Abstract
Background and aims: Characterisation of the underlying molecular mechanisms that promote Barrett’s progression may ultimately lead to identification of potential predictive genetic markers that classify patients’ malignant risk. In an attempt to understand these causative pathways, fluorescence in situ hybridisation (FISH) was used in this study to determine when specific genetic alterations arise during Barrett’s associated neoplastic progression.
Methods: Endoscopic cytology brushings were obtained from 28 patients with Barrett’s metaplasia, 28 with dysplasia (20 low grade dysplasia (LGD) and eight with high grade dysplasia (HGD)), and seven with adenocarcinoma, together with paired control brushings from regions of normal proximal squamous cell epithelium. The exfoliated epithelial cells were washed and deposited onto slides. Probes specific for the centromeres of chromosomes 4, 8, 20, and Y, and locus specific probes for the tumour suppressor genes p16, p53, and Rb were subsequently hybridised.
Results: Aneuploidy was found early in progression, with metaplastic tissues displaying increased copy numbers of chromosomes 4 and 8. Chromosome 4 hyperploidy was found in 89%, 90%, 88%, and 100% of metaplasias, LGD, HGD, adenocarcinomas, respectively, while chromosome 8 hyperploidy occurred in 71%, 75%, 100%, and 100% of patients with the respective staging. Loss of the p16 tumour suppressor gene also presented in metaplastic epithelium (7%) but most other genetic aberrations were only seen in HGD.
Conclusions: Genetic instability arises well before dysplasia in Barrett’s oesophagus, with chromosome 4 and 8 hyperploidy representing the earliest and most common alterations identified. As these aberrations are widespread at all the premalignant stages, there may be genes on chromosomes 4 and 8 that are involved in both the initiation and progression of Barrett’s oesophagus.
Keywords: Barrett’s oesophagus, chromosome 4, hybridisation, hyperploidy, interphase cytogenetics
Barrett’s oesophagus is a premalignant lesion that is strongly associated with chronic gastro-oesophageal reflux1 and has recently been defined as the presence of any length of columnar epithelia containing goblet cells within the distal oesophagus.2 The condition predisposes patients to oesophageal adenocarcinoma, however, due to self treatment for heartburn and diminished sensitivity of the columnar mucosa to refluxed acid, such adenocarcinomas are often only diagnosed at an advanced stage. The incidence of this malignancy is rapidly rising in the USA and Western Europe, currently varying from 3.2 to 5.2 cases/100 000/year3–5 and is reputed to be very aggressive due to both early invasion and metastasis, which consequently result in low five year survival rates of just 10%.6
Barrett’s metaplasia is known to progress through low grade dysplasia (LGD) and high grade dysplasia (HGD) prior to adenocarcinoma but the underlying molecular mechanisms involved in Barrett’s neoplastic progression are still unclear. Genetic abnormalities involving a variety of oncogenes, tumour suppressor genes, cell adhesion molecules, and growth factors have been identified7–9 but the functional involvement of these alterations in driving tumorigenesis has yet to be defined.
Currently, endoscopic surveillance coupled with histopathological evaluation for dysplasia remains the “gold standard” for assessing the neoplastic potential of Barrett’s oesophagus (with HGD employed as the marker for subsequent adenocarcinoma development). Unfortunately, several problems exist with this current regimen. Firstly, the progression rate of HGD to adenocarcinoma is highly variable, approximately 60% of patients with HGD will develop adenocarcinoma within five years,10 but 41% of these patients may already have an in situ carcinoma at first diagnosis of HGD.11 Secondly, the patient benefits and cost effectiveness of the current biopsy protocols have come into question because of the low proportion of Barrett’s cases that progress to adenocarcinoma.12–14 Finally, subjective grading by pathologists results in intra and interobserver variation, and together with biopsy sampling error can result in misdiagnosis.15 These areas of concern clearly indicate a need for improved diagnostic markers to classify patient risks for cancer development.
An understanding of the molecular mechanisms responsible for neoplastic progression is needed before genetic markers for risk assessment can be identified. Elucidation of such underlying pathways requires characterisation of the genetic alterations that accumulate at each stage of progression. However, the current chromosomal data available for Barrett’s oesophagus lack consistency and implicate a wide variety of genetic aberrations, possibly due to analysis mainly limited to tissues from oesophageal adenocarcinoma (known to be genetically unstable). Initial cytogenetic studies in our laboratory employed comparative genomic hybridisation (CGH) to distinguish genetic alterations that may predict the malignant potential of Barrett’s patients.16 Amplifications of chromosomes 4 and 8 were found in HGD but not in LGD or metaplasia. As CGH involves bulk analysis of sample tissues, aberrations must be present in a high proportion of cells within the sample to be detected. Consequently, early changes present in a low percentage of cells would be overlooked. Hence in this study fluorescence in situ hybridisation (FISH), which involves single cell analysis, was used. The sensitivity of the FISH technique allows the detection of low frequency aberrations such as those that may be present in premalignant lesions.
