Abstract
Background: Prophylactic efficacy against colitis following lactobacillus consumption in interleukin 10 (IL-10) knockout (KO) mice has been reported. Whether this applies equally to other probiotic strains is unknown, and the mechanism is unclear.
Aims: (1) To compare the effect of feeding Lactobacillus salivarius subspecies salivarius 433118 and Bifidobacterium infantis 35624 against placebo on enterocolitis, the intestinal microflora, and (2) to compare the systemic immunological response to in vitro microbial challenge in probiotic fed and control IL-10 KO mice.
Methods: Three groups of 10 IL-10 KO mice were fed fermented milk products containing Lb salivarius 433118 at 109 CFU/ml, B infantis 35624 at 108 CFU/ml, and unmodified milk, respectively, for 19 weeks. Faecal samples were taken at regular intervals to confirm gut transit, recovery of fed probiotics, and to assess the impact on the microflora. At sacrifice, the bowels were histologically scored. Cytokine production from Peyers’ patches and splenocytes was measured in vitro by ELISA.
Results: Faecal recovery of probiotics was confirmed in all probiotic fed mice but not in controls. Colonic and caecal inflammatory scores were significantly decreased in both groups of probiotic fed mice (p<0.05). Proinflammatory cytokine production by Peyers’ patches and splenocytes was significantly reduced in probiotic fed animals whereas transforming growth factor β (TGF-β) levels were maintained.
Conclusion: Both Lactobacillus salivarius 433118 and Bifidobacterium infantis 35624 significantly attenuate colitis in this murine model. Attenuation of colitis is associated with a reduced ability to produce Th1-type cytokines systemically and mucosally, while levels of TGF-β are maintained.
Keywords: Crohn’s disease, probiotics, interleukin 10, knockout mice, cytokines
The enteric microflora is implicated in driving the abnormal inflammatory response seen in individuals with inflammatory bowel disease.1 Certain murine models develop spontaneous colitis similar to that of inflammatory bowel disease. In these models, the influence of the enteric flora on gut inflammatory activity has been demonstrated, as germ free animals do not develop disease.2,3 One such murine model is the interleukin 10 (IL-10) knockout (KO) mouse.4
Probiotic bacteria offer a means of modifying the enteric microflora. Probiotics are live microorganisms that confer health benefits through a number of mechanisms, such as therapeutic modification of the enteric flora.5–7 We and others have previously shown that consumption of probiotic lactobacilli can attenuate colitis seen in IL-10 KO mice.8–11 It is not known whether this effect also applies to other bacteria. In addition, the probiotic mechanism of action is not well understood. In our previous study, we demonstrated a significant alteration in the microbial flora of probiotic fed mice, with a reduction in Clostridia perfringens.8 Alteration in the enteric flora is one possible explanation for the reduction seen in the severity of gut inflammation. There is also evidence that commensals signal to the gut epithelium and, in doing so, can influence the mucosal immune response.9,10 The IL-10 KO mouse is a useful model in which to further study a probiotic effect on the immune response as intestinal inflammation is associated with apparently dysregulated production of T helper 1 (Th1)-type proinflammatory cytokines, similar to that of Crohn’s disease.12–14 IL-12 and interferon γ (IFN-γ) are key mediators responsible for inducing enterocolitis, with IL-12 being necessary for sustaining the proliferation of chronically activated Th1 cells.13
The purpose of the present study was: (i) to assess two different probiotic strains for their impact on colitis in IL-10 KO mice; in particular, to investigate whether the beneficial effect noted for lactobacilli in colitis is also seen with bifidobacteria. In addition, (ii) we assessed the impact of probiotic bacteria on cytokine production, both at the mucosal level and systemically.
METHODS
IL-10 KO mice
Thirty 129 Ola×C57BL/6-IL10 KO female mice, aged 7–9 weeks of age, were used in this study (B&K Universal Ltd, East Yorkshire, UK). These mice were maintained on a homozygous background and were housed under specific pathogen free conditions. Mice were housed in a 12 hour light/dark cycle. Following initiation of this study, all mice consumed a standard non-sterile diet. In our specific pathogen free animal house, KO mice of this genetic background do not develop colitis until ⩾20 weeks. Thus this is a preventative study rather than a treatment study.
