Abstract
Background and aims: Colorectal epithelial cells are prone to malignant transformation. Therefore, identification of differences in gene expression in the process from normal colonic crypts to adenomas with low grade dysplasia is essential for further insights into early tumorigenesis. To achieve this goal, a novel gene expression analysis strategy, screening for expressed transcripts in small histologically defined tissue samples, was performed.
Methods: First, laser mediated microdissection was used to isolate normal and adenomatous crypts from colonic cryosections. Then, nested RNA arbitrarily primed polymerase chain reaction (RAP-PCR) for differential display was performed to screen mRNA populations and to generate hybridisation probes for cDNA expression arrays. After evaluation of cDNA expression arrays, differential expression was confirmed at the protein level by immunohistochemistry.
Results: Evaluation of gene expression profiles of normal versus adenomatous colonic crypts of six different patients revealed, in general, dysregulation of up to 11% of all analysed genes (total number n=588): specifically, p21-rac1 was upregulated in four of six patients, mitogen activated protein kinase (MAPK) p38α in three of six patients, and interferon γ receptor in three of six patients. Conversely, FAST kinase was found to be downregulated in three of six patients, p53 in three of six patients, and thrombospondin 2 in three of six patients.
Conclusions: For the first time, distinct gene expression profiles of dysplastic areas within colonic adenomas, using the combination of laser mediated microdissection with RAP-PCR and cDNA expression array, were shown. In these samples, upregulation of proliferation associated genes (ras-oncogene related p21-rac1 and MAPK p38α) as well as downregulation of apoptosis related genes (FAST kinase and p53) most likely reflects specific alterations in adenomas with low grade dysplasia. Based on upregulation of p21-rac1 and MAPK p38α, activation of the MAPK pathway appears to be an early event in colonic carcinogenesis.
Keywords: laser mediated microdissection, gene expression pattern, adenomas, low grade dysplasia, RAS/MAPK pathway
Colon cancer is one of the most common causes of cancer death in industrialised countries. Although diagnosis and treatment have made great progress in the last decade, there is still the need for developing novel diagnostic systems for detection of the early stages of colon carcinogenesis.
Fearon and Vogelstein1 proposed a model of multistep carcinogenesis based on the development of colon cancer through the “adenoma-carcinoma sequence”. Different key tumour suppressor genes as well as oncogenes have been associated with this model. Adenomatous polyposis coli gene inactivation or loss, and hypermethylation occur early in the “adenoma-carcinoma sequence”, followed by sequential loss of other tumour suppressor genes (deleted in colon cancer DCC, p53) and activation of cellular oncogenes (k-ras).1 However, our knowledge of gene regulation and growth regulating signal transduction pathways in the early steps of colon carcinogenesis is still very limited.
Therefore, identification of differences in gene expression in the process from normal colonic crypts to adenomas with low grade dysplasia is essential to provide further insights into tumorigenesis.
At present, the specificity of gene expression studies in tissue samples is mostly limited by mixed cell populations, and this problem of cell heterogeneity has been a significant barrier for the detailed molecular analysis of normal or diseased tissue. Thus the possibility of preparing exactly defined microscopic tissue samples by introduction of laser mediated microdissection (LMM)2–4 has the advantage of more specific and precise analyses.
In this report, a novel gene expression analysis strategy5 combining the differential display approach of RNA arbitrarily primed polymerase chain reaction (RAP-PCR) fingerprinting and cDNA expression array has been used to identify and validate gene expression profiles of distinct colonic crypts of adenomas with low grade dysplasia prepared by LMM.
MATERIALS AND METHODS
Tissue sections
Fresh tissue from normal colonic biopsies and adenomatous polyps during routine colonoscopy was immediately snap frozen in liquid nitrogen. All cryoblocks were stored at −80°C until cutting.
Glass slides were membrane mounted (PEN membrane; PALM, Wolfratshausen, Germany) and poly-l-lysine coated (0.2% poly-l-lysine in sterile DEPC treated H2O) under RNase free conditions. Cryosections (5–8 μm) were prepared in a cryostat, dehydrated with ethanol/acetic acid (20/1) at −20°C for 10 minutes, and stained with haematoxylin and eosin.
