDT results for IBD, CD, and UC phenotypes in the British families are shown in table 2 ▶. The association of CD with IBD5 was replicated. The strength of association, as measured by the transmitted/untransmitted allele ratio for IGR2060a_1, was weaker in the British cohort (1.42) than reported in 139 Canadian trios (2.38, χ21=4.8, p=0.03). No evidence for association of IBD5 with UC was observed, and this locus appeared to be CD specific.
Table 2.
Transmission disequilibrium test analysis of the IBD5 Crohn’s disease risk haplotype in Caucasian families*
| IBD5 (IGR2060a_1) | |||
| Phenotype | Trios | T/U | p Value |
| IBD | 544 | 272/243 | 0.23 |
| UC | 252 | 105/124 | 0.24 |
| CD | 282 | 162/114 | 0.0066 |
*Transmitted (T)/untransmitted (U) minor alleles from heterozygous parents to affected offspring shown, with bootstrap p values.
CD, Crohn’s disease; UC, ulcerative colitis; IBD, inflammatory bowel disease.
The weaker association of the IBD5 haplotype in the British (mean age at diagnosis of affected offspring 22.4 years) versus Canadian (16.3 years) CD affected offspring raised the possibility that the IBD5 haplotype may have an effect on age at diagnosis of CD. To test this hypothesis, we performed Kaplan-Meier analysis of the effect of IBD5 haplotype status on age at diagnosis in all CD individuals from the British cohort. IBD5 haplotype status did not have a significant effect on age at diagnosis in log rank comparisons between groups (data not shown). Mean age at diagnosis in individuals not possessing the IBD5 risk haplotype was 26.5 (SEM 1.0), heterozygotes 25.4 (SEM 0.7), and homozygotes 25.2 (SEM 0.8) years.
Genotype stratified association studies of CARD15, IBD5, and TNF−857 in Caucasian CD
Stratified TDT results for the CD phenotype in the British families are shown in table 3 ▶. The association, in this cohort, of the Arg702Trp and Leu1007fsinsC CARD15 variants with CD, and of TNF−857C with CD in individuals not possessing CARD15 variants, has been reported elsewhere.9,16
Table 3.
Stratified transmission disequilibrium test analysis of the IBD5 haplotype, CARD15, and TNF−857 variants in Caucasian Crohn’s disease
| Variant | Phenotype* | T/U‡ | Ratio | p Value |
| IBD5 | CD | 162/114 | 1.4 | 0.0066 |
| CD-CARD15-0 | 106/66 | 1.6 | 0.0033 | |
| CD-CARD15-1+2 | 52/45 | 1.2 | 0.55 | |
| CD-TNF857-CT/TT | 15/9 | 1.7 | 0.31 | |
| CD-TNF857-CC | 145/103 | 1.4 | 0.0088 | |
| CARD15 Arg702Trp | CD | 65/37 | 1.8 | 0.0059 |
| CD-IBD5-0 | 17/12 | 1.4 | 0.45 | |
| CD-IBD5-1+2 | 47/24 | 2.0 | 0.0064 | |
| CD-TNF857-CT/TT | 11/4 | 2.8 | 0.12 | |
| CD-TNF857-CC | 53/32 | 1.7 | 0.021 | |
| CARD15 Leu1007fsinsC | CD | 47/9 | 5.2 | <10−5 |
| CD-IBD5-0 | 19/4 | 4.8 | 0.0067 | |
| CD-IBD5-1+2 | 27/5 | 5.4 | 0.00019 | |
| CD-TNF857-CT/TT | 7/0 | 0.016 | ||
| CD-TNF857-CC | 40/9 | 4.4 | <10−4 | |
| TNF−857C† | CD | 40/27 | 1.5 | 0.16 |
| CD-IBD5-0 | 8/11 | 0.7 | 0.61 | |
| CD-IBD5-1+2 | 31/16 | 1.9 | 0.026 | |
| CD-CARD15–0 | 27/12 | 2.3 | 0.030 | |
| CD-CARD15–1+2 | 13/14 | 0.9 | 1.0 |
*CARD15 (or IBD5 or TNF−857)-0, wild type; -1+2 carrying one or two disease associated variants (for CARD15 any of Arg702Trp, Gly908Arg, Leu1007fsinsC).
†Note the common TNF−857C allele is associated with inflammatory bowel disease.
