Abstract
Background: The RET gene encodes a tyrosine kinase receptor involved in different human neurocristopathies, such as specific neuroendocrine tumours and Hirschsprung disease (HSCR). Gene expression is developmentally regulated and the RET transcript is undetectable in most adult cells, including lymphocytes. The impossibility of performing functional studies on RET mRNA has to date limited the detection and characterisation of an indefinite proportion of gene anomalies that cannot be identified by conventional DNA genomic screening in HSCR cases.
Aims: Development of a protocol suitable to activate RET expression in RET negative cell lines and therefore to investigate directly RET mRNA, extending the conventional gene mutation analysis to detection of splicing anomalies and impaired expression of the RET gene.
Methods: The effect of sodium butyrate (NaB), a histone deacetylase inhibitor, on rescuing RET expression was tested by one round of reverse transcription- polymerase chain reaction from total RNA of treated lymphoblasts from both HSCR patients and control individuals.
Results: Analysis of RET expression was possible by NaB treatment of RET negative cells, such as lymphoblasts. This treatment allowed us to detect impaired RET expression as well as a splicing defect in two HSCR patients previously believed to be devoid of any gene abnormality.
Conclusions: The full application of the proposed protocol in most of the unexplained HSCR cases will allow us to establish the precise role of RET not only in causing but also in predisposing to HSCR pathogenesis.
Keywords: Hirschsprung disease, RET proto-oncogene, sodium butyrate, molecular screening, expression defects
The RET proto-oncogene (REarranged during Transfection) encodes a tyrosine kinase receptor predominantly expressed during neural crest development and involved in different human neurocristopathies.1 Germline RET mutations have been found in association with both inherited cancer syndromes, including multiple endocrine neoplasia type 2A (MEN2A), type 2B (MEN2B), and familial medullary thyroid carcinoma, and Hirschsprung disease (HSCR), a developmental disorder characterised by the absence of enteric neurones in variable lengths of the distal gastrointestinal tract.2 Loss of function, low penetrant RET mutations have been identified in only 10–40% of total HSCR cases while a small proportion of patients (5–10%) show alterations in other genes, generally related to the developmental programme of neural crest cells.3,4 By using sibpair analysis, a recent study has recognised three HSCR susceptibility loci at 3p21, 10q11, and 19q12 which may clarify the molecular basis of a great proportion of unexplained HSCR cases.5 The locus in 10q11 probably corresponds to RET, confirming the central role played by this gene in HSCR pathogenesis. Taking into account the persistent failure to detect RET coding sequence mutations in a proportion of familial cases found to be linked to the gene, it has recently been suggested that genetic defects in introns and in the promoter region can contribute in some cases to the HSCR phenotype.5–7
Moreover, several common polymorphisms of the RET proto-oncogene have been associated with a variable risk of HSCR development. In particular, in different populations RET single nucleotide polymorphism (SNP) alleles are present with different frequencies in HSCR patients with respect to controls,8,9 while specific haplotypes have been demonstrated to display either protective or predisposing effects, and/or to modulate the severity of the resulting phenotype.10,11
To date, no RET SNP allele has been proved to have any effect on Ret function; on the other hand, the functional variants responsible for the predisposing or protective effect displayed by specific RET haplotypes, and expected in linkage disequilibrium with them, have not yet been identified. These latter variants may be represented by genetic defects in regulatory regions, contributing to HSCR by decreasing the expression level of correct and active Ret receptor.
Unfortunately, the search for non-coding RET mutations associated with HSCR disease has been hampered by: (i) the size of the gene (52.4 kb); (ii) the presence of large introns (intron 1 spans more than 20 kb); (iii) the abundance of DNA repeats; and (iv) the lack of mutational hotspots. As already proved for other genes such as ATM (ataxia-telangiectasia mutated) and NF1 (neurofibromatosis type 1),12,13 direct investigation on splicing and expression would be very useful to detect uncovered RET molecular defects. At present, such a study is not possible as the transcript is undetectable in the majority of adult human tissues, including peripheral blood lymphocytes.14,15 Indeed, it is well known that RET expression is mostly confined to specific stages during embryogenesis when the gene is transcribed in tissues derived from embryonic neuroectoderm and the developing urogenital system.16,17
In the past few years, we and others have approached the study of the functional effect of RET splicing mutations (synonymous mutations or splice site alterations) by using RNA obtained from illegitimate transcription coupled with nested reverse transcription-polymerase chain reaction (RT-PCR).18–20 Indeed, in immortalised lymphoblastoid cells a minimal amount of RET product can be detected after two rounds of PCR (for a total of 70 cycles).21 However, illegitimate transcription does not reflect physiological gene expression and false positive products can be obtained as a result of its use. These considerations compelled us to confirm results from RT-PCR with mini gene constructs, cloning genomic fragments of the RET gene in the expression vector pSPL3.18–20 Unfortunately, such a time consuming and expensive approach cannot easily be used for rapid screening of RET splicing anomalies or to assess defective allelic expression.