Microdissection
LMM was performed as described previously5 using a Robot Microbeam laser microscope (PALM). The tissue area of interest was positioned and cut out using a focused pulsed laser beam. Dissected areas were collected in the cap of a microcentrifuge tube via laser pressure catapulting. The cap with the procured tissue was placed immediately on the microfuge tube containing 300 μl of RLT lysis buffer of the RNeasy spin column purification kit (Qiagen, Hilden, Germany) and lysed by mixing.
RNA extraction
RNA was extracted by silica gel binding using the RNeasy spin column purification kit (Qiagen). To remove the remaining genomic DNA, total RNA was treated with DNase I (Qiagen) within the extraction column at room temperature for 30 minutes. Extraction was performed according to the manufacturer’s instructions, and finally RNA was eluted with 30 μl of RNase free water. The eluate was applied repeatedly to the column and spun through to increase the yield of RNA.5 The final concentration ranged from 0.8 to 2.3 ng/μl.
Atlas cDNA expression array
Atlas cancer cDNA arrays (Clontech, Heidelberg, Germany) containing 588 known cancer related genes designed for the evaluation of different molecular expression patterns were used.
Preparation of hybridisation probe
The hybridisation probe was generated by amplification of the cDNA via nested RAP- PCR. Nested RAP-PCR was performed by amplification in two steps, as described previously.5 Briefly, reverse transcription for first strand synthesis was performed with total RNA using a 10mer arbitrary primer (OPN23 5′-CAGGGGCACC-3′). RNA (2 ng in a final volume of 30 μl) was mixed with 20 μl of reverse transcriptase mixture for a final 50 μl reaction containing 50 mM Tris/HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2, 20 mM DTT, 0.2 mM dNTP, 2 μM primer, and 100 U MuLV reverse transcriptase (Promega, Madison, Wisconsin, USA). To exclude DNA contamination, a reverse transcriptase free control reaction was included in the RAP-PCR experiments. The reaction was carried out at 37°C for 60 minutes, after a five minute ramp from 25°C to 37°C; inactivation of the reaction was done at 68°C for 15 minutes. Synthesised cDNA was purified by precipitation and resolved in 30 μl of nuclease free water.
PCR for second strand synthesis was performed using a nested PCR strategy. In the first step, preamplification was achieved by 15 cycles with a 10mer arbitrary primer (OPN21 5′- ACCAGGGGCA-3′). The cDNA (30 μl) was mixed with 20 μl of PCR mixture for final concentrations of 10 mM Tris/HCl, pH 8.3, 10 mM KCl, 4 mM MgCl2, 0.2 mM dNTPs, 4 μM primer, and 2.5 U AmpliTaq DNA polymerase Stoffel fragment (Perkin Elmer, Norwalk, Connecticut, USA). Cycling conditions were 94°C for 30 seconds, 35°C for 30 seconds, and 72°C for 60 seconds.
The PCR product was purified by precipitation and resolved in 30 μl of nuclease free water. A sample of the product of the first amplification step (10 μl) was used for the second amplification step with a nested 10mer arbitrary primer (OPN21nested 5′-CCAGGGGCAC- 3′). The reaction was carried out in a volume of 20 μl containing 10 mM Tris/HCl, pH 8.3, 10 mM KCl, 4 mM MgCl2, 0.2 mM dNTPs (without dATP), 4 μM primer, 2.5 U AmpliTaq DNA polymerase Stoffel fragment (Perkin Elmer), and 28 μCi [α-32P]dATP (3000 Ci/mmol, 2.8 μl; Amersham, Freiburg, Germany). Cycling conditions were 35 cycles of 94°C for 30 seconds, 35°C for 30 seconds, and 72°C for 60 seconds. All reactions were carried out in duplicate to test the reproducibility and stability of the RNA fingerprint.
After confirmation of the success of the RAP-PCR (via gel electrophoresis and autoradiography5), the PCR reaction was purified from unincorporated 32P labelled nucleotides and small DNA fragments (<0.1 kb) by column chromatography using NucleoSpin columns (Clontech), as outlined by the manufacturer.
