Abstract
Background: A severe form of iron overload with the clinicopathological features of haemochromatosis inherited in an autosomal dominant manner has been described in the Solomon Islands. The genetic basis of the disorder has not been identified. The disorder has similarities to type 4 haemochromatosis, which is caused by mutations in ferroportin1.
Aims: The aims of this study were to identify the genetic basis of iron overload in a patient from the Solomon Islands.
Patient and methods: Genomic DNA was isolated from peripheral blood leucocytes of a Solomon Islands man with severe iron overload. The entire coding region and splice sites of the ferroportin1 gene was sequenced.
Results and conclusions: A novel missense mutation (431A>C; N144T) was identified in exon 5 of the ferroportin1 gene. A novel restriction endonuclease based assay which identifies both the N144T and N144H mutations was developed which will simplify the diagnosis and screening of patients for iron overload in the Solomon Islands and other populations. This is the first identified mutation associated with haemochromatosis in the Solomon Islands population.
Keywords: hereditary haemochromatosis, ferroportin1
Hereditary haemochromatosis is one of the most common inherited disorders and is associated with an increase in iron absorption and iron deposition in the body. Untreated, this accumulation of iron leads to tissue damage including cirrhosis, diabetes mellitus, arthropathy, cardiomyopathy, endocrine abnormalities, and hepatocellular carcinoma.1 The most common form of haemochromatosis (type 1) is caused by mutations in the HFE gene.2 This is inherited as an autosomal recessive trait and affects approximately 1 in 200 people of Northern European origin.
Other non-HFE related forms of iron overload have been described. Juvenile haemochromatosis (JH or type 2) is inherited as an autosomal recessive trait. Recently, two forms of JH have been recognised: one mapping to chromosome 1q3 and one to chromosome 19.4 Mutations in the gene encoding the antimicrobial peptide hepcidin have been implicated in the chromosome 19 form.4 However, the gene responsible for chromosome 1 JH has not yet been identified.3 Mutations in the transferrin receptor 2 gene have been implicated in another form of haemochromatosis (type 3).5
A locus for an autosomal dominant form of haemochromatosis (type 4) was recently identified. The gene responsible was shown to be ferroportin1.6,7 The ferroportin1 gene, also known as SLC11A3, IREG1, and MTP1,8–10 encodes a multiple transmembrane domain protein responsible for iron export from cells. Three mutations in the ferroportin1 gene have been reported, N144H