Lactase-phlorizin hydrolase (LPH), an intestinal microvillus membrane enzyme that hydrolyses lactose, is a critical enzyme for neonatal nutrition. The developmental pattern of lactase expression in the human fetus is distinct from that of similar digestive enzymes. Before week 24 of gestation, intestinal lactase activity is low. It then begins to increase, and during the third trimester lactase activity increases markedly until levels in term neonates are at or above those of infants aged 2–11 months.1 Lactase exhibits a characteristic proximal to distal pattern of expression in the small intestine; enzyme activity is greatest in the mid- jejunum, with decreasing activity both proximally and distally, resulting in minimal activity in the proximal duodenum and the terminal ileum.2
In most human populations, lactase activity decreases during mid-childhood (about five years of age), resulting in low levels from that age onwards. This pattern is similar to that seen in all other mammals examined, with a reduction in intestinal lactase activity at weaning to a fraction of that found in the suckling newborn. In striking contrast, a minority of the human population, especially people of Northern European extraction and a few other racial groups, retain high levels of activity throughout adult life.3 Persistence of elevated lactase activity is thought to be a relatively recent human evolutionary development, arising within the last 10 000 years, coincident with the development of dairying.4 A small number of subjects with lactase non-persistence have been demonstrated to have an abnormality in the intracellular processing of newly synthesised LPH protein, indicating post-transcriptional control of non-persistence.5 However, it is now clear that in humans, as in all mammals studied, the primary mechanism of both the persistence and non-persistence phenotypes is regulation of gene transcription.6–8 Considerable effort has been devoted to the elucidation of the molecular mechanisms involved in the transcriptional regulation responsible for these two human phenotypes.
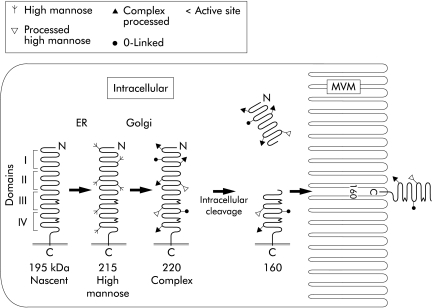
The gene for human LPH, located on chromosome 2q21, comprises 17 exons and covers approximately 49 kb, giving rise to a messenger RNA (mRNA) of slightly more than 6 kb. From initiation codon to stop codon, human LPH mRNA encodes 1927 amino acids forming the complete translation product.9 Sequence comparisons indicate that the coding region is comprised of four homologous parts, leading to the suggestion that the gene is the product of two duplication events during evolution.10 The nascent protein is heavily glycosylated so that the final translation product is about 220 kDa (fig 1 ▶). This high molecular mass glycoprotein undergoes intracellular cleavage, dividing regions I and II from regions III and IV. The protein consisting of regions III and IV contains the two active sites and is inserted into the microvillus membrane of the enterocyte as a mature enzyme of approximately 160 kDa.11 The proximal portion encompassing regions I and II has no enzymatic activity, but has been shown to function in correct folding of the enzyme.12 Initial analyses of the gene identified several single base polymorphisms (SNPs) within both the coding region and the 5′ flanking region. None was considered to have functional significance.9
Figure 1.
Model of the molecular forms of lactase-phlorizin hydrolase during synthesis and processing in the human villus enterocyte. The early changes in apparent molecular size are due to glycosylation, as indicated in the diagram. Note that the two active sites are located in domains III and IV. The subsequently removed domains I and II are important for correct folding of the nascent protein. Although not indicated on this drawing, the enzyme forms a homodimer during processing. The final N terminal cleavage of a small segment is depicted by the elimination of the terminal loop in the microvillus form of the enzyme.