Hybridisation
The RAP-PCR product was hybridised to the Atlas human cDNA expression array membranes, as published recently.5
Wash
Filters were washed three times in wash solution 1 (2× standard saline citrate (SSC) and 2% sodium dodecyl sulphate (SDS)) for 30 minutes at 68°C each. Two washing steps were performed with wash solution 2 (0.1× SSC and 0.5% SDS) at 68°C for 20 minutes and one step for five minutes at room temperature in 2× SSC and then exposed to a Phosphor-Imager- Screen (Molecular Dynamics, Sunnyvale, California, USA) for approximately five days.
Data analysis
Analysis of the scanned Phosphor-Imager-Screen was performed using Ambis software (ImageQuant, Molecular Dynamics) and gene expression evaluation was performed using AtlasImage 2.0 software which was developed specifically for analysis of the Atlas cDNA expression arrays (Clontech).
Confirmation of differential expression
The first step for confirmation of differential expression was achieved by performing each experiment in duplicate (microdissection), in two parallel reactions (amplification and hybridisation) under the same conditions, and only those hybridisation results which indicated differential expression (twofold or more dysregulation) in both reactions were regarded to be of further interest.
In the second step, immunohistochemistry was performed with polyclonal antibodies against p21-Rac1 (Santa Cruz Biotechnology, Santa Cruz, California, USA) and monoclonal antibodies against interferon gamma receptor (IFGR; Santa Cruz Biotechnology) using the AEC (3-amino-9-ethylcarbazole) substrate kit for peroxidase (Vector Laboratories, Burlingame, California, USA) to confirm the differential expressed genes at the protein level. Snap frozen sections were cut (4–6 μm), fixed for five minutes in acetone, and covered with a 4% non-fat dry milk 2% normal goat serum buffer for 30 minutes at room temperature to block non-specific binding. After rinsing in Tris NaCl, pH 7.6, the slides were incubated for 45 minutes at room temperature with the primary antibodies diluted 1:50–1:200 in Tris NaCl containing 2% non-fat dry milk. The slides were rinsed in Tris NaCl and incubated for 30 minutes at room temperature with a secondary biotinylated antimouse and antirabbit IgG antibody, respectively (Pharmingen, San Diego, California, USA) in a 1:400 dilution in Tris NaCl containing 2% non-fat dry milk. Incubation with the AEC substrate (Vector Laboratories) was performed for 30 minutes at room temperature. Colour development (varying from 10 to 30 minutes) was stopped under microscopic examination by washing with water. After staining with haematoxylin, the slides were mounted immediately (Gel Mount, Biomeda, Foster City, California, USA).
RESULTS
Histology and LMM
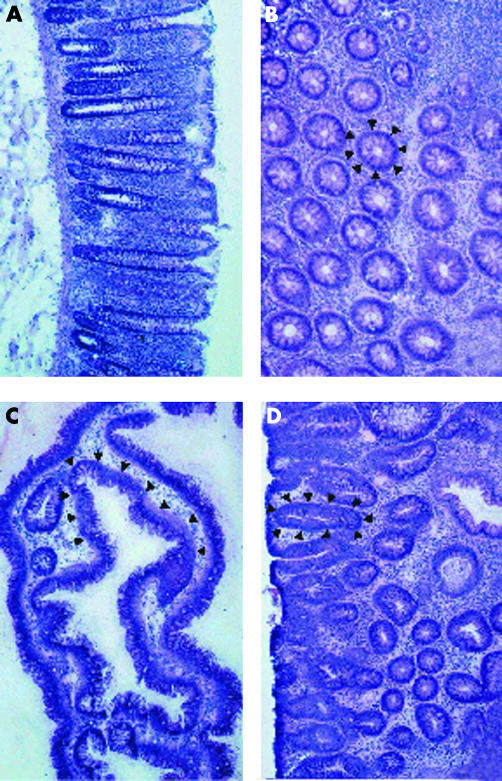
LMM was used to isolate colonic crypts of normal and adenomatous (low grade dysplasia, D1) mucosa of six different patients. Characteristics for classification as normal or adenomatous crypt, respectively, were the following: crypt size, crypt shape, thickness of the epithelial layer, quantity of cytoplasm, and existence of goblet cells (fig 1 ▶).