‡Transmitted (T)/untransmitted (U).
The association between CD and the IBD5 haplotype was strongest in the CARD15 negative group, between CD and Leu1007fsinsC equally strong in both groups after IBD5 stratification, and between CD and TNF−857C only in the CARD15 negative group. The association between CD and Arg702Trp was strongest in the IBD5 haplotype possession group. Transmission/non-transmission ratios (a statistic related to genotype relative risk) between groups possessing and not possessing variants were similar, and simple χ2 tests of allelic transmission between groups were not significant after any stratification (table 3 ▶ and data not shown). It is noteworthy that the TDT only uses a proportion of the dataset (because only allele transmissions from heterozygous parents are counted), and is not therefore a powerful method to test for possible epistatic interactions.
Numbers of CD trios in the TNF−857CT/TT stratified group were small and the results from this analysis need to be interpreted with caution.
Log linear analyses of potential interactions between variants in British Crohn’s disease
The most appropriate statistical method to assess the significance of potential interactions between variants using family based data has not been established. We therefore reduced the family data to case and control alleles (sample size equivalent to a study of 230 case and 230 control individuals). We performed a log linear analysis using the haplotype relative risk association statistic. Single locus models were first tested in which only the effect of a single genetic variant on phenotype (that is, presence of absence of CD) was analysed. Two (and similarly three) locus models allowed the main effects of multiple genetic variants to be analysed, with the addition of a higher order term(s) in the model to account for possible non- additive interactions between variants (epistasis). These models therefore allowed testing for genetic heterogeneity (that is, association still present after accounting for all non-additive interactions) and epistasis.
The Gly908Arg CARD15 variant did not show a significant association with CD, and was too rare in our population to test for interactions.16 As before, no association between non-stratified CD and TNF−857C was observed.9
The IBD5 haplotype and the Arg702Trp/Leu1007fsinsC CARD15 variants were associated with CD (table 4 ▶), as observed in the TDT analysis. Analysis of the main effects was then performed using two locus models, in which association was tested after conditioning for higher order effects from potential gene-gene interactions (tables 5 ▶, 6 ▶). Significant association with CD remained for the Arg702Trp and Leu1007fsinsC CARD15 variants and the IBD5 haplotype after conditioning, providing direct evidence for independent main effects (heterogeneity). Finally, no evidence for gene-gene interactions (epistasis) were noted in two or three locus models performed using combinations of CARD15 variants, TNF−857C, and the IBD5 haplotype, as demonstrated by similar frequencies of allelic combinations in both case and control groups (table 5 ▶) and a p value of >0.05 for all models (data not shown).
Table 4.
Log linear analysis of variants in British Crohn’s disease. Single locus model: allele frequencies and tests of association
| Allele frequency | ||||
| Control | Case | χ21 | p Value | |
| Crohn’s disease association | ||||
| IBD5 (IGR2060a_1) | 39.3% | 48.2% | 8.0 | 0.005 |
| CARD15 (Arg702Trp) | 5.4% | 9.8% | 5.9 | 0.02 |
| CARD15 (Leu1007fsinsC) | 1.3% | 7.7% | 16.7 | <0.0001 |
| TNF−857C | 93.5% | 95.0% | 1.0 | 0.3 |
Table 5.
Log linear analysis of variants in British Crohn’s disease. Two locus model: allele frequencies
| Cohort | Frequencies of observed allelic combinations | |||
| Arg702 | Arg702 | 702Trp | 702Trp | |
| IBD5- | IBD5+ | IBD5- | IBD5+ | |
| Control | 56.4% | 37.9% | 3.5% | 2.2% |
| Case | 47.9% | 42.1% | 6.0% | 4.0% |
| Leu1007 | Leu1007 | 1007fsinsC | 1007fsinsC | |
| IBD5- | IBD5+ | IBD5- | IBD5+ | |
| Control | 59.1% | 39.4% | 1.5% | 0% |
| Case | 47.3% | 45.3% | 4.9% | 2.5% |
| TNF-857T | TNF-857T | TNF-857C | TNF-857C | |
| IBD5- | IBD5+ | IBD5- | IBD5+ | |
| Control | 5.5% | 1.5% | 55.0% | 38.0% |
| Case | 2.8% | 1.5% | 49.0% | 46.8% |
| Arg702 | Arg702 | 702Trp | 702Trp | |
| TNF-857T | TNF-857C | TNF-857T | TNF-857C | |
| Control | 5.9% | 88.5% | 0.7% | 4.8% |
| Case | 4.4% | 85.7% | 0.4% | 9.5% |
| Leu1007 | Leu1007 | 1007fsinsC | 1007fsinsC | |
| TNF-857T | TNF-857C | TNF-857T | TNF-857C | |
| Control | 4.3% | 87.8% | 0.7% | 7.2% |
| Case | 6.5% | 92.2% | 0.0% | 1.3% |
Table 6.