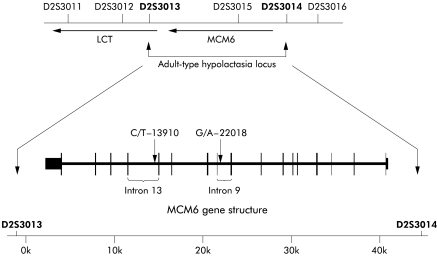
Subsequent analysis has led to the identification of additional SNPs and several other features unique to the human gene that may be of relevance to the mechanisms of LPH persistence/non-persistence. The first 100 bp of the proximal promoters of the mammals analysed to date (rat, mouse, pig, and human) are virtually identical and appear to be similarly regulated.13–15 Studies in transgenic animals have indicated that approximately 1 kb of the 5′ flanking sequence in the pig, and 2 kb of the 5′ flanking sequence in the rat, are sufficient to direct appropriate tissue, cell, and villus expression, as well as the developmental decline at weaning.16–18 Comparable studies in humans have not been carried out. In contrast with the other mammals analysed, the 5′ flanking region of the human LPH gene contains five inserted stretches of repetitive DNA, two Alu sequences of approximately 300 bp each, and three other short repetitive sequences, making a direct comparison of the more distal regulatory region to those of other mammals difficult. Whether or not these inserted repetitive DNA segments affect LPH expression is currently unknown. Furthermore, exon 17 of MCM6, a cell cycle regulatory gene, ends 3.5 kb from the start site of the human LPH gene.19 The transcriptional start site of the MCM6 gene lies approximately 39 kb 5′ of the LPH transcriptional start site. The two genes are close together but the available evidence indicates that their regulation is independent. Two polymorphisms associated with LPH non- persistence originally identified by Enattah and colleagues 20 and examined further here, lie within introns 13 and 9 of the MCM6 gene (fig 2 ▶).
Figure 2.
Schematic representation of the region between the two highlighted genetic markers that is associated with adult-type hypolactasia (Enattah and colleagues20). Vertical bars within the MCM6 gene represent the exons, the widths indicating relative size. The two associated single base polymorphisms (SNPs) lie within the indicated introns of the MCM6 gene which is located 5′ to the LCT (lactase-phlorizin hydrolase) gene on human chromosome 2. The arrows indicate the direction of gene transcription (the centromeric region of the chromosome is located to the left). The scale bar at the bottom of the figure indicates the size of the DNA region occupied by the MCM6 gene. The numbers at introns 13 and 9 indicate the positions of the SNPs relative to the transcriptional start site of lactase (redrawn from Enattah and colleagues20).
The report by Enattah and colleagues 20 mapped the DNA changes responsible for lactase persistence (or its converse, adult-type hypolactasia) to a region 13–22 kb upstream of the LPH gene. Using traditional linkage analysis, they first narrowed the region to approximately 3.4 million bases between genetic markers named D2S114 and D2S2385. They then hypothesised that the allele causing lactase persistence arose once in the recent past on a particular chromosome. In this scenario, recombination events in the subsequent history of the population would separate the persistence allele from alleles in other parts of the chromosome, but in the immediate vicinity the persistence alleles would still be inherited together (in linkage disequilibrium) with nearby alleles from the ancestral chromosome. Thus recombination events can be used to narrow the region of interest.
To identify this signal of linkage disequilibrium, they typed several additional markers within the critical 3.4 Mb region. They identified a 47 kb region containing LPH and upstream sequences in which all individuals with lactase persistence carried the same alleles. The localisation was based in part on data from two chromosomes that differ from the ancestral chromosome at only a single marker. These two chromosomes could therefore be derived from a recent mutation at that one marker rather than from a recombination event, meaning that localisation to the 47 kb region might be premature. Nevertheless, they re- sequenced the entire 47 kb region, including the entire LPH gene, and identified only one variant (a C>T SNP, located 13910 bases upstream of LPH) that was perfectly associated with lactase persistence. All 99 individuals with low lactase activity were homozygous for a C at this SNP whereas all 137 individuals with lactase persistence carried either C/T or T/T. A similar but not quite perfect association was found with a G>A SNP at −22018. No other variants were as tightly associated with lactase persistence as were these two SNPs. Interestingly, other haplotypes had previously been associated with lactase persistence and non-persistence.21
A report in this issue of Gut by the same group22 extends these studies by testing whether these SNPs are associated with decreased expression of LPH mRNA levels [see page 647]. As expected, higher levels of LPH mRNA and lactase activity were found in intestinal biopsy samples of subjects whose DNA contained a T at the −13910 SNP and an A at the −22018 SNP. This correlation is perhaps not surprising given the previous very tight correlation between these alleles and lactase persistence 20 and the tight correlation between lactase persistence and high lactase mRNA levels, as first reported in 1992.6,7 However, they also used a clever technique whereby SNPs in the coding region were used to distinguish the transcripts synthesised from the two LPH alleles in an individual heterozygous for one of these coding SNPs. By using allele specific reverse transcription-polymerase chain reaction directed at the coding SNPs, they were able to quantify not only the total levels of LPH mRNA but also the relative levels of expression from the two different transcripts. By this method, they showed that LPH mRNA transcripts are less abundant when synthesised from a chromosome carrying the C at −13910 and G at −22018 than from chromosomes carrying a T and an A at these two sites. Thus, in this population, levels of LPH mRNA were confirmed to be correlated with these SNP patterns.