Figure 1.
Haematoxylin-eosin stained sections of colon mucosa with normal (A, B) and adenomatous villous (C) or tubulous (D) colonic crypts (low grade dysplasia, D1). Arrows mark objects for laser capture microdissection (B, C, D).
cDNA array hybridisation and evaluation of differential gene expression
Each array experiment revealed approximately 55–120 hybridisation events (out of 588 possible events). Using the AtlasImage 2.0 software, these events could be clearly coordinated with the cDNA spots.
Evaluation of the cDNA expression arrays of all six patients revealed reproducible differential expression of 68 genes (11.6% of all 588 analysed genes) comparing adenomatous versus normal colonic crypts; 7.7% of all 588 analysed genes were only differentially expressed in one of the six analysed patients. The remaining 3.9% of the differentially expressed genes were dysregulated in 2–4 patients (table 1 ▶).
Table 1.
Number and percentage of identically dysregulated genes analysing adenomatous (low grade dysplasia, D1) versus normal colonic crypts
| Identically dysregulated genes | 2 patients | 3 patients | 4 patients |
| Upregulation | 13 (2.2%) | 2 (0.3%) | 1 (0.2%) |
| Downregulation | 4 (0.7%) | 3 (0.5%) | — |
Percentages were calculated as follows: differentially regulated genes divided by all analysed genes, n=588.
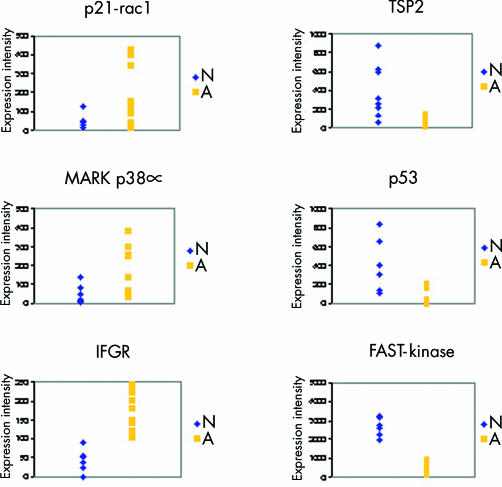
Only those genes which were dysregulated in at least three of six dysplastic compared with normal colonic crypts of a patient were further examined (1% of all analysed 588 genes, marked in table 1 ▶). The genes were: p21-rac1, mitogen activated protein kinase (MAPK) p38, IFGR, thrombospondin 2, p53, and FAST kinase. In table 2 ▶, expression levels of the dysregulated genes are listed. Figure 2 ▶ demonstrates differences in expression intensities in normal and adenomatous colonic mucosa.
Table 2.
Differentially expressed genes in adenomatous (low grade dysplasia, D1) versus normal colonic samples (differentially regulated in at least 3/6 patients)
| Gene (accession No) | Upregulated (fold increase) | Downregulated (fold decrease) | Regulation |
| p21-rac1; ras related protein TC25 (M29870, M31467) | 2.4 | ↑ 4 (6)* | |
| 3.4 | |||
| 3.0 | |||
| 4.5 | |||
| (M=3.3) | |||
| MAPK p38 (L35253, L35263) | 2.8 | ↑ 3 (6) | |
| 3.3 | |||
| 3.5 | |||
| (M=3.2) | |||
| IFGR (A09781) | 3.7 | ↑ 3 (6) | |
| 4.0 | |||
| 2.8 | |||
| (M=3.5) | |||
| TSP2 (L12350) | 8.3 | ↓ 3 (6) | |
| 8.7 | |||
| 9.8 | |||
| (M=8.9) | |||
| p53 cellular tumour antigen (M14694, M14695) | 2.5 | ↓ 3 (6) | |
| 3.2 | |||
| 28.2 | |||
| (M=11.3) | |||
| FAST kinase (X86779) | 4.6 | ↓ 3 (6) | |
| 9.9 | |||
| 2.5 | |||
| (M=5.7) |
Values represent the mean up- or downregulation of all experiments in each patient.