Probiotic strains
Lactobacillus salivarius subspecies salivarius UCC118 and Bifidobacterium infantis 35624 were originally isolated from the ileal-caecal region of a human adult undergoing reconstructive surgery. Both probiotic strains were isolated on the basis of having desirable probiotic properties.7,15,16 Briefly, these properties included being of human origin, non- pathogenic, resistant to intestinal acid and bile, ability to adhere to human epithelial cells, and ability to temporarily colonise and to be metabolically active within the human gastrointestinal tract. Lb salivarius UCC118 was routinely cultured in de Man, Rogosa, Sharpe (MRS) broth (Oxoid, UK) at 37°C in an anaerobic environment for 24 hours. B infantis 35624 was cultured in MRS broth (Oxoid) enriched with cysteine at 37°C in an anaerobic environment for 48 hours. Spontaneous rifampicin resistant variants of both strains were isolated, prior to initiation of this study, in order to facilitate uncomplicated identification of these bacteria from all other lactobacilli and bifidobacteria.
Feeding trial
The 30 IL-10 KO mice were randomised to one of three groups. The lactobacillus fed group consumed Lb salivarius subspecies salivarius UCC118 at 1×109 organisms/ml of milk, the bifidobacterium fed group consumed B infantis 35624 at 1×108 organisms/ml of milk, while the control group consumed unmodified pasteurised milk vehicle only. Mice were monitored daily; each mouse consumed 4–7 ml of milk per day. Mice in the lactobacillus fed group therefore consumed 4–7×109 organisms per day and mice in the bifidobacterium fed group consumed 4–7×108 organisms per day. The probiotic Lb salivarius UCC118 was initially grown to a 10 ml volume in MRS broth (Oxoid) by incubating overnight at 37°C under anaerobic conditions. A 1% inoculum (vol/vol) was transferred to 400 ml of fresh MRS broth and incubated as before. Lb Salivarius UCC118 was pelleted by centrifugation and resuspended at 1×109 cells/ml in 10% pasteurised skim milk. B infantis 35624 was also grown to a 10 ml volume in MRS broth enriched with 10% cysteine by incubating for 48 hours at 37°C under anaerobic conditions. Again, a 1% inoculum (vol/vol) was transferred to 400 ml of fresh MRS broth with cysteine and incubated under the same conditions. B infantis was pelleted by centrifugation and resuspended at 1×108 cells/ml in 10% pasteurised milk. Both probiotic containing milk solutions and the unmodified pasteurised milk were administered respectively to the groups of IL-10 KO mice in water bottles and mice consumed the solutions ad libitum.
Murine faecal samples were collected at weekly intervals over the trial period. The trial was completed after 19 weeks of feeding, at which time all surviving mice were sacrificed by cervical dislocation. The ileum, caecum, and colon were fixed in formalin for histopathological analysis. Spleens were removed from each mouse at sacrifice and splenocytes isolated for in vitro culturing.
Histopathology
At sacrifice, the ileum, caecum, proximal colon (ascending and transverse colon), and distal colon (descending colon, rectum, and anal canal) of all mice were fixed in 10% formalin and assessed histologically. Two blinded independent observers, using a histological index ranging from 0 to 4 as previously published,8 graded the severity of inflammation at each site within the murine gastrointestinal tract. This index was based on the degree of epithelial layer erosion, goblet cell depletion, and inflammatory cell infiltrate (0=normal; 1=minimal evidence of inflammatory infiltrate; 2=significant evidence of inflammatory infiltrate (cryptitis, crypt abscesses); 3=significant evidence of inflammatory infiltrate with goblet cell depletion; 4=significant evidence of inflammatory infiltrate with erosion of the mucosa).