In this study, Barrett’s epithelial cells were exfoliated with endoscopic cytology brushes as this cell collection method permitted the generation of interphase cell preparations (necessary for cytogenetic investigation) from oesophageal tissues. FISH was subsequently performed to determine at which premalignant stage chromosome 4 and 8 aneuploidy and other commonly detected aberrations in HGD and adenocarcinoma first arise.
METHODS
Patients
Ethics approval was obtained for sample collection over a two year period from consenting patients at Morriston District General Hospital, Swansea, UK. Patients attended an endoscopy clinic with a special interest in Barrett’s oesophagus, which sought referrals for additional patients with dysplasia, in particular HGD. Samples were in the form of cytology brushings taken from 63 consecutive patients in total during the sampling period: 28 with Barrett’s metaplasia, 28 with dysplasia (20 LGD and eight HGD), and seven patients with oesophageal adenocarcinoma. The study group male:female ratio was 3:1, while the age range was 38–90 years (median 64). All the patients were Caucasian.
Endoscopic cytology brushings
Endoscopic cytology brushes were used to obtain two epithelial samples from each patient: one was of normal proximal squamous oesophageal tissue while the second was taken within each patient’s Barrett’s segment. The brushes were immediately placed into cold ETN buffer (0.1 M EDTA, 0.01 M Tris-HCl, 0.02 M NaCl, pH 7) and agitated to generate a cell suspension. Several biopsies were also taken from the Barrett’s lesions (around the brushing site) for histological grading. As brushing collects cells from a larger area than biopsies, they are less likely to be subject to sampling error and so were graded according to the most severe degree of dysplasia recorded in the patient clinical notes.
Interphase cell preparations
The resultant cell suspension was washed three times by centrifugation/re-suspension in ETN buffer. Following cleansing, the cell pellet was resuspended in 500 μl of the buffer solution, and Cytospin4 (ThermoShandon, Cheshire, UK) was used to generate a single layer of interphase cells on pre-cleaned glass slides. The cytodot cell densities were examined under a light microscope and their respective cell suspensions were diluted/concentrated accordingly. Cell preparations were fixed in 90% methanol at −20°C, left to air dry, and then stored at −20°C. On average, 3–5 slides were generated from each brushing.
Slide pretreatment
The interphase cell preparations were treated with 300 μl/ml 0.01 M HCl pepsin, pH 2.7–3 (Sigma, Dorset, UK) at 37°C for seven minutes to remove cytoplasmic proteins, hence improving probe penetration.17 Two five minute washes in phosphate buffered saline (PBS), followed by PBS/50 mM MgCl2 (both at ambient temperature) arrested the enzymatic treatment, and the slides were then dehydrated in an ethanol series to prepare them for FISH.
FISH
Chromosome enumeration probes (CEP) for the centromeres of chromosomes 4, 8, 20, and Y and locus specific identifier (LSI) probes for p53, 13q14 (Rb locus), and dual probe 9p21/CEP9 (p16 locus) were utilised (Vysis, Surrey, UK).
Due to the limited number of slides available for each sample, two probes were simultaneously hybridised in each reaction. The dual probe 9p21/CEP9 was hybridised to one slide, CEPs 8 and 20 were applied to a second slide, and the LSIp53 and 13q14 probes were hybridised to a third. Once scored, the p53 and 13q14 probes were washed off the slide by rinsing in 1×phosphate buffered detergent at room temperature for five minutes, followed by 1×standard saline citrate (SSC) at 90°C for six minutes. To verify complete probe removal, these slides were examined under an epifluorescence microscope prior to a second round of probe hybridisation involving CEPs 4 and Y.
FISH was performed according to the manufacturer’s instructions, with slight modifications. Briefly, 5 μl of probe mixture (consisting of 3.5 μl hybridisation buffer, 0.5 μl each probe, and water, to a 5 μl volume) were applied to each cytodot. The sample and probe were co-denatured on a 75°C hot plate for two minutes, and the slides were incubated for 30 minutes or 16 hours (for CEP or LSI probes, respectively) at 37°C. Following hybridisation, slides were washed in 0.4×SSC/0.3% Nonidet P-40 at 73°C for two minutes, 2×SSC/0.1% Nonidet P-40 at room temperature for 30 seconds, and allowed to air dry. DAPI II 10 μl (Vysis, Surrey, UK) was subsequently applied to counterstain the nuclei.