*4 (6) indicates 4 of 6 patients regulated differentially.
M, median; MAPK p38, mitogen activated protein kinase p38; IFGR, interferon γ receptor; TSP2, thrombospondin 2; FAST kinase, Fas activated serine/threonine kinase.
Figure 2.
Expression intensities of p21-rac1, mitogen activated protein kinase (MAPK) p38a, interferon gamma receptor (IFGR), thrombospondin 2 (TSP2), p53, and FAST kinase. Each symbol represents one expression intensity of one array hybridisation. All patients are included, but there is large overlap of expression values resulting in a lower number of visible symbols. N, normal colonic crypts; A, adenomatous colonic crypts (low grade dysplasia, D1).
Validation of array data with immunohistochemistry
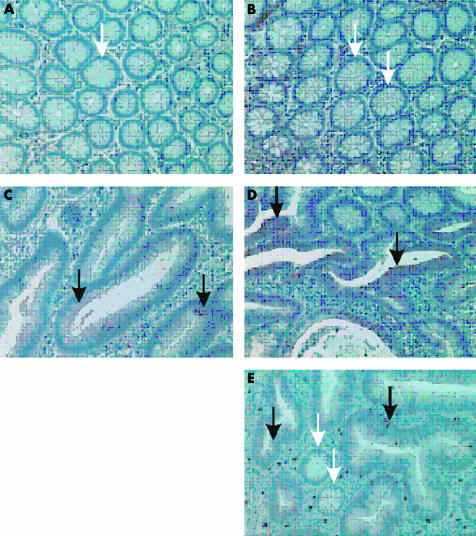
To further confirm the reliability of our array data, we performed immunohistochemical analysis of IFGR and p21-rac1 protein using tissue from the same patients. Similar to the differential expression pattern observed with the cDNA array, normal crypt cells showed no or minimal immunoreactivity for IFGR (fig 3A ▶) or p21-rac1 (fig 3C ▶) whereas crypt cells of D1 dysplastic lesions showed strong immunoreactivity for these proteins (fig 3B, D, E ▶). These results further support the reliability of our array data and demonstrate the specific location of IFGR and p21-rac1 expression in early adenoma.
Figure 3.
Immunohistochemical staining using antibodies against interferon gamma receptor (IFGR) (A, B) and against p21-rac1 (C, D, E). (A, C) Normal colonic crypts; (B, D, E) mainly adenomatous colonic crypts with low grade dysplasia (D1). Black arrows show positive staining (red-brown), and white arrows show crypts without staining; negative controls showed no staining. Original magnification 200-fold.
DISCUSSION
Using the combination of LMM, RAP-PCR,6,7 and cDNA expression array,5,8,9 we showed for the first time distinct gene expression profiles of areas of low grade dysplasia within colonic adenomas. This strategy provides an unique opportunity to relate gene expression of subpopulations of cells to their actual tissue environment. Moreover, using LMM, genetic analysis of pure and very small cell populations is possible and the limitations associated with in vivo and in vitro studies can be minimised.
When we analysed six individual patients, distinct alterations in gene expression of normal versus low grade dysplastic adenomas (D1) were demonstrated. Upregulation of proliferation associated genes (ras-oncogene related p21-rac1 and MAPK p38α) as well as downregulation of apoptosis related genes (FAST kinase and p53) was demonstrated at the mRNA level in adenomas with low grade dysplasia.
However, thrombospondin 2, whose expression is generally associated with tumour defence via regulation of angiogenesis,10 was found to be downregulated in low grade dysplastic lesions. This downregulation may also be involved in adenoma/tumour development.