Microbial analysis
Faecal samples were collected weekly, weighed, and dispersed in 10 ml of PBS. Microbial analysis of the faecal samples involved enumeration of Lb salivarius subspecies salivarius UCC118, B infantis 35624, total lactobacilli, total bifidobacteria, coliforms, and C perfringens. This analysis was performed by pour plating or spread plating onto MRS agar (pH 5.5) plus rifampicin; MRS agar supplemented with 0.05% cysteine hydrochloride (Sigma, St Louis, Missouri, USA) plus rifampicin; MRS agar; MRS agar supplemented with 5% sheep blood, 0.2% lithium chloride (BDH Laboratory Supplies, Poole, UK), 0.3% sodium propionate (Sigma), and 0.05% cysteine hydrochloride (Sigma); violet red bile agar (VRBA); and C perfringens selective medium OPSP with supplements A SR 76 and B SR 77, respectively (all Oxoid, UK, unless otherwise stated). VRBA plates were incubated aerobically for 24 hours while all other plates were incubated anaerobically for 48 hours at 37°C. Anaerobic environments were created using CO2 generating kits (Anaerocult A; Merck, Damstadt, Germany) in sealed gas jars (BBL Becton Dickinson, Cockeysville, Maryland, USA).
DNA was extracted from faecal samples for microbial community profiling by denaturing gradient gel electrophoresis (DGGE). DNA extraction from faecal cells was performed using the BIO 101 FastDNA Spin kit for soil (BIO 101, Vista, California, USA) according to the manufacturer’s instructions with the following exceptions. The faecal pellet (10–30 mg) was resuspended in 978 μl sodium phosphate buffer and 122 μl MT buffer from the BIO 101 kit. Bead beating of samples was performed at 5000 rpm for 3×1 minute in a mini-bead beater (Biospec Products, Bartlesville, Oklahoma, USA), with a one minute chilling in ice between treatments. Following bead beating, samples were centrifuged for four minutes; 100 μl of kit supplied DES buffer (DNA elution solution; ultra pure water) was used to elute DNA from the matrix.
The total predominant bacterial community was profiled by targeting the V6-V8 region of the 16S ribosomal DNA and separating the resultant amplicons by DGGE using a gradient of 38–48% (100% denaturing solution contains 7 M urea and 40% (v/v) formamide) as previously described.15 The Lactobacillus population was profiled using nested polymerase chain reaction (PCR) with Bact-0124-GCf and Univ-5125r primers on previously generated products from amplification with Bact-0011f and Lab-0677r.16 Bifidobacterium specific DGGE-PCR was performed using Bif164-f and Bif662-GC-r primers, as previously described.17 In an attempt to enhance the DGGE-PCR detection sensitivity, the whole 16S rDNA was first amplified with primers Bact-0011f and 1492r,18 and the resultant purified amplicons used as templates in a nested PCR with species specific primers.
After electrophoresis, gels were silver stained.19 DGGE gels were scanned at 400 dpi and profiles compared using the Molecular Analyst 1.12 software (Biorad, Hemel Hempstead, Hertfordshire, UK). Similarity indices were calculated from densitometric curves of the scanned DGGE profiles using the Pearson product moment correlation coefficient.20
Cytokine production by splenocytes
The spleens of all mice were removed at the time of sacrifice. Each spleen was immediately placed in serum free DMEM. The spleen was sieved through a sterilised metal filter into 5 ml of ammonium chloride, which lyses red blood cells. The cell suspension was centrifuged twice at 100 g. Cells were resuspended in DMEM (10% fetal calf serum) and diluted to 1×106 cells/ml for in vitro culturing. The isolated lymphocytes were cocultured with the proinflammatory bacterium Salmonella typhimurium (1×106 cells/ml) for 72 hours at 37°C. Cell supernatants were isolated and stored at −80°C. Cytokine analysis was performed on the supernatants using ELISA (R&D, Abingdon, UK). Cytokines analysed were tumour necrosis factor α (TNF-α), IFN-γ, IL-12, and transforming growth factor β (TGF-β).