Signal visualisation
An Olympus BX50 epifluorescence microscope with single and multiple bandpass filters was used to view the FISH slides. All images were captured with a CCD camera coupled to the MacProbe version 4.1 software (Applied Imaging, Newcastle Upon Tyne, UK).
Scoring criteria
Slides were coded and 400 nuclei were analysed per cytodot. Nuclei were only included in the analysis if they had at least one signal from each probe utilised in order to avoid inclusion of results caused by inadequate hybridisation. Overlapping, damaged, or smeared nuclei and those covered with bacteria were excluded from the analysis.
As all matched samples generated from the squamous oesophageal tissues were normal, they were used as controls to establish the background hybridisation variation. The cut off levels for diagnosis of abnormal deletion/amplification were therefore defined by the mean percentage of cells +3 SD of the signal losses/gains displayed in the control samples evaluated.18,19 As no signal gains were seen in the control samples with any of the probes utilised, all such amplifications found within Barrett’s samples were considered aberrant. The mean +3 SD percentage of nuclei with loss of one signal was 2.3%, 2.4%, 1.4%, 3.0%, 2.6%, 2.4%, 4.5%, and 3.0% for CEP4, CEP8, CEP9, CEP20, CEPY, LSI 9p21, LSIp53, and LSI13q14, respectively, and therefore signal loss was defined as abnormal when higher than these cut off percentages.
Reproducibility
To verify protocol reproducibility, two slides from the same brushing sample were hybridised with CEP8 and CEP20. In addition, the whole battery of probes was applied to slides originating from two separate brushings of the same Barrett’s site. No significant difference was found between the data generated (according to the χ2 test with 95% confidence limits) thus demonstrating acceptable scoring reproducibility.
Histological correlation of brushings
Previous investigations have reported a positive correlation between results from cytology brushings and biopsies taken from the same Barrett’s segment.20,21 However, there are existing reports that have found brush cytology to be less sensitive and specific for histological diagnosis.22,23 In this study, to correlate the FISH data generated from the brushings with histology, biopsies taken from the brushed Barrett’s region of five patients were halved. One half was sent to pathology for grading (as metaplasia, indefinite for dysplasia, LGD, HGD, or adenocarcinoma) by a panel of three histopathologists while FISH using centromeric probes for chromosomes 4 and 8 was performed on the other biopsy half. Chromosome 4 and/or 8 aneuploidy identified in the samples generated by the brushings were also detected in 90% of the corresponding biopsies, suggesting histological grading of biopsies reliably correlates with data obtained from cytology brushes.
RESULTS
The significant numerical abnormalities (involving the chromosomes and loci examined) that were detected during this analysis are catalogued in table 1 ▶, while fig 1 ▶ illustrates some examples of these aberrant nuclei.
Table 1.
Proportion of cells in each sample displaying significant signal amplifications or deletions of each fluorescence in situ hybridisation probe utilised
| Patient | Pathology | % 4L | % 4G | % 8L | % 8G | % 9L | % 9G | % p16L | % p16G | % RbL | % RbG | % p53L | % p53G | % 20L | % 20G | % YL |