One of the pathways thought to be activated in colonic tumorigenesis is the RAS/MAPK pathway which plays an important role in regulation of proliferation and transcription. At the end of the MAPK signalling cascade, activation of transcription factors such as ATF2, c-Jun, c-Fos, CHOP/GADD153, Max, or MEF211 is essential. Different genes in this signalling cascade were identified as being upregulated in low grade dysplastic adenomas: p21-rac1 (see table 2 ▶ and fig 2 ▶), MAPK p38α (see table 2 ▶ and fig 2 ▶), rho-GDP dissociation inhibitor 2 (see table 3 ▶, upregulated in 2/6 patients), p21-activated kinase α, and MAPK 6 (data not shown, both upregulated in 1/6 patients).
Table 3.
Differentially expressed genes in adenomatous (low grade dysplasia, D1) versus normal colonic samples (differentially regulated in at least 2/6 patients)
| Gene (accession No) | Upregulated (fold increase) | Downregulated (fold increase) | Regulation |
| DNA-PK; DNA-PKCS (U35835) | 4.4 | ↑ 2 (6)* | |
| 11.5 | |||
| (M=7.95) | |||
| DNA topoisomerase II alpha (170kDa) (J04088) | 2.6 | ↑ 2 (6) | |
| 23 | |||
| (M=12.8) | |||
| rho GDP dissociation inhibitor 2; LY-GDI; ARHGDIB; GDID4 (L20688) | 3.0 | ↑ 2 (6) | |
| 24.0 | |||
| (M=13.5) | |||
| Laminin beta 1 (M61916) | 4.4 | ↑ 2 (6) | |
| 5.5 | |||
| (M=4.95) | |||
| Catenin (cadherin associated protein) beta 1 (88 kDa) (X87838) | 2.2 | ↑ 2 (6) | |
| 27.6 | |||
| (M=14.9) | |||
| Cyclin dependent kinase 6 X66365) | 6.8 | ↑ 2 (6) | |
| 12.1 | |||
| (M=9.45) | |||
| CDKN1A; melanoma differentiation associated protein 6; CIP1; WAF1 (U09579) | 29.5 | ↑ 2 (6) | |
| 42.5 | |||
| (M=36.0) | |||
| v-erb-b2 avian erythroblastic leukaemia viral oncogene homolog 3 (M29366) | 13.5 | ↑ 2 (6) | |
| 4.0 | |||
| (M=8.75) | |||
| BARD1 (U76638) | 2.6 | 5.4 | ↑ 2 (6)/↓ 2 (6) |
| 40.0 | 7.4 | ||
| (M=21.3) | (M=6.4) | ||
| Hepatoma derived growth factor (high mobility group protein 1- like) (D16431) | 2.8 | ↑ 2 (6)/↓ 1 (6) | |
| 7.8 | 7.5 | ||
| (M=5.3) | |||
| Inhibitor of growth 1 family member 1 (AF001954) | 9.45 | ↑ 1 (6)/↓ 2 (6) | |
| 29.7 | 5.6 | ||
| (M=7.53) | |||
| Integrin alpha 6; VLA6, CD49F antigen (X53586) | 2.9 | ↑ 1 (6)/↓ 2 (6) | |
| 2.25 | 51.0 | ||
| (M=26.95) | |||
| Urokinase-type plasminogen activator receptor GPI-anchored form precursor (U-PAR); monocyte activation MO3; CD87 antigen (U08839) | 24.7 | ↓ 2 (6) | |
| 2.6 | |||
| (M=13.65) | |||
| Type II cytoskeletal 8 keratin (KRT8); cytokeratin 8 (K8; CK8) (M34225) | 2.4 | ↓ 2 (6) | |
| 3.3 | |||
| (M=2.85) |
Values represent the mean up- or downregulation of all experiments in each patient.
*2 (6) indicates 2 of 6 patients regulated differentially.