The reported perfect correlation between T at −13910 and lactase persistence is extremely suggestive, and this paper indicates that lactase persistence is largely or completely explained by a cis acting effect on mRNA levels that is due to either the −13910 SNP or an SNP in perfect linkage disequilibrium with this SNP. In this study, all of the chromosomes carrying a T at −13910 also had an A at −22018. It would be of interest to apply this same technique to individuals in whom these two alleles had been separated (for example, a C at −13910 but an A at −22018) to determine which of these SNPs was more tightly correlated with the cis acting effect on mRNA levels. One could also imagine using this assay in vitro to try to determine whether the −13910 SNP is truly causative or whether a more distant genetic variant might be responsible for the persistence of lactase activity into adulthood.
Except for rare cases of congenital lactase deficiency, reported to be due to a separate gene,23 every human infant has high levels of LPH expression. If the polymorphisms regulate LPH expression, it is unclear how to account for both universal elevated expression in infants and the later development of lactase persistence/non-persistence in different individuals. While the correlation of the polymorphisms with the LPH phenotypes presented here is excellent, it does not demonstrate causation. The discussion indicates that the polymorphisms are both located within repetitive DNA sequences. As the Alu sequences are unique to primates, this is consistent with a mechanism for LPH persistence unique to humans. However, post-weaning LPH non-persistence is common to all mammals. It is unclear how both pre- and post-weaning human patterns can be accounted for by one or both of these polymorphisms.
In contrast with the previous publication, in which it was suggested that the polymorphisms altered a transcription factor binding site, no mechanism is presented here. The discussion implies that the two SNPs may identify LPH enhancers. Experiments to test this hypothesis should be straightforward to carry out. At present, it remains unclear whether the polymorphisms directly affect expression of LPH or are simply markers for LPH persistence or non-persistence.