In a previous report, we demonstrated that sodium butyrate (NaB), a histone deacetylase inhibitor, upregulated RET transcription in RET positive cell lines.22 In particular, NaB treatment increased RET transcription in IMR32 cells, already displaying low levels of its mRNA, while it had no effect on MTC-TT cells, characterised by a plateau level of RET mRNA.22 Accordingly, we also demonstrated that NaB treatment increased the endogenous level of RET histones acetylation (generally associated with transcriptionally active chromatin) in IMR32 but not in MTC-TT cells. These data strongly suggest that NaB treatment mimics a physiological regulatory mechanism of the RET gene.
In the present study, we show that NaB also stimulates a detectable level of transcription in two RET negative cell lines, SK-N-MC and lymphoblastoid cells. We started from such an observation to develop an original methodological approach to study the molecular basis of HSCR and to perform functional tests on RET mRNA obtained from cell sources easily available and routinely used for genomic DNA extraction, such as lymphocytes or lymphoblasts. The full application of the proposed NaB treatment protocol to unexplained HSCR cases (still representing the majority of all cases) will allow us to establish the precise role of RET in the pathogenesis of HSCR.
METHODS
Cells and NaB treatment
The human neuroblastoma cell line SK-N-MC and lymphoblasts from HSCR patients and controls were grown in RPMI medium (Hyclone Laboratories, Logan, Utah, USA), supplemented with 10% fetal calf serum (Gibco BRL, Life Technologies Italia Srl, Milano, Italy), 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin. NaB (B 5887; Sigma-Aldrich Srl, Milano, Italy) was dissolved in water (1 M stock solution) and delivered to cells at a final concentration of 5 mM. Treatment was performed overnight. The day after, cells were given fresh medium and NaB was delivered for another night. RNA extraction was performed using commercially available TRIzol (Gibco BRL).
RNA analysis
Random hexamer primed cDNA was prepared from 500 ng of total RNA. The human housekeeping G3PDH gene (glucose 3-phosphate dehydrogenase) was amplified with forward primer 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ and reverse primer 5′- GCAGAGATGATGACCCTTTTGGCTC-3′. The human RET gene was amplified with forward IP18F primer 5′-GGATTTCGGCTTGTCCCGAG-3′ and reverse IP20R primer 5′- CCATGTGGAAGGGAGGGCTC-3′.
We analysed RNA from NaB treated lymphoblasts of eight individuals (both HSCR patients and controls). Individuals 1–4 carry different exon 14 genotypes. Random hexamer primed total cDNA (3 μl) and 200 ng of genomic DNA of each individual were used to perform PCR reactions with the exonic primers 14F (5′-CTCCTCATCGTGGAGTACGC-3′) and 14R (5′-CTCGGCCAGATACTGCATCC-3′) (38 cycles). Restriction enzyme and single strand conformation polymorphism (SSCP) analysis were performed, as previously described.11,23 Individual No 5 is a normal control while individual No 6 is a HSCR patient carrying an intronic mutation, IVS12+19 C>T. RT-PCR from NaB induced lymphoblast RNA of these two latter individuals was performed as previously described.20
Individuals belonging to the family reported below were genotyped using the intragenic polymorphisms of RET exons 2, 13, and 19, as described by Griseri and colleagues.11,23 Individuals 7 (control) and 8 (HSCR patient) were both heterozygous for the 2307T>G SNP of exon 13. RT-PCR was performed from total cDNA with primers 4F (5′-GAAAAGTGGTCAAGGCAA-3′) and 18R (5′-GAATCTTCATCTTCCGCCC-3′) (38 cycles), and products digested with TaqI to verify the presence of the two alleles in the transcripts.