Log linear analysis of variants in British Crohn’s disease. Two locus model: tests of association after conditioning on potential gene-gene interactions
| Crohn’s disease association (main effects) | χ21 | p Value |
| IBD5, conditioned on | ||
| CARD15 (Arg702Trp) | 6.9 | 0.008 |
| CARD15 (Leu1007fsinsC) | 6.3 | 0.01 |
| TNF−857C | 7.1 | 0.008 |
| CARD15 (Arg702Trp), conditioned on | ||
| IBD5 | 4.5 | 0.03 |
| TNF−857C | 6.1 | 0.01 |
| CARD15 (Leu1007fsinsC), conditioned on | ||
| IBD5 | 9.0 | 0.003 |
| TNF−857C | 15.2 | <0.0001 |
| TNF−857C, conditioned on | ||
| IBD5 | 1.7 | 0.2 |
| CARD15 (Arg702Trp) | 1.4 | 0.2 |
| CARD15 (Leu1007fsinsC) | 1.5 | 0.2 |
Association studies of IBD5 variants in the Japanese population
In the Japanese population, three healthy controls and two Crohn’s disease cases were heterozygous for IGR2060a_1, and IGR3096a_1 was not polymorphic in either Japanese cohort (all wild-type). No individuals were homozygous for the IBD5 haplotype. The rarity of these variants (<1% allele frequency) in the Japanese precluded tests for association.
DISCUSSION
Much progress has been made in our understanding of the genetic susceptibility to IBD. Linkage studies have identified regions of the human genome likely to contain susceptibility loci, and more recently disease associated variants have been reported from three distinct loci (CARD15/NOD2, chromosome 5q31 cytokine cluster/IBD5 risk haplotype, and TNF).5,7,9,10 The association between CD and CARD15 variants has now been widely confirmed in Caucasian populations,16,21–24 and replication studies of the other two loci are awaited.
We sought first to study genetic variants from the 5q31 cytokine gene cluster in a British IBD cohort. This region contains a cluster of immunoregulatory cytokines potentially important in Th1/Th2 differentiation. However, the presence of strong linkage disequilibrium means the genetic approach alone has so far been unable to pinpoint the disease causing mutation.10 We confirmed the association of the IBD5 risk haplotype with CD previously reported in Canadian families. However, the strength of association was weaker in the British cohort. Evidence for both linkage and association in the Canadian cohort was greatest in early onset CD,10 and mean age at diagnosis was 4.1 years younger in the Canadian cohort than in the current British cohort. However, no effect of the IBD5 haplotype on age at diagnosis was observed in the British cohort. Other differences in phenotype (for example, disease location or behaviour) might explain the stronger association in the Canadian cohort, although these comparisons could not be made as this information was not provided in the published Canadian study.10
The role of the IBD5 risk haplotype in UC has not previously been investigated. No evidence for association was observed in our British cohort, adding to the molecular evidence for heterogeneity between CD and UC obtained from studies of CARD15.
We genotyped IBD5 haplotype variants in Japanese CD and healthy controls, and found that in contrast with the Caucasian population, the variants were extremely rare. This difference is too large to be explained by the minor demographic and phenotypic differences between the populations. The two variants tested are from extreme ends of the “Caucasian” CD risk haplotype, and it therefore seems likely that the disease causing variant carried on the “Caucasian” haplotypic background does not play a role in Japanese CD (although until the variant is known this cannot be directly tested). It is also possible that other Japanese specific disease causing mutations have arisen independently in the same gene that carries the Caucasian specific mutation. A comprehensive study is now necessary to determine whether there are variants in this region specifically associated with Japanese CD, and to assess the population specific pattern of linkage disequilibrium in the 5q31 region. If CD associated variants are found in the Japanese, this may provide an alternative method for functional studies for identifying the disease causing Caucasian IBD5 mutation. It is interesting that common Caucasian CARD15 variants have also been found to be rare in both Japanese and African American populations.25–27 It thus seems likely that different genetic defects contribute to CD susceptibility in different ethnic populations.