| 2 | BM | 3.3 | 3.5 | 3.7 | ||||||||||||
| 3 | BM | 7.1 | 1.8 | 3.2 | 4.9 | |||||||||||
| 4 | BM | 22.8 | 1.3 | |||||||||||||
| 5 | BM | 61.3 | 7.1 | 6.4 | ||||||||||||
| 8 | BM | 4.5 | 1.0 | 2.8 | 1.5 | |||||||||||
| 11 | BM | |||||||||||||||
| 16 | BM | 4.7 | 2.8 | |||||||||||||
| 19 | BM | 8.5 | 1.3 | F | ||||||||||||
| 20 | BM | 5.6 | 4.5 | 3.8 | ||||||||||||
| 33 | BM | 13.5 | 4.7 | F | ||||||||||||
| 35 | BM | 9.7 | F | |||||||||||||
| 46 | BM | 7.8 | F | |||||||||||||
| 49 | BM | 9.0 | ||||||||||||||
| 51 | BM | 1.8 | 7.0 | |||||||||||||
| 63 | BM | 14.5 | 4.2 | 19.8 | ||||||||||||
| 73 | BM | 2.8 | 1.3 | F | ||||||||||||
| 86 | BM | 25.3 | 3.8 | |||||||||||||
| 95 | BM | 13.9 | 2.3 | F | ||||||||||||
| 100 | BM | 37.3 | ||||||||||||||
| 105 | BM | 5.5 | 7.8 | |||||||||||||
| 107 | BM | 3.3 | 5.5 | |||||||||||||
| 109 | BM | 3.3 | 2.5 | F | ||||||||||||
| 111 | BM | 4.5 | 3.8 | F | ||||||||||||
| 117 | BM | 5.8 | 6.8 | F | ||||||||||||
| 121 | BM | 19.8 | 2 | 5.3 | 52.3 | 33.3 | ||||||||||
| 127 | BM | 4.3 | 3.3 | 26.8 | ||||||||||||
| 131 | BM | 8.8 | 3.5 | 12.5 | ||||||||||||
| 135 | BM | 9.3 | 16.8 | 1.0 | ||||||||||||
| 9 | LGD | 55.9 | 5.4 | 3.7 | 1.4 | |||||||||||
| 18 | LGD | 5.4 | 38.7 | |||||||||||||
| 21 | LGD | 12.5 | 3.4 | 2.2 | 6.6 | 8.1 | ||||||||||
| 27 | LGD | 14.3 | ||||||||||||||
| 29 | LGD | 29.0 | 92.8 | F | ||||||||||||
| 31 | LGD | 2.8 | ||||||||||||||
| 36 | LGD | 2.4 | 4.5 | 15.8 | 7.3 | |||||||||||
| 40 | LGD | 28.5 | ||||||||||||||
| 53 | LGD | 8.5 | 3.7 | 1.5 | F | |||||||||||
| 55 | LGD | 1.5 | 4.3 | 4.8 | 4.8 | |||||||||||
| 57 | LGD | 1.8 | 2.3 | 2.0 | 1.3 | 10.5 | ||||||||||
| 65 | LGD | 2.5 | 2.3 | 2.75 | 6 | |||||||||||
| 69 | LGD | 5.3 | 10.4 | 4.5 | 17.25 | |||||||||||
| 80 | LGD | 6.2 | 1.8 | 5.0 | ||||||||||||
| 89 | LGD | 47.3 | 8.0 | 2.0 | 9.8 | 6.3 | ||||||||||
| 98 | LGD | 2.0 | 3.2 | 5.0 | ||||||||||||
| 123 | LGD | 8.0 | 3.3 | |||||||||||||
| 129 | LGD | 6.3 | 6.5 | F | ||||||||||||
| 133 | LGD | 7.8 | 6.5 | |||||||||||||
| 141 | LGD | 4.5 | 8.1 | 2.7 | F | |||||||||||
| 1 | HGD | 4.7 | 1.7 | 6.5 | 1 | 11.3 | ||||||||||
| 10 | HGD | 25.4 | 20.8 | 18.7 | 34.9 | 2.0 | 11.5 | 9.3 | 66.8 | |||||||
| 12 | HGD | 9.6 | 1.3 | 21.5 | 8.5 | 7.8 | ||||||||||
| 25 | HGD | 26.7 | 1.5 | 45.3 | 6.3 | 10.3 | ||||||||||
| 42 | HGD | 10.2 | 2.8 | 2.5 | 5.8 | 5.5 | ||||||||||
| 91 | HGD | 50.6 | 20.0 | 11.0 | 5.5 | 12 | F | |||||||||
| 102 | HGD | 25.5 | 48.0 | 38.0 | 36.0 | 11.4 | ||||||||||
| 139 | HGD | 9.5 | 18.5 | 8.8 | 9.5 | 5.8 | 10.2 | |||||||||
| 15 | OA | 32.4 | 2.7 | 3.0 | 1.5 | 4.3 | 5.8 | 7.7 | 4.2 | F | ||||||
| 17 | OA | 16.5 | 63.8 | 1.3 | 5.0 | 1.0 | 5.1 | 2.2 | 2.2 | 1.3 | 63.3 | |||||
| 22 | OA | 54.0 | 12.0 | 2.2 | 8.5 | 10.0 | 4.6 | 3.0 | 2.3 | 7.8 | 1.4 | 15.8 | 10.3 | |||
| 48 | OA | 86.0 | 5.5 | 11.0 | 2.3 | 5.8 | 11.8 | 4.0 | 15.8 | 10.8 | 9.0 | 16.5 | F | |||
| 83 | OA | 65.8 | 5.3 | 7.0 | 28.0 | 8.0 | 4.0 | 43.3 | ||||||||
| 88 | OA | 13.3 | 11.5 | 6.0 | 6.0 | 36.0 | 5.3 | 1.0 | 7 | |||||||
| 97 | OA | 11.0 | 8.5 | 6.8 | 2.3 | 14.3 | 1.3 | 4.0 | 6.0 | 1.0 | 7 |
G, signal gains; L, signal losses; F, female patients.
BM, metaplasia; LGD, low grade dysplasia; HGD, high grade dysplasia; OA, oesophageal adenocarcinoma.
All blank cells indicate samples that were normal (that is, did not display signal losses/gains above the cut off limits).