Cytokine production by intestinal immunocytes (Peyers’ patches)
A subsequent separate group of IL-10 KO mice were analysed for cytokine production by immunocytes at Peyers’ patches. One group consumed B infantis 35624 at 1×108 organisms/ml of milk while the control group consumed unmodified pasteurised milk vehicle only (n=8 per group). The small intestines of the mice were removed by dissection and the lymphoid follicles of the Peyers’ patches carefully removed from the intestinal serosal side with curved scissors. Seven to 10 Peyers’ patches were obtained per mouse, collected in 5 ml of phosphate buffered saline containing 1 mM EDTA and 1 mM collagenase (Sigma; C-6885, type II) and placed in a shaking oven at 37°C for 20 minutes. The collected patches were then placed between two sterile glass slides and smashed. Immunocytes from the follicles were released into the medium. This cell suspension was centrifuged (100 g×10 minutes) and the pellet was resuspended and diluted in DMEM, 25 mM glucose supplemented with 1% penicillin-streptomycin, and 40 μg/ml gentamicin (Faulding Pharmaceuticals Plc, UK) to a concentration of 106 cells/ml.
Peyers’ patches cells were seeded under duplicate conditions in 24 well tissue culture plates (Costar, Corning, New York, USA) at 1×106 cells per well. Following a 72 hour incubation period (5% CO2 and 37°C humidified atmosphere) all supernatants were harvested for analysis of TNF-α and IFN-γ cytokines. Cytokine production was measured by ELISA (R&D Systems, Abingdon, UK and BD, Pharmingen, Oxford, UK).
Statistical analysis
Analysis of variance for differences between groups in inflammatory activity was carried out using ANOVA while differences between groups in microbial numbers were estimated using area under the curve analysis. A value of p<0.05 was considered to be statistically significant.
RESULTS
Histopathology
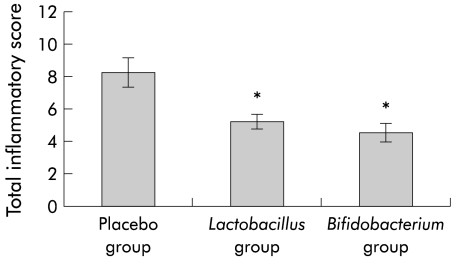
Post sacrifice, a score of 0–4 was awarded to each examined area of bowel: caecum, proximal colon (ascending and transverse colon), and distal colon (descending colon, rectum, and anal canal) with a total possible score of 12 awarded to each mouse gut. When all sections had been examined, the mean score for each group was calculated. Mean inflammatory score for the control group was 8.25 (SEM 0.94). Mean inflammatory scores for the Lb salivarius group and the B infantis groups were 5.25 (0.5) and 4.6 (0.6), respectively. Both probiotic fed groups showed a significant reduction compared with the control group (p<0.05) (fig 1 ▶). In addition, four mice in the control group were found to have pancolitis, with the entire proximal and distal colon equally affected. No cases of pancolitis were seen in either of the probiotic fed groups.
Figure 1.
Gastrointestinal inflammatory score, including the ileum, caecum, and colon, in probiotic and control fed groups (*p<0.05; n=10 each group). Results are expressed as mean (SEM) inflammatory score.
Microbial analysis
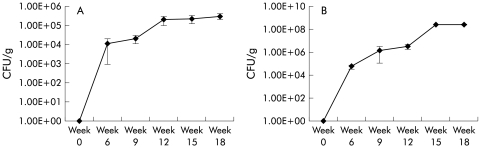
Faecal samples from all mice were analysed to assess transit of the probiotic strains. The probiotic strains were recovered in faeces from all mice in both test groups, within one week of feeding, confirming survival and transit of both the probiotics in the test mice. Lb salivarius UCC118 was recovered in stool samples at 1×108 colony forming units/g (CFU/g) by week 15 of feeding. Stool recovery of B infantis 35624 was approximately 1×105 CFU/g by week 12 of feeding (fig 2 ▶). The probiotic strains were not isolated from any of the mice in the placebo group.
Figure 2.
Recovery of Bifidobacteria infantis 35624 (A) and Lactobacillus salivarius 118 (B) in stool from probiotic fed mice. Results are expressed as mean (SEM) colony forming units/g (CFU/g). No probiotics were isolated from mice in the placebo group.