Epistasis, or non-additive interaction between disease causing genetic variants, is well recognised in Drosophilia and mouse models,28,29 and has been demonstrated more recently in several human diseases. In IBD, a genome wide search for colitis susceptibility loci in the interleukin 10 deficient mouse demonstrated complex epistatic interactions between loci.15 Conversely, it is possible to knockout, over express, or mutate distinct genes involved in diverse pathways and produce a similar colitis phenotype in mouse models. Now that associated variants have been determined from three separate loci in human IBD, it is possible to assess whether epistasis plays a major role or whether heterogeneity exists between the variants. No significant interactions were found between the CARD15 variants, TNF−857, and the IBD5 haplotype in either stratified TDT or log linear analyses. This finding is supported by the lack of interaction between CARD15 variants and the IBD5 haplotype reported in a smaller Canadian cohort,10,24 although only stratified TDT analyses were used. Furthermore, direct evidence for genetic heterogenity in the British CD cohort was provided in a novel analysis of the main effects and interactions. Strong association persisted between CD and the CARD15 variants/IBD5 haplotype after conditioning on possible interactions between variants. Different interactions may however be present between other loci, and it seems likely that both genetic heterogeneity and epistasis may coexist in human IBD. Indeed, preliminary evidence for epistasis has already been reported between the 1p36 and IBD1 loci.30
We conclude that the IBD5 cytokine cluster risk haplotype does not influence susceptibility to UC and plays a lesser role in genetic susceptibility to CD in the British than in the Canadian population. Our data support the existence of genetic heterogeneity and population specific differences in the inherited susceptibility to human CD. Further studies are now needed to address the relative importance of IBD susceptibility genes in different populations and in different disease subphenotypes. The possibility of epistatic interactions in IBD will need to be reassessed if associated variants are identified from other susceptibility loci.
Acknowledgments
We thank Joseph Terwilliger for advice on the heterogeneity analysis. This work was supported by the British Medical Research Council (DAvH, DPMcG, LINK Grant), the Wellcome Trust (LRC), the Hamanako Symposium (KN), Oxagen Ltd, and NIH grant EY15652 (LRC). We thank all participating families, our research nurses, the National Association for Crohn’s and Colitis, consultants, and general practitioners who helped ascertain families.
Abbreviations
CD, Crohn’s disease
UC, ulcerative colitis
IBD, inflammatory bowel disease
TDT, transmission disequilibrium test
TNF, tumour necrosis factor α
NFκB, nuclear factor κB
PCR, polymerase chain reaction
SNP, single nucleotide polymorphism
REFERENCES
- 1.Orholm M, Binder V, Sorensen TI, et al. Concordance of inflammatory bowel disease among Danish twins. Results of a nationwide study. Scand J Gastroenterol 2000;35:1075–81. [DOI] [PubMed] [Google Scholar]
- 2.Thompson NP, Driscoll R, Pounder RE, et al. Genetics versus environment in inflammatory bowel disease: results of a British twin study. BMJ 1996;312:95–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tysk C, Lindberg E, Järnerot G, et al. Ulcerative colitis and Crohn’s disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut 1988;29:990–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Heel DA, Satsangi J, Carey AH, et al. Inflammatory bowel disease: Progress toward a gene. Can J Gastroenterol 2000;14:207–18. [DOI] [PubMed] [Google Scholar]
- 5.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001;411:603–6. [DOI] [PubMed] [Google Scholar]
- 6.Hampe J, Cuthbert A, Croucher PJ, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet 2001;357:1925–8. [DOI] [PubMed] [Google Scholar]
- 7.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001;411:599–603. [DOI] [PubMed] [Google Scholar]
- 8.Inohara N, Ogura Y, Chen FF, et al. Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J Biol Chem 2001;276:2551–4. [DOI] [PubMed] [Google Scholar]