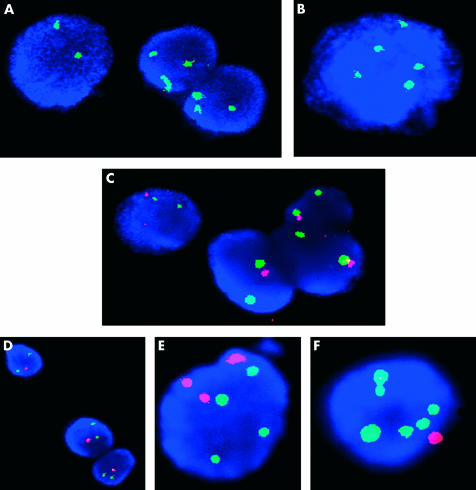
Figure 1.
Examples of a variety of fluorescence in situ hybridisation (FISH) images obtained from Barrett’s interphase cell preparations. (A) Two of three nuclei exhibiting chromosome 4 trisomy (cells originating from a female patient). (B) Male nucleus displaying chromosome 4 hyperploidy and loss of chromosome Y. (C) Nuclei all monoallelic for p16 (red) despite retaining two copies of chromosome 9 (green). (D) Nuclei containing the normal compliment of the Rb gene (green signal) but were monoallelic for p53 (red). (E) Nucleus that was trisomic for chromosome 20 (red) and tetrasomic for chromosome 8 (green signals). (F) Chromosome 8 hyperploidy (green) and chromosome 20 monosomy (red) are exhibited in this nucleus.
Chromosome 4
Chromosome 4 hyperploidy was the most prominent alteration found in Barrett’s metaplasia with 89% of patients displaying the aberration. As illustrated in fig 2 ▶, an increased chromosome 4 copy number persisted into LGD and HGD where it was present in 90% and 88% of cases, respectively, as well as in all adenocarcinomas examined (fig 1A ▶, B). The average percentage of cells displaying chromosome 4 hyperploidy also increased with neoplastic progression from 12% (range 1–61%) in metaplasia to 14% (range 2–56%) in LGD, to 19% (range 2–51%) and 40% (range 11–66%) of cells in HGD and adenocarcinomas, respectively. Although most cells with this aberration contained three copies of chromosome 4, up to five copies were seen in some samples, but as they all displayed either loss or a single copy of chromosome Y, the cells were probably not polyploid, but aneuploid for chromosome 4.
Figure 2.
Percentage of metaplastic, dysplastic, and oesophageal adenocarcinoma samples that had chromosome 4/8 hyperploidy, monoallelic p16, or lacked their Y chromosome. The results are derived from analysis of 28 patients with metaplasia (BM), 20 with low grade dysplasia (LGD), eight with high grade dysplasia (HGD), and seven with oesophageal adenocarcinoma (OA).
Chromosome 8
Chromosome 8 hyperploidy was another predominant genetic alteration present in 71% of metaplastic Barrett’s patients (fig 1E ▶, F). This alteration was maintained during progression, occurring in 75% of LGD and 100% of both HGD and adenocarcinomas (fig 2 ▶). An increased chromosome 8 copy number was detected in almost every sample that displayed chromosome 4 hyperploidy (table 1 ▶) but chromosome 8 aneuploidy generally developed in a lower proportion of cells in each sample: 4% of cells in metaplastic samples (range 1–17%), 5% (range 2–10%) in LGD, 18% (range 1–48%) in HGD, and 17% (range 3–64%) in oesophageal adenocarcinomas. Three copies was also the most predominant chromosome 8 complement found in these cells but up to six copies were present in some. Cells generally displayed differing chromosome 8 and 20 copy numbers, and so again these cells appeared to be aneuploid and not polyploid.
Chromosome 20
Both losses and gains in chromosome 20 copy number were evident during progression of Barrett’s oesophagus (fig 1E ▶, F). Fourteen per cent and 15% of metaplastic and LGD samples, respectively, displayed a gain of chromosome 20 while 11% and 10%, respectively, presented loss of the chromosome. In HGD, chromosome 20 hyperploidy was far more prevalent, occurring in 63% of samples compared with only 13% with loss of the chromosome.
Chromosome Y
Only four males with metaplasia (21%) displayed loss of chromosome Y but this increased to 38% in LGD, 71% in HGD, and finally to 100% in oesophageal adenocarcinomas bearing the aberration (fig 2 ▶).