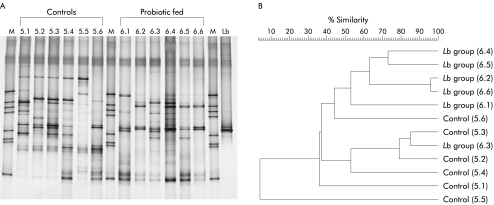
There was no significant difference in culturable bacteria when total lactobacilli, total bifidobacteria, coliforms, and C perfringens were counted in faecal samples, unlike in the previous trial.8 However, comparison of culture independent molecular profiles (16s rDNA DGGE) of faecal flora from control fed mice and probiotic fed mice indicated differences in the dominant microflora of the groups (fig 3A ▶). Each mouse had a distinct individual profile for the dominant microbiota, although there were bands in common. The profiles within each study group were more similar than when compared across groups. The accompanying dendrogram (fig 3B ▶) shows similarity indices for the dominant flora, as assessed by image analysis of the gel. The banding profiles of the control mice clustered together in the dendrogram and were distinct from the profiles generated from the Lactobacillus fed mice. Thus ingestion of Lb salivarius UCC118 influenced the dominant microbiota of the mice. Similarly, the dominant microflora profiles seen in Bifidobacterium fed mice differed from control mice, but lactobacillus and bifidobacterium specific profiles were not altered significantly (data not shown).
Figure 3.
Culture independent molecular profile (16s rDNA DGGE) of faecal flora from control fed mice and probiotic fed mice (A). Lane M, molecular marker; lanes 5.1–5.6, control fed mice; lanes 6.1–6.6, Lactobacillus salivarius subspecies salivarius UCC118 fed mice; Lane Lb, pure culture of Lb salivarius UCC118. The accompanying dendrogram (B) shows similarity indices for the dominant flora, as assessed by image analysis of the gel. The banding profiles for the two groups of mice are distinct and cluster separately.
Cytokine production
Splenocytes
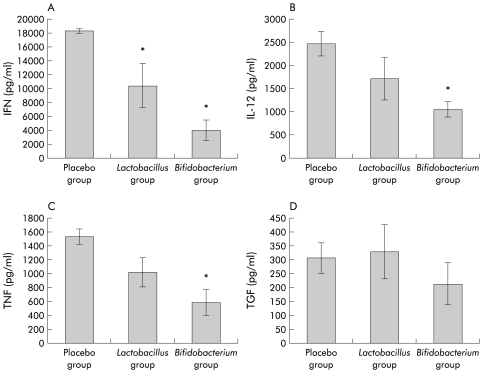
Cytokine analysis was performed on splenocyte supernatants by ELISA following stimulation in vitro with the proinflammatory bacterium S typhimurium. The proinflammatory cytokines IFN-γ, TNF-α, and IL-12 were significantly reduced in the group fed B infantis 35624. In contrast, production of the anti-inflammatory cytokine TGF-β was maintained in all groups (fig 4 ▶). Changes in proinflammatory cytokine production were greater in the bifidobacterium fed group than in the lactobacillus fed group, and this reached significance in the case of IFN-γ. Resting levels of IL-12 in splenocyte supernatants were reduced in both probiotic fed groups compared with the control group but did not reach statistical significance. TNF-α, IFN-γ, and TGF-β levels were not detectable in non-stimulated cultures.
Figure 4.
Cytokine production by stimulated splenocytes in placebo fed and probiotic fed mice. Results are expressed as mean (SEM) cytokine levels per group. There was a significant reduction in the proinflammatory cytokines (interferon γ (IFN) (A), tumour necrosis factor α (TNF) (C), and interleukin 12 (IL-12) (B)) with maintenance of the anti- inflammatory transforming growth factor β (TGF) (D) (*p<0.05; n=10). Resting levels of TNF-α, IFN-γ, and TGF-β were not detectable in non-stimulated cultures. Resting IL-12 levels were lower in both probiotic fed groups compared with the control group but did not reach statistical significance.