RESULTS
NaB treatment: rescue of gene expression in RET negative cells
RET upregulation obtained with NaB treatment of RET positive cell lines compelled us to also test RET negative cells, such as lymphoblasts and neuroblastoma SK-N-MC cells, for NaB dependent expression of the gene.
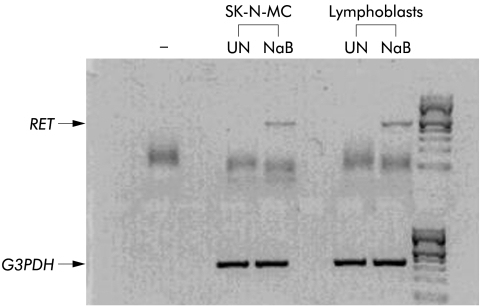
As shown in fig 1 ▶, in total RNA from untreated cells, in agreement with previous data,24,25 RET expression was not detectable while in NaB treated cell lines, both lymphoblasts and neuroblastoma cells, we were able to detect RET transcript after a single round of PCR.
Figure 1.
Appearance of RET transcript in RET mRNA negative cells after sodium butyrate (NaB) treatment. Results obtained after a single round of RET amplification (38 cycles) from retrotranscribed diluted samples of total RNA obtained from untreated (UN) and NaB treated SK-N-MC and lymphoblastoid cell cultures. Glucose 3-phosphate dehydrogenase (G3PDH) expression was used as an internal standard and the minus symbol indicates the reaction blank control.
Comparison of RET analysis between genomic DNA and NaB induced cDNA
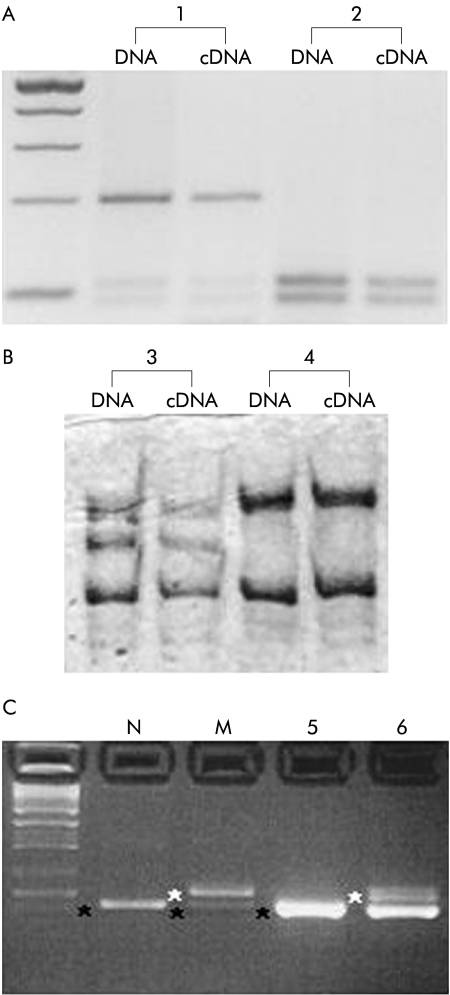
As a first application of the NaB treatment to lymphoblasts, we assessed whether RET cDNA analysis could reproduce results from genomic DNA. In fig 2A ▶ we report the migration pattern of endonuclease digestion of a known polymorphism in exon 14 of the RET gene, 2508C>T (S836S), from both cDNA and genomic DNA of a heterozygote for the common T variant (individual No 1), and of a CC control (individual No 2). Moreover, fig 2B ▶ shows the results of SSCP analysis of RET exon 14 from both cDNA and genomic DNA in two additional individuals, a normal control (individual No 4) and a HSCR patient (individual No 3). In individual No 4, only wild-type alleles are present, while individual No 3 carries the 2436C>T missense mutation (R813W), previously described in an unrelated HSCR patient.18 In all of these samples both the amount and migration pattern of PCR products from genomic DNA were comparable with those obtained from NaB induced cDNA. This result represents the basis of developing a rapid primary screening of RET unknown mutations in which the entire cDNA is used as a direct substrate for traditional mutation detection methods such as denaturing high performance liquid chromatography (DHPLC), SSCP, etc.
Figure 2.