Tumour suppressor genes
Of the tumour suppressor genes analysed, hemizygous deletion of p16 was the most common alteration (fig 1C ▶), first arising in metaplasia (7% of patients) and increasing in prevalence with progression, as illustrated in fig 2 ▶. Loss of the Rb tumour suppressor gene was a very rare event during progression of Barrett’s oesophagus, only being detected in a single patient with HGD and in 2/7 of those with oesophageal adenocarcinoma. p53 loss was also relatively uncommon in the earlier stages of Barrett’s neoplastic progression. Only 7% and 5% of metaplastic and LGD samples, respectively, displayed losses of p53 but this increased to 38% of HGD patients and 71% of adenocarcinomas.
DISCUSSION
We have shown that coupling brush cytology with interphase FISH is an effective and sensitive way of analysing chromosomal alterations that arise during Barrett’s associated neoplastic progression. FISH was selected for this purpose as it involves single cell analysis and thus has the potential to detect rare alterations such as those present in premalignant lesions. Meanwhile, brush cytology exfoliates a large number of epithelial cells over a broad area and hence is less prone to sampling errors.
Our data verify previous CGH data produced by our group that chromosomes 4 and 8 are frequently amplified in Barrett’s oesophagus.16 The earliest and most prominent alteration identified in this study was chromosome 4 hyperploidy which was present in 89% of metaplastic samples. As this change is widespread in metaplasia, it may be involved in the initiation of Barrett’s oesophagus. An increased chromosome 4 copy number persisted in the majority of LGD and HGD (90% and 88%, respectively) and was present in all oesophageal adenocarcinomas examined. This suggests that the aberration may also confer a cellular growth advantage that is selected for in Barrett’s tissues and may consequently be involved in promoting the progression of Barrett’s oesophagus. Nevertheless, as only 10% of patients with Barrett’s metaplasia progress to adenocarcinoma,24 chromosome 4 aneuploidy alone cannot be sufficient to drive neoplastic progression.
Investigations using CGH have reported both losses and gains of chromosome 4 in oesophageal adenocarcinoma.16,25,26 However, data published by Persons and colleagues27 also demonstrated the presence of chromosome 4 hyperploidy in Barrett’s associated adenocarcinoma using FISH, although it was not as prevalent as our findings. This discrepancy could be due to the different sample preparations utilised. The investigation by Persons et al was performed on paraffin embedded tissue sections, which require considerably higher cut off percentages (to define signal losses/gains as abnormal) due to nuclei truncation caused by the sectioning procedure. The Persons et al study had to ignore alterations present in <25% of cells, thus small abnormal clones would have been overlooked. By performing FISH on a population of undamaged nuclei, we were able to identify low level alterations, hence chromosome 4 hyperploidy was detected at a comparatively higher frequency. Although chromosome 4 contains cell cycle control genes including CENPE (4q24-q25) and MAD2 (4q27) and oncogenes such as c-kit (4q12) and GRO2 (4q12-q13), the target genes associated with chromosome 4 aneuploidy are unknown and require further study.
An increased chromosome 8 copy number was also identified in this study as an early abnormality (present in 71% of metaplastic samples) that persisted into LGD (75%) and all HGD and adenocarcinomas. Chromosome 8 hyperploidy occurred in a lower proportion of cells compared with chromosome 4 hyperploidy, but its prevalence suggests this aberration, in conjunction with other genetic alterations, may result in a phenotype that promotes neoplastic progression. Previously, several investigations have detected chromosome 8 amplifications in a high proportion of oesophageal adenocarcinomas25,26,28 and one study has identified the alteration in premalignant Barrett’s tissue.29 The oncogene c-myc (8q24) has been implicated as a potential target for this amplification by these previous analyses and a recent investigation has further highlighted its amplified expression in Barrett’s metaplasia and adenocarcinoma.30
Of the tumour suppressor genes examined, hemizygous deletion of p16 (9p21) was the most common, first arising in a small proportion of the metaplastic samples examined and then increasing in prevalence with progression. There was a very low rate of p53 deletion in premalignant Barrett’s oesophagus, while Rb loss was only found in a single HGD and two oesophageal adenocarcinoma samples, indicating that deletion of these two latter genes are not very important events in the progression of Barrett’s oesophagus. Other means of p53 and Rb inactivation (for example, mutation, CpG methylation) may play more significant roles.31 32
A general increase in the percentage of aberrant nuclei per sample can be seen as metaplasia progresses through dysplasia to adenocarcinoma. When examining the number of alterations that are present in samples at each stage of progression, it is clear that metaplasia displays the least number of abnormalities while adenocarcinomas exhibit the widest range. This observation, previously noted in our laboratory16 and by several others,25,29,33 demonstrates the increasing genetic instability that accumulates and may be responsible for driving neoplastic progression in Barrett’s oesophagus. The proportion of cells per sample harbouring a particular abnormality also increased with neoplastic progression. For example, the average percentage of cells per sample displaying amplifications of chromosome 4 copy number gradually increased from 12% in metaplasias to 40% in adenocarcinoma samples, therefore providing further evidence for the role of clonal selection and expansion in the progression of Barrett’s oesophagus.34
In conclusion, this study has identified genetic instability in Barrett’s metaplastic tissues with chromosome 4 and 8 aneuploidy representing the most prominent and earliest alterations. The implications of these alterations are as yet unclear but as they are so widespread in all of the premalignant Barrett’s stages, it is possible that the amplification points to key genes on these chromosomes that may be mechanistically involved in the initiation and progression of the lesion. This study has also demonstrated brush cytology coupled to FISH analysis as a suitable technique for identification of early genetic abnormalities. Although follow up investigation of these patients has yet to be performed, this approach may prove to be a useful surveillance tool for identification of aberrations in premalignant lesions that increase a patient’s neoplastic potential.