REFERENCES
- 1.Grand RJ, Watkins JB, Torti FM. Development of the human gastrointestinal tract. A review. Gastroenterology 1976;70:790–810. [PubMed] [Google Scholar]
- 2.Newcomer AD, McGill DB. Distribution of disaccharidase activity in the small bowel of normal and lactase-deficient subjects. Gastroenterology 1966;51:481–8. [PubMed] [Google Scholar]
- 3.Rings EH, Grand RJ, Buller HA. Lactose intolerance and lactase deficiency in children. Current Opin Pediatric 1994;6:562–7. [DOI] [PubMed] [Google Scholar]
- 4.Flatz G. Genetics of lactose digestion in humans. Adv Hum Genet 1987;16:1–77. [DOI] [PubMed] [Google Scholar]
- 5.Witte J, Lloyd M, Lorenzsonn V, et al. The biosynthetic basis of adult lactase deficiency. J Clin Invest 1990;86:1338–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escher JC, de Koning ND, van Engen CG, et al. Molecular basis of lactase levels in adult humans. J Clin Invest 1992;89:480–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lloyd M, Mevissen G, Fischer M, et al. Regulation of intestinal lactase in adult hypolactasia. J Clin Invest 1992;89:524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krasinski SD, Estrada G, Yeh KY, et al. Transcriptional regulation of intestinal hydrolase biosynthesis during postnatal development in rats. Am J Physiol 1994;267:G584–94. [DOI] [PubMed] [Google Scholar]
- 9.Boll W, Wagner P, Mantei N. Structure of the chromosomal gene and cDNAs coding for lactase-phlorizin hydrolase in humans with adult-type hypolactasia or persistence of lactase. Am J Hum Genet 1991;48:889–902. [PMC free article] [PubMed] [Google Scholar]
- 10.Mantei N, Villa M, Enzler T, et al. Complete primary structure of human and rabbit lactase-phlorizin hydrolase: implications for biosynthesis, membrane anchoring and evolution of the enzyme. EMBO J 1988;7:2705–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naim H Y, Jacob R, Naim H, et al. The pro region of human intestinal lactase- phlorizin hydrolase. J Biol Chem 1994;269:26933–43. [PubMed] [Google Scholar]
- 12.Jacob R, Peters K, Naim HY. The prosequence of human lactase-phlorizin hydrolase modulates the folding of the mature enzyme. J Biol Chem 2002;277:8217–25. [DOI] [PubMed] [Google Scholar]
- 13.Fang R, Santiago NA, Olds LC, et al. The homeodomain protein Cdx2 regulates lactase gene promoter activity during enterocyte differentiation. Gastroenterology 2000;118:115–27. [DOI] [PubMed] [Google Scholar]
- 14.Krasinski SD, Van Wering HM, Tannemaat MR, et al. Differential activation of intestinal gene promoters: functional interactions between GATA-5 and HNF-1 alpha. Am J Physiol 2001;281:G69–84. [DOI] [PubMed] [Google Scholar]
- 15.Mitchelmore C, Troelsen JT, Spodsberg N, et al. Interaction between the homeodomain proteins Cdx2 and HNF1alpha mediates expression of the lactase-phlorizin hydrolase gene. Biochem J 2000;346:529–35. [PMC free article] [PubMed] [Google Scholar]
- 16.Troelsen JT, Mehlum A, Olsen J, et al. 1 kb of the lactase-phlorizin hydrolase promoter directs post-weaning decline and small intestinal-specific expression in transgenic mice. FEBS Lett 1994;342:291–6. [DOI] [PubMed] [Google Scholar]
- 17.Krasinski SD, Upchurch BH, Irons SJ, et al. Rat lactase-phlorizin hydrolase/human growth hormone transgene is expressed on small intestinal villi in transgenic mice. Gastroenterology 1997;113:844–55. [DOI] [PubMed] [Google Scholar]
- 18.Lee SY, Wang Z, Lin C-K, et al. Regulation of intestine-specific spatiotemporal expression by the rat lactase promoter. J Biol Chem 2002:13099–105. [DOI] [PubMed]
- 19.Harvey CB, Wang Y, Darmoul D, et al. Characterisation of a human homologue of a yeast cell division cycle gene, MCM6, located adjacent to the 5′ end of the lactase gene on chromosome 2q21. FEBS Lett 1996;398:135–40. [DOI] [PubMed] [Google Scholar]
- 20.Enattah NS, Sahi T, Savilahti E, et al. Identification of a variant associated with adult- type hypolactasia. Nat Genet 2002;30:233–7. [DOI] [PubMed] [Google Scholar]
- 21.Hollox EJ, Poulter M, Zvarik M, et al. Lactase haplotype diversity in the Old World. Am J Hum Genet 2001;68:160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuokkanen M, Enattah NS, Oksanen A, et al. Transcriptional regulation of the lactase-phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut 2003;52:647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarvela I, Sabri Enattah N, Kikkonen J, et al. Assignment of the locus for congenital lactase deficiency to 2q21, in the vicinity of but separate from the lactase-phlorizin hydrolase gene. Am J Hum Genet 1998;63:1078–85. [DOI] [PMC free article] [PubMed] [Google Scholar]