Intestinal immunocytes (Peyers’ patches)
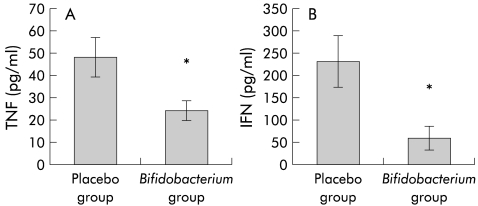
Cytokine analysis was performed on supernatants of Peyers’ patches cells in a separate group fed B infantis 35624 and contemporaneous controls. Spontaneous production of TNF-α and IFN-γ was significantly reduced in animals fed B infantis 35624 compared with controls (fig 5 ▶).
Figure 5.
Resting in vitro production of tumour necrosis factor α (TNF) and interferon γ (IFN) by intestinal mononuclear cells from Peyers’ patches in placebo fed and probiotic fed mice (Bifidobacterium infantis 35624). Results are expressed as mean (SEM) cytokine levels per group (*p<0.05; n=8).
DISCUSSION
The probiotics used in this study (Lactobacillus salivarius subspecies salivarius UCC118 and Bifidobacterium infantis 35624) are well characterised and have been demonstrated to be bile tolerant and acid resistant, and to survive passage through the gastrointestinal tract.21,22 They are non-pathogenic and have been tested in human trials of healthy volunteers and patients with Crohn’s disease7 (unpublished). The results of the present study confirm survival of both probiotics following murine gastrointestinal transit and show that both strains attenuate colitis in the IL-10 KO model. The probiotic effect on mucosal inflammation was reflected in a reduction in the proinflammatory cytokines TNF-α and IFN-γ, secreted by intestinal Peyers’ patches cells. This attenuation of colitis is consistent with our previous study involving the lactobacillus strain8 and is the first report of a beneficial effect using bifidobacteria in this model. The beneficial probiotic impact on mucosa was also associated with changes in systemic cytokine production in vitro. Following challenge (in vitro with S typhimurium), there was a reduction in Th1 cytokines (TNF-α, IFN-γ, and IL-12) in the probiotic groups whereas production of the immunoregulatory (Th3) cytokine TGF-β was maintained.
Probiotic consumption also altered the culture independent molecular profiles of the predominant microflora in the murine gastrointestinal tract. Although there were no detectable differences in the culturable bacteria examined, differences in the DGGE banding profiles representing the dominant microflora in the three groups indicated changes in unidentified bacterial groups in the probiotic fed animals.
A variety of mechanisms may be responsible for the observed probiotic attenuation of colitis. The effects of probiotic consumption on host immune function have previously been discussed.23–25 Attenuation of colitis in the IL-10 KO model suggests that mediators other than IL-10 are involved in the demonstrable probiotic efficacy. Alteration in cytokine profiles observed is important as inflammation in both Crohn’s disease and this murine model is associated with a predominance of Th1 cytokines, whereas Th3 cells, producing TGF-β, are involved in suppressing inflammatory responses and promoting tolerance.10,26–28 The results suggest that the probiotic effect may be mediated by altering the balance between proinflammatory and anti-inflammatory or regulatory cytokines. Secretion of cytokines by isolated splenocytes in vitro is dependent on the stimulus used and the cytokine milieu the cells were isolated from. In this study, the stimulus used was identical in each group. This suggests that it is the cytokine environments from which these cells were isolated which differ.
S typhimurium was chosen as the in vitro challenge because it is a reliable and relevant biological stimulus, being a Gram negative bacteria containing lipopolysaccharide endotoxin. The probiotic linked reduction of inflammatory cytokines in response to the in vitro challenge does not however imply that these mice are more susceptible to infection with this pathogen. Using the same probiotic organisms, we have shown a benefit in mice infected with the salmonella organism with significantly reduced pathogen survival and translocation in mice fed probiotics (O’Mahony et al, in press).