- 9.Van Heel DA, Udalova IA, De Silva AP, et al. Inflammatory bowel disease is associated with a TNF polymorphism that affects an interaction between the OCT1 and NF- kappaB transcription factors. Hum Mol Genet 2002;11:1281–9. [DOI] [PubMed] [Google Scholar]
- 10.Rioux JD, Daly MJ, Silverberg MS, et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet 2001;29:223–8. [DOI] [PubMed] [Google Scholar]
- 11.Nelson MR, Kardia SL, Ferrell RE, et al. A combinatorial partitioning method to identify multilocus genotypic partitions that predict quantitative trait variation. Genome Res 2001;11:458–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie MD, Hahn LW, Roodi N, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet 2001;69:138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zubenko GS, Hughes HB 3rd, Stiffler JS. D10S1423 identifies a susceptibility locus for Alzheimer’s disease in a prospective, longitudinal, double-blind study of asymptomatic individuals. Mol Psychiatry 2001;6:413–19. [DOI] [PubMed] [Google Scholar]
- 14.Carrasquillo MM, McCallion AS, Puffenberger EG, et al. Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat Genet 2002;32:237–44. [DOI] [PubMed] [Google Scholar]
- 15.Farmer MA, Sundberg JP, Bristol IJ, et al. A major quantitative trait locus on chromosome 3 controls colitis severity in IL-10-deficient mice. Proc Natl Acad Sci U S A 2001;98:13820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Heel D, McGovern D, Cardon L, et al. Fine mapping of the IBD1 locus did not identify Crohn’s disease associated NOD2 variants: implications for complex disease genetics. Am J Med Genet 2002;111:253–9. [DOI] [PubMed] [Google Scholar]
- 17.Abecasis GR, Cookson WO. GOLD—graphical overview of linkage disequilibrium. Bioinformatics 2000;16:182–3. [DOI] [PubMed] [Google Scholar]
- 18.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Gen 1993;52:506–16. [PMC free article] [PubMed] [Google Scholar]
- 19.Abecasis GR, Cherny SS, Cookson WO, et al. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002;30:97–101. [DOI] [PubMed] [Google Scholar]
- 20.Terwilliger JD, Ott J. A haplotype-based ‘haplotype relative risk’ approach to detecting allelic associations. Hum Hered 1992;42:337–46. [DOI] [PubMed] [Google Scholar]
- 21.Cuthbert AP, Fisher SA, Mirza MM, et al. The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology 2002;122:867–74. [DOI] [PubMed] [Google Scholar]
- 22.Murillo L, Crusius JB, van Bodegraven AA, et al. CARD15 gene and the classification of Crohn’s disease. Immunogenetics 2002;54:59–61. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad T, Armuzzi A, Bunce M, et al. The molecular classification of the clinical manifestations of Crohn’s disease. Gastroenterology 2002;122:854–66. [DOI] [PubMed] [Google Scholar]
- 24.Vermeire S, Wild G, Kocher K, et al. CARD15 Genetic variation in a Quebec population: prevalence, genotype-phenotype relationship, and haplotype structure. Am J Hum Genet 2002;71:74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamazaki K, Takazoe M, Tanaka T, et al. Absence of mutation in the NOD2/CARD15 gene among 483 Japanese patients with Crohn’s disease. J Hum Genet 2002;47:469–72. [DOI] [PubMed] [Google Scholar]
- 26.Inoue N, Tamura K, Kinouchi Y, et al. Lack of common NOD2 variants in Japanese patients with Crohn’s disease. Gastroenterology 2002;123:86–91. [DOI] [PubMed] [Google Scholar]
- 27.Bonen DK, Nicolae DL, Moran T, et al. Racial differences in NOD2 variation: Characterization of NOD2 in African-Americans with Crohn’s disease. Gastroenterology 2002;122:A29 (abstract 249). [Google Scholar]
- 28.Nadeau J. Modifier genes in mice and humans. Nat Rev Genet 2001;2:165–73. [DOI] [PubMed] [Google Scholar]
- 29.Mackay T. Quantitative trait loci in Drosophilia. Nat Rev Genet 2002;2:11–20. [DOI] [PubMed] [Google Scholar]
- 30.Cho JH, Nicolae DL, Gold LH, et al. Identification of novel susceptibility loci for inflammatory bowel disease on chromosomes 1p, 3q and 4q: evidence for epistasis between 1p and IBD1. Proc Nat Acad Sci U S A 1998;95:7502–7. [DOI] [PMC free article] [PubMed] [Google Scholar]