Acknowledgments
SH Doak was funded by a Tenovus Studentship and the work was supported in part by the European Union PEPFAC project.
Abbreviations
CEP, chromosome enumeration probe
CGH, comparative genomic hybridisation
FISH, fluorescence in situ hybridisation
HGD, high grade dysplasia
LGD, low grade dysplasia
LSI, locus specific identifier
PBS, phosphate buffered saline
SSC, standard saline citrate
REFERENCES
- 1.Lagergren J, Bergstrom R, Lindgren A, et al. Symptomatic gastroesophageal reflux as a risk factor for esophageal adenocarcinoma. N Engl J Med 1999;340:825–31. [DOI] [PubMed] [Google Scholar]
- 2.Sampliner RE. Practice guidelines on the diagnosis, surveillance, and therapy of Barrett’s esophagus. Am J Gastroenterol 1998;93:1028–31. [DOI] [PubMed] [Google Scholar]
- 3.Devesa SS, Blot WJ, Fraumeni JF. Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer 1998;83:2049–53. [PubMed] [Google Scholar]
- 4.Conio M, Cameron AJ, Romero Y, et al. Secular trends in the epidemiology and outcome of Barrett’s oesophagus in Olmsted County, Minnesota. Gut 2001;48:304–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solaymani Dodaran M, Silcocks PB, Logan RFA. Continuing rise in incidence of oesophageal adenocarcinoma in England and Wales. Gut 2001;48(suppl 1):110.11115831 [Google Scholar]
- 6.Farrow DC, Vaughan TL. Determinants of survival following the diagnosis of esophageal adenocarcinoma (United States). Cancer Causes Control 1996;7:322–7. [DOI] [PubMed] [Google Scholar]
- 7.Jenkins GJS, Doak SH, Parry JM, et al. Barrett’s metaplasia: the genetic pathways involved in its progression to adenocarcinoma. Br J Surg 2002;89:824–38. [DOI] [PubMed] [Google Scholar]
- 8.Jankowski JA, Wright NA, Meltzer SJ, et al. Molecular evolution of the metaplasia- dysplasia-adenocarcinoma sequence in the esophagus. Am J Pathol 1999;154:965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald RC, Triadafilopoulos G. Recent developments in the molecular characterization of Barrett’s esophagus. Dig Dis 1998;16:63–80. [DOI] [PubMed] [Google Scholar]
- 10.Montgomery E, Goldblum JR, Greenson JK, et al. Dysplasia as a predictive marker for invasive carcinoma in Barrett esophagus: A follow up study based on 138 cases from a diagnostic variability study. Hum Pathol 2001; 32:379–88. [DOI] [PubMed] [Google Scholar]
- 11.Edwards MJ, Gable DR, Lentsch AB, et al. The rationale for esophagectomy as the optimal therapy for Barrett’s esophagus with high-grade dysplasia. Ann Surg 1996;223:585–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spechler SJ. Barrett’s esophagus: an overrated risk factor. Gastroenterology 2000;119:587–9. [DOI] [PubMed] [Google Scholar]
- 13.Macdonald CE, Wicks AC, Playford RJ. Final results from 10 year cohort of patients undergoing surveillance for Barrett’s oesophagus: Observational study. BMJ 2000;321:1252–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van der Burgh A, Dees J, Hop WCJ, et al. Oesophageal cancer is an uncommon cause of death in patients with Barrett’s oesophagus. Gut 1996;39:5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hulscher JBF, Haringsma J, Benraadt J, et al. Comprehensive cancer centre Amsterdam Barrett advisory committee: first results. Neth J Med 2001;58:3–8. [DOI] [PubMed] [Google Scholar]
- 16.Croft J, Parry EM, Jenkins GJS, et al. Analysis of the premalignant stages of Barrett’s oesophagus through to adenocarcinoma by comparative genomic hybridisation. Eur J Gastroenterol Hepatol 2002;14:1179–86. [DOI] [PubMed] [Google Scholar]
- 17.Ramirez MJ, Surralles J, Galofre P, et al. FISH analysis of 1cen-1q12 breakage, chromosome 1 numerical abnormalities and centromeric content of micronuclei in buccal cells from thyroid cancer and hyperthyroidism patients treated with radioactive iodine. Mutagenesis 1999;14:121–7. [DOI] [PubMed] [Google Scholar]