Even though there was no difference in the histological score between the two probiotic groups, it is noteworthy that there was a strong trend towards lower levels of the systemic Th1 cytokines in the bifidobacteria group compared with the lactobacilli group. However, this only reached statistical significance in the case of IFN-γ. Whether this reflects a more potent anti-inflammatory effect is unknown at this stage. The effect may not be proportional to the dose of organism but related to variations in mechanism.
Probiotic bacteria are attractive alternatives for the treatment of gastrointestinal inflammation due to their effects on the composition of the gut flora and activity of the immune system.29 Administration of the probiotic organisms in this study attenuated inflammation and altered the systemic immune response of IL-10 KO mice. The probiotic effect is modest but is consistent and not limited to lactobacilli. However, results cannot be extrapolated to all lactobacilli or bifidobacteria strains. Further study of the therapeutic role of these probiotic bacteria in human trials is warranted. It is tempting to suggest that a combination of probiotics would be synergistic but this cannot be assumed; some combinations may even be antagonistic.30 Probiotic cocktails should therefore be carefully studied and the individual bacterial constituents evaluated before routine use in clinical settings. Several studies are currently underway to assess these issues, including an EU funded, multicentre, placebo controlled trial testing both Lb salivarius and B infantis in the setting of Crohn’s disease and ulcerative colitis.
Acknowledgments
The authors are funded, in part, by the Health Research Board of Ireland, the Higher Education Authority, and the European Union (PROGID QLK-2000–00563).
Conflict of interest: LO’M, BK, GO’S, JKC, and FS are members of a multidepartmental university campus research company (Alimentary Health Ltd) which investigates host-flora interactions and the therapeutic manipulation of these interactions in various human and animal diseases. The content of this article was neither affected nor constrained by this fact.
Abbreviations
IL-10, interleukin 10
KO, knockout
Th1, T helper 1
IFN-γ, interferon γ
MRS broth, de Man, Rogosa, Sharpe broth
VRBA, violet red bile agar
DGGE, denaturing gradient gel electrophoresis
PCR, polymerase chain reaction
TNF-α, tumour necrosis factor α
TGF-β, transforming growth factor β
CFU, colony forming units
REFERENCES
- 1.Shanahan F. Probiotics and Inflammatory Bowel disease: Is there a scientific rationale? Inflamm Bowel Dis 2000;6:107–15. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn R, Lohler J, Rennick D, et al. Interleukin-10 deficient mice develop chronic enterocolitis. Cell 1993;75:263–74. [DOI] [PubMed] [Google Scholar]
- 3.Shanahan F. Gene-targeted immunologic knockouts: new models of inflammatory bowel disease. Gastroenterology 1994;17:312–14. [PubMed] [Google Scholar]
- 4.Rennick D, Fort M. Lessons from genetically engineered animal models XII. IL- 10- deficient (IL-10−/−) mice and intestinal inflammation. AJP-Gastrointest Liver Physiol 2000;278:G289–33. [DOI] [PubMed] [Google Scholar]
- 5.Fuller R. Probiotics in human medicine. Gut 1991;32:439–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bengmark S. Ecological control of the gastrointestinal tract. The role of probiotic flora. Gut 1998;42:2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunne C, Murphy L, Flynn S, et al. Probiotics from myth to reality. Demonstration of functionality in animal models of disease and human clinical trials. Antonie van Leeuwenhoek 1999;76:279–92. [PubMed] [Google Scholar]
- 8.O’Mahony L, Feeney M, O’Halloran S, et al. Probiotic impact on microbial flora, inflammation, and tumour development in IL-10 knockout mice. Aliment Pharmacol Ther 2001;15:1219–25. [DOI] [PubMed] [Google Scholar]