M, median; DNA-PK, DNA dependent protein kinase; DNA-PKCS, DNA-PK catalytic subunit; CDKN1A cyclin dependent kinase inhibitor 1 A; CIP1, CDK-interacting protein 1; BARD1, BRCA1 associated RING domain.
p21-rac1 is a member of the RAS superfamily of the small GTP binding proteins and can activate protein kinases such as JNK, SAPK, or MAPK p38. MAPK p38α, in turn, is an activator of different transcription factors.11
Based on upregulation of p21-rac1 and MAPK p38α in low grade dysplastic adenomas, activation of the MAPK pathway appears to be an early event in colonic carcinogenesis. This activation seems to be characteristic for low grade dysplastic lesions as Wang and colleagues12 have shown that MAPK p38, ERK1, ERK2, and JNK, all members of the MAPK pathway, are downregulated in carcinomas, which appear late in colon carcinogenesis.
The results of the present study also provide the basis for a more general use of laser microdissection in colon cancer histomolecular analysis. We believe that examination of adenomas with low grade dysplasia not only reveals the initial steps towards established colon carcinoma but most likely will facilitate identification of subgroups of patients at high risk for more rapid development of malignancy. Moreover, understanding of these initial steps may also provide the rationale for therapeutic strategies in more advanced stages of the disease—for example, the development of specific drugs following colon surgery with a high risk for local or distant metastases based on severe gene dysregulation already operative in macroscopically non-visible premalignant lesions (for example, aberrant crypt foci).
We have shown that the combination of RAP-PCR and cDNA expression array together with LMM offers the possibility of gaining insight into the early molecular events in colon carcinogenesis. Our findings indicate that p21-rac1 and MAPK p38 are interesting candidate genes potentially involved in key mechanisms leading to malignant transformation in the colonic crypt.
Acknowledgments
The authors wish to thank Olga Wiesner, Wibke Ballhorn, and Elena Neumann for excellent technical assistance. This research was funded by the Deutsche Forschungsgemeinschaft (Ku 1024/6-3, Mu 1383/1-3 and Mu 1383/3-3).
Abbreviations
LMM, laser mediated microdissection
RAP-PCR, RNA arbitrarily primed polymerase chain reaction
MAPK, mitogen activated protein kinase
IFGR, interferon gamma receptor
SSC, standard saline citrate
SDS, sodium dodecyl sulphate
AEC, 3-amino-9-ethylcarbazole
REFERENCES
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67. [DOI] [PubMed] [Google Scholar]
- 2.Emmert-Buck MR, Bonner RF, Smith, et al. Laser capture microdissection. Science 1996;274:998–1001. [DOI] [PubMed] [Google Scholar]
- 3.Simone NL, Bonner RF, Gillespie JW, et al. Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet 1998;14:272–6. [DOI] [PubMed] [Google Scholar]
- 4.Suarez-Quian CA, Goldstein SR, Pohida T, et al. Laser capture microdissection of single cells from complex tissues. Biotechniques 1999;26:328–35. [DOI] [PubMed] [Google Scholar]
- 5.Lechner S, Müller-Ladner U, Neumann E, et al. Use of simplified transcriptors for the analysis of gene expression profiles in laser-microdissected cell populations. Lab Invest 2001;81:1233–42. [DOI] [PubMed] [Google Scholar]
- 6.McClelland M, Mathieu-Daude F, Welsh J. RNA fingerprinting and differential display using arbitrarily primed PCR. Trends Genet 1995;11: 242–6. [DOI] [PubMed] [Google Scholar]
- 7.Welsh J, Chada K, Dalal SS, et al. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res 1992;20:4965–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trenkle T, Welsh J, Jung B, et al. Non-stoichiometric reduced complexity probes for cDNA arrays. Nucleic Acids Res 1998;26:3883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neumann E, Kullmann F, Judex M, et al. Identification of differentially expressed genes in rheumatoid arthritis by a combination of cDNA array and RAP-PCR. Arthritis Rheum 2002;46:52–63. [DOI] [PubMed] [Google Scholar]
- 10.Streit M, Riccardi L, Velasco P, et al. Thrombospondin-2: a potent endogenous inhibitor of tumor growth and angiogenesis. Proc Natl Acad Sci U S A 1999;96:14888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev 2001;81:807–69. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, Ding Q, Dong Z, et al. Downregulation of mitogen-activated protein kinases in human colon cancers. Anticancer Res 2000;20:75–83. [PubMed] [Google Scholar]