- 18.Debiec-Rychter M, Lasota J, Sarlomo-Rikala M, et al. Chromosomal aberrations in malignant gastrointestinal stromal tumors: correlation with c-KIT gene mutation. Cancer Genet Cytogenet 2001;128:24–30. [DOI] [PubMed] [Google Scholar]
- 19.Huang S-F, Hsu H-C, Fletcher JA. Investigation of chromosomal aberrations in hepatocellular carcinoma by fluorescence in situ hybridisation. Cancer Genet Cytogenet 1999;111:21–7. [DOI] [PubMed] [Google Scholar]
- 20.Geisinger KR. Endoscopic biopsies and cytologic brushings of the esophagus are diagnostically complementary. Am J Clin Pathol 1995;103:295–9. [DOI] [PubMed] [Google Scholar]
- 21.Tsai TT, Bongiorno PF, Orringer MB, et al. Detection on p53 nuclear protein accumulation in brushings and biopsies of Barrett’s esophagus. Cancer Detect Prev 1997;21:326–31. [PubMed] [Google Scholar]
- 22.Wang HH, Sovie S, Zeroogian JM, et al. Value of cytology in detecting intestinal metaplasia and associated dysplasia at the gastroesophageal junction. Hum Pathol 1997;28:465–71. [DOI] [PubMed] [Google Scholar]
- 23.Falk GW, Chittajallu R, Goldblum JR, et al. Surveillance of patients with Barrett’s esophagus for dysplasia and cancer with balloon cytology. Gastroenterol 1997;112:1787–97. [DOI] [PubMed] [Google Scholar]
- 24.Reid BJ, Levine DS, Longton G, et al. Predictors of progression to cancer in Barrett’s esophagus: Baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol 2000;95:1669–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riegman PHJ, Vissers KJ, Alers JC, et al. Genomic alterations in malignant transformation of Barrett’s esophagus. Cancer Res 2001;61:3164–70. [PubMed] [Google Scholar]
- 26.Van Dekken H, Vissers CJ, Tilanus HW, et al. Clonal analysis of a case of multifocal oesophageal (Barrett’s) adenocarcinoma by comparative genome hybridisation. J Pathol 1999;188:263–6. [DOI] [PubMed] [Google Scholar]
- 27.Persons DL, Croughan WS, Borelli KA, et al. Interphase cytogenetics of esophageal adenocarcinoma and precursor lesions. Cancer Genet Cytogenet 1998;106:11–17. [DOI] [PubMed] [Google Scholar]
- 28.Moskaluk CA, Hu J, Perlman EJ. Comparative genomic hybridisation of esophageal and gastroesophageal adenocarcinomas shows consensus areas of DNA gain and loss. Genes Chromosomes Cancer 1998;22:305–11. [PubMed] [Google Scholar]
- 29.Walch AK, Zitzelsberger HF, Bruch J, et al. Chromosomal imbalances in Barrett’s adenocarcinoma and the metaplasia-dysplasia-carcinoma sequence. Am J Pathol 2000;156:555–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tselepis C, Morris CD, Wakelin D, et al. Upregulation of the oncogene c-myc in Barrett’s adenocarcinoma: induction of c-myc by acidified bile acid in vitro. Gut 2003;52:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coggi G, Bosari S, Roncalli M, et al. p53 protein accumulation and p53 gene mutation in esophageal carcinoma. Cancer 1997;79:425–32. [DOI] [PubMed] [Google Scholar]
- 32.Coppola D, Schreiber RH, Mora L, et al. Significance of Fas and retinoblastoma protein expression during the progression of Barrett’s metaplasia to adenocarcinoma. Ann Surg Oncol 1999;6:298–304. [DOI] [PubMed] [Google Scholar]
- 33.Menke-Pluymers MBE, Van Drunen E, Vissers KJ, et al. Cytogenetic analysis of Barrett’s mucosa and adenocarcinoma of the distal esophagus and cardia. Cancer Genet Cytogenet 1996;90:109–17. [DOI] [PubMed] [Google Scholar]
- 34.Barrett MT, Sanchez CA, Prevo LJ, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet 1999;22:106–9 [DOI] [PMC free article] [PubMed] [Google Scholar]