Analysis of RNA from patients with Hirschsprung disease (HSCR) and controls. Lymphoblastoid cells from six individuals were used for cDNA synthesis and transcripts analysis. (A) Restriction analysis of RET exon 14 from genomic DNA and cDNA is shown. Individual No 1 is heterozygous for the 2508C>T polymorphic variant while only wild-type alleles are present in individual No 2. (B) Single strand conformation polymorphism profiles of individuals carrying the 2436C>T mutation (individual No 3) and the wild-type control (individual 4) are compared. (C) The intronic RET mutation IVS12+19C>T was analysed from two RNA sources. In the first two lanes, cDNA was obtained from COS7 cells transfected with pSPL3 constructs carrying the wild-type (N) and mutated (M) alleles.20 Lanes 5 and 6 refer to reverse transcription-polymerase chain reaction samples from sodium butyrate treated cells of one control (individual No 5) and the patient carrying the splicing variant (individual No 6). Both correctly processed and aberrant transcripts are shown. An additional band corresponding to the heteroduplex product is present in the last lane.
Detection of RET abnormal splicing in a HSCR patient
To determine if NaB treatment allowed for efficient detection of RET anomalous transcripts, we performed RT-PCR analysis on total RNA from a HSCR patient carrying the intronic RET mutation IVS12+19C>T, already demonstrated to cause aberrant RNA processing.20 We previously showed, using mini gene constructs, that such an intronic mutation introduces a novel splice donor site, recognised and actively used during RNA splicing, leading to an alternative transcript bearing 17 additional nucleotides retained from the 5′ end of intron 12. In the present work, we assessed whether lymphoblastoid cells treated with NaB could maintain the same pattern of alternative splicing previously demonstrated in vitro. As shown in fig 2C ▶, an expected fragment of 187 bp, corresponding to the normally spliced transcript, was present in both the patient and control cDNA samples while an additional larger product of 204 bp, corresponding to the aberrantly spliced transcript, was detected only in the HSCR individual, in agreement with the splicing pattern obtained from the use of mini gene constructs (see legend to fig 2C ▶). This result suggests that our approach can be used for primary screening of unknown RET splicing anomalies, undetectable with standard genomic analysis.
Loss of heterozygosity of the RET cDNA in a familial case of HSCR disease
In HSCR disease, RET coding mutations account for only a small proportion of cases, suggesting the existence of non-coding variations in the RET gene and/or additional genetic risk factors.5
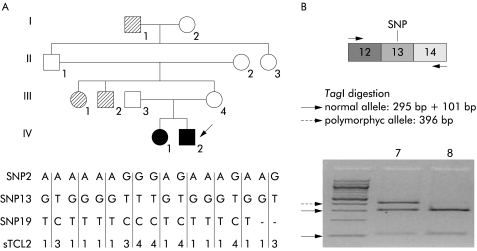
In order to identify RET expression defects otherwise undetectable, we performed NaB treatment on lymphoblastoid cells from a HSCR patient affected with a familial long form of congenital megacolon. Linkage analysis previously showed that the disease phenotype recurring in this family was closely linked to RET gene markers (fig 3 ▶).26 However, screening of all 21 exons and the intronic junctions did not show any type of mutation. To investigate the possible presence of a RET deletion, we characterised genomic DNA for three SNPs distributed along the whole gene. However, the heterozygous status of the proband for SNPs 2, 13, and 19 excluded the presence of genomic deletions involving these polymorphic loci. Lymphoblasts from the proband and a control individual, both heterozygous for a known polymorphism in exon 13 (2307G>T), were treated with NaB. Endonuclease digestion of RT- PCR products was used to identify the two alleles in RNA: the transversion G>T eliminates the Taq1 recognition site leading to a variant fragment of 396 bp. As shown in fig 3 ▶, the different products expected from digestion of variant and normal alleles (396 bp+295 bp+101 bp) were present in the transcript obtained from the control sample (individual No 7) while only the bands corresponding to the normal allele (295 bp+101 bp) were observed in the patient (individual No 8). These data clearly indicate loss of expression of one RET allele, which segregates with the disease in the pedigree, and confirm the hypothesis that non-coding mutations can significantly contribute to RET haplo insufficiency and therefore to the pathogenesis of HSCR.
Figure 3.