- 9.Shanahan F. Therapeutic manipulation of gut flora. Science 2000;289:1311–2. [DOI] [PubMed] [Google Scholar]
- 10.Hooper LV, Wong MH, Gordon JI, et al. Molecular analysis of commensal host/ microbial relationships in the intestine. Science 2001;291:881–4 [DOI] [PubMed] [Google Scholar]
- 11.Madsen KL, Doyle JS, Jewell LD, et al. Lactobacillus sp prevents development of enterocolitis in interleukin-10 deficient mice. Gastroenterology 1999;116:1107–14. [DOI] [PubMed] [Google Scholar]
- 12.Berg DJ, Davidson NJ, Kuhn R, et al. Enterocolitis and colon cancer in interleukin- 10-deficient mice are associated with aberrant cytokine production and CD4+ TH1-like responses. J Clin Invest 1996;98:1010–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson NJ, Hudak SA, Lesley RE, et al. IL-12, but not IFN- γ, plays a major role in sustaining the chronic phase of colitis in IL-10-deficient mice. J Immunol 1998;161:3143–9. [PubMed] [Google Scholar]
- 14.Davidson NJ, Fort M, Muller W, et al. Chronic colitis in IL-10 −/− mice: Insufficient counter regulation of a Th1 response. Intern Rev Immunol 2000;19:91–121. [DOI] [PubMed] [Google Scholar]
- 15.Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host- specific communities of active bacteria. Appl Environ Microbiol 1998;64:3854–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heilig HG, Zoetendal EG, Vaughan EE, et al. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol 2002;68:114–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satokari RM, Vaughan EE, Akkermans ADL, et al. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol 2001;67:504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane DJ. 16S/23S rDNA sequencing, In: Stackebrandt E, Goodfellow M, eds. Nucleic acid techniques in bacterial systematics. Chichester, UK: J Wiley and Sons, 115–75. *
- 19.Sanguinetti CJ, Dias Neto E, Simpson AJG. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 1994;17:915–19. [PubMed] [Google Scholar]
- 20.Hane BG, Jager K, Drexler H. The Pearson product-moment correlation coefficient is better suited for identification of DNA fingerprint profiles than band matching algorithms. Electrophoresis 1993;14:967–72. [DOI] [PubMed] [Google Scholar]
- 21.Dunne C, O’Mahony L, Murphy L, et al. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr 2001;73:386–92S. [DOI] [PubMed] [Google Scholar]
- 22.Murphy L, Dunne C, Kiely B, et al. In vivo assessment of potential probiotic Lactobacillus salivarius strains: evaluation of their establishment, persistence, and localisation in the murine gastrointestinal tract. Micro Ecol Health Dis 1999;11:149–57. [Google Scholar]
- 23.Dugas B, Mercenier A, Lenoir-Wijnkoop I, et al. Immunity and Probiotics. Immunol Today 1999;20:387–90. [DOI] [PubMed] [Google Scholar]
- 24.O’Mahony L, Feeney M, Dunne C, et al. Probiotic bacteria and the human immune system. In: Buttress J, ed. Functional foods II: claims and evidence. Royal Society of Chemistry, 2000:63–70.*
- 25.Yasui H, Shida K, Matsuzaki T, et al. Immunomodulatory function of lactic acid bacteria. Antonie Van Leeuwenhoek 1999;76:383–9. [PubMed] [Google Scholar]
- 26.Plevy SE, Landers CJ, Prehn J, et al. A role for TNF- α and mucosal T helper-1 cytokines in the pathogenesis of Crohn’s disease. J Immunol 1997;159:6276–82. [PubMed] [Google Scholar]
- 27.Romagnani P, Annunziato F, Bacceri Mc, et al. T cells and cytokines in Crohn’s disease. Curr Opin Immunol 1997;9:793–9. [DOI] [PubMed] [Google Scholar]
- 28.Strober W, Kelsall B, Fuss I, et al. Reciprocal IFN-γ and TGF-β responses regulate the occurrence of mucosal inflammation. Immunol Today 1997;18:6–14. [DOI] [PubMed] [Google Scholar]
- 29.Schultz M, Sartor RB. Probiotics and inflammatory bowel disease. Am J Gastroenterology 2000;95:S19–21. [DOI] [PubMed] [Google Scholar]
- 30.Murphy L, Byrne F, Collins JK, et al. Evaluation and characterisation of probiotic therapy in the CD45Rbhi transfer model of colitis. Gastroenterology 1999;116:G3382. [Google Scholar]