Detection of a RET expression defect. (A) Haplotype reconstruction at the RET locus in a family with Hirschsprung disease (HSCR); filled symbols represent histologically confirmed long segment HSCR patients while hatched symbols represent individuals showing a disease status, not studied by histological examination but clinically documented, as previously described.26 The two alleles of single nucleotide polymorphisms (SNP)2, SNP13, and SNP19, and alleles of the microsatellite sTCL2 (previously reported as corresponding to RET by Yin and collegues26) are indicated. (B) cDNA analysis from the proband (individual No 8, corresponding to individual IV-2 of the pedigree) and a control (individual No 7), both heterozygous for a known polymorphism in exon 13 (2307G>T). A schematic representation of the polymerase chain reaction products and the subsequent TaqI digestion is shown. Arrows indicate the normal and polymorphic alleles.
DISCUSSION
It is well established that histone acetylation is an important mechanism that modulates gene expression by altering accessibility of transcription factors to DNA. Switch from inactive to active chromatin is often accompanied by histone hyperacetylation of critical sites in gene regulatory regions.27 In a previous paper, we demonstrated that the histone deacetylase inhibitor NaB increased RET transcription in cells displaying low mRNA levels while it had no effect in cells already expressing high mRNA levels.22 Moreover, we identified two RET upstream regions sensitive to NaB treatment, the SOX10/Pax3 enhancer and the minimal promoter, supporting the view that acetylation of RET histones is a crucial and physiological regulatory mechanism of RET transcription.22 Here we have shown that NaB treatment induces the appearance of RET transcript in RET negative cells, such as lymphoblasts and the SK-N-MC cell line. Starting from such an observation we have developed a novel and useful approach to analyse the RET transcription product. In particular, we have demonstrated that, after NaB treatment, RET cDNA can be analysed directly through either restriction enzyme digestion (normally used for typing RET polymorphisms) or SSCP analysis (a common method to investigate RET mutations). Indeed, we found a similar intensity and migration pattern of RT-PCR products obtained after NaB treatment compared with PCR products from genomic DNA. These data clearly indicate that RET analysis normally used for genomic DNA assay (SSCP, DHPLC, sequencing, and others) can now also be applied to the full length cDNA.
Splicing alterations are believed to represent 15% of all gene defects but they are rarely detected when located far from the splice canonical sites.28 For this reason, it has been generally suggested that gene mutation analysis should also cover introns, an approach applicable only to a small number of genes characterised by reduced number and size of intronic regions. Analysis of gene transcripts is a useful alternative to detect the effect of splicing mutations, as suggested by the high number of such anomalies demonstrated in genes normally expressed in lymphoblasts, such as ATM and NF.12,13 Another important achievement of the present pilot study is the demonstration that NaB treatment allows direct detection of RET anomalous transcripts in mRNA obtained from lymphoblasts of HSCR patients, without inducing aberrant splicing per se, as shown in fig 2B ▶. Lymphoblasts from both HSCR patients and controls were treated with NaB but only patient carrying the IVS12+19C>T mutation showed an aberrant spliced product. These data confirm our method as a rapid primary screening for splicing mutations, provided positive results obtained by NaB treated lymphoblasts are followed by specific genomic analysis to avoid the possibility of false positives such as, for instance, those derived from tissue specific patterns of splicing.
The potency of NaB treatment combined with the fact that it seems to reproduce a physiological mechanism of transcription convinced us to undertake a NaB based approach for the study of possible RET expression defects.
As hypothesised, we demonstrated defective RET expression in a familial case of HSCR showing no mutation in the coding region of the gene. The observed loss of heterozygosity in the cDNA might be caused by an as yet unidentified defect in the RET transcription regulatory region (promoter and 5′ region) and/or in post-transcriptional control (splicing defect or instability of allele specific RET mRNA), both impairing correct RET expression. Characterisation of the functional variant responsible for this effect is currently under investigation. This result is in agreement with the hypothesis that non-coding RET variations have been underestimated and this could explain the discrepancy between the proportion of HSCR families linked to RET (88%) and the proportion of identified mutations (<40%).5 Hence NaB treatment could allow the detection of a wide spectrum of RET defects which we were unable to identify by standard genomic analysis.
As a further useful application, NaB treatment could be used to investigate additional elements possibly involved in HSCR susceptibility such as the potential functional effect of RET polymorphic variants and the amount of the two terminal isoforms, RET9 and RET51.10,11,29,30 Indeed, the full application of the proposed NaB treatment protocol may allow dissection of the possible functional contribution of SNPs, such as the 135G>A in exon 2 of the gene or the newly identified SNPs of the RET promoter.9,11 In addition, the expression pattern of the RET9 and RET51 isoforms, recently demonstrated to have different biological and signalling properties, may be correlated with the development of HSCR disease and allied related pathologies such as MEN2A and MEN2B.29,30
In conclusion, our observation that NaB treatment activates RET expression in cultured cells represents a novel powerful tool to test the relative amount and quality of the RET transcript in cell sources easily available, such as lymphocytes or lymphoblasts. The full application of the proposed NaB treatment protocol to unexplained HSCR cases (still representing the majority of all cases) and other RET related pathologies will allow the establishment of the precise role the RET proto-oncogene plays in their development.
Acknowledgments
We are grateful to Mrs L Velo for secretarial assistance and to B Pesce, F Lantieri, and G Santamaria for technical support. P Griseri, G Patrone, and F Puppo are supported by a Fellowship awarded by the Fondazione Italiana per la Ricerca sul Cancro (FIRC). The financial support of Italian Telethon (grant No E791), European Community (Contract No QLG1-2001-01646) and CEBR-Center of Excellence at the University of Genova, are gratefully acknowledged.
Abbreviations
ATM, ataxia-telangiectasia mutated
DHPLC, denaturing high performance liquid chromatography
G3PDH, glucose 3-phosphate dehydrogenase
HSCR, Hirschsprung disease
MEN, multiple endocrine neoplasia
NaB, sodium butyrate
NF1, neurofibromatosis type 1
RET, REarranged during Transfection
RT-PCR, reverse transcription-polymerase chain reaction
SNP, single nucleotide polymorphism
SSCP, single strand conformation polymorphism
REFERENCES
- 1.Manie S, Santoro M, Fusco A, et al. The RET receptor: function in development and dysfunction in congenital malformation. Trends Genet 2001;17:80–9. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi M, Asai N, Iwashita T, et al. Mechanisms of development of multiple endocrine neoplasia type 2 and Hirschsprung’s disease by ret mutations. Recent Results Cancer Res 1998;54:229–36. [DOI] [PubMed] [Google Scholar]
- 3.Chakravarti A, Lyonnet S. Hirschsprung disease. In: Scriver CS, et al, eds. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill, Medical Publishing Division, 2002:6231–55.
- 4.Wakamatsu N, Yamada Y, Yamada K, et al. Mutations in SIP1, encoding Smad interacting protein 1, cause a form of Hirschsprung disease. Nat Genet 2001;27:369–70. [DOI] [PubMed] [Google Scholar]
- 5.Gabriel SB, Salomon R, Pelet A, et al. Segregation at three loci explains familial and population risk in Hirschsprung disease. Nat Genet 2002;31:89–93. [DOI] [PubMed] [Google Scholar]
- 6.Bolk S, Pelet A, Hofstra RMW, et al. A human model for multigenic inheritance: phenotypic expression in Hirschsprung disease requires both the RET gene and a new 9q31 locus. Proc Natl Acad Sci U S A 2000;97:268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrasquillo MM, McCallion AS, Puffenberger EG, et al. Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat Genet 2002;32:237–44. [DOI] [PubMed] [Google Scholar]
- 8.Borrego S, Saez ME, Ruitz A, et al. Specific polymorphisms in the RET proto- oncogene are over-represented in patients with Hirschsprung disease and may represent loci modifying phenotypic expression. J Med Genet 1999;65:1469–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitze G, Cramer J, Ziegler A, et al. Association between c135G/A genotype and RET proto-oncogene germline mutations and phenotype of Hirschsprung’s disease. Lancet 2002;359:1200–5. [DOI] [PubMed] [Google Scholar]
- 10.Borrego S, Wright FA, Fernandez RM, et al. A founding locus within the RET proto- oncogene may account for a large proportion of apparently sporadic Hirschsprung disease and a subset of sporadic medullary thyroid carcinoma. Am J Hum Genet 2002;72:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griseri P, Pesce B, Patrone G, et al. A rare haplotype of the RET proto-oncogene is a risk-modifying allele in Hirschsprung disease. Am J Hum Genet 2002;71:969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teraoka SN, Telatar M, Becker-Catania S, et al. Splicing defects in the ataxia- telangiectasia gene, ATM: underlying mutations and consequences. Am J Hum Genet 1999;64:1617–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomson SA, Wallace MR. RT-PCR splicing analysis of the NF1 open reading frame. Hum Genet 2002;110:495–502. [DOI] [PubMed] [Google Scholar]
- 14.Takaya K, Yoshimasa T, Arai H, et al. Expression of the RET proto-oncogene in normal human tissues, pheochromocytomas, and other tumors of neural crest origin. J Mol Med 1996;74:617–21. [DOI] [PubMed] [Google Scholar]
- 15.Gattei V, Celetti A, Cerrato A, et al. Expression of the RET receptor tyrosine kinase and GDNFR-alpha in normal and leukemic human hematopoietic cells and stromal cells of the bone marrow microenvironment. Blood 1997;89:2925–37. [PubMed] [Google Scholar]
- 16.Pachnis V, Mankoo B, Costantini F. Expression of the c-RET proto-oncogene during mouse embryogenesis. Development 1993;119:1005–17. [DOI] [PubMed] [Google Scholar]
- 17.Tsuzuki T, Takahashi M, Asai N, et al. Spatial and temporal expression of the RET proto-oncogene product in embryonic, infant and adult rat tissue. Oncogene 1995;10:191–8. [PubMed] [Google Scholar]
- 18.Auricchio A, Griseri P, Carpentieri L, et al. Double heterozygosity for a RET substitution interfering with splicing and a EDNRB missense mutation in Hirschsprung disease. Am J Hum Genet 1999;64:1216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griseri P, Sancandi M, Patrone G, et al. Decreased frequency of a single nucleotide polymorphism of the RET proto-oncogene in sporadic Hirschsprung disease. Eur J Hum Genet 2000;8:721–4. [DOI] [PubMed] [Google Scholar]
- 20.Griseri P, Mishto M, Priolo M, et al. An intronic nucleotide variant of the RET proto- oncogene causes Hirschsprung disease by interfering with RNA splicing. Gene Funct Dis 2000;1:184–8. [Google Scholar]
- 21.Kaplan JC, Kahn A, Chelly J. Illegitimate transcription: its use in the study of inherited disease. Hum Mutat 1992;1:357–60. [DOI] [PubMed] [Google Scholar]
- 22.Puppo F, Griseri P, Fanelli M, et al. Cell-line specific chromatin acetylation at the Sox10-Pax3 enhancer site modulates the RET proto-oncogene expression. FEBS letters 2002;523:123–7. [DOI] [PubMed] [Google Scholar]
- 23.Ceccherini I, Hofstra R, Luo Y, et al. DNA polymorphisms and conditions for SSCP analysis of the exons of the RET proto-oncogene. Oncogene 1994;9:3025–9. [PubMed] [Google Scholar]
- 24.Patrone G, Puppo F, Scaranari M, et al. Cell-line specific transcription rates of the RET gene and functional domains in its minimal promoter. Gene Funct Dis 2000;3:1–8. [Google Scholar]
- 25.Takahashi M, Buma Y, Taniguchi M. Identification of the RET proto-oncogene products in neuroblastoma and leukemia cells. Oncogene 1991;6:297–301. [PubMed] [Google Scholar]
- 26.Yin L, Ceccherini I, Pasini B, et al. Close linkage with the RET proto-oncogene and boundaries of deletion mutations in autosomal dominant Hirschsprung disease. Hum Mol Genet 1994;2:1803–8. [DOI] [PubMed] [Google Scholar]
- 27.Emerson BM. Specificity of gene regulation. Cell 2002;109:267–70. [DOI] [PubMed] [Google Scholar]
- 28.Krawzczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet 1992;90:41–54. [DOI] [PubMed] [Google Scholar]
- 29.De Graaf E, Sriniva S, Kilknny C, et al. Differential activities of the RET tyrosine- kinase receptor isoforms during mammalian embryogenesis. Genes Dev 2001;15:2433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsui-Pierchala BA, Ahrens RC, Crowder RJ, et al.. The long and short isoforms of Ret function as independent signalling complexes. J Biol Chem 2002;277:34618–25. [DOI] [PubMed] [Google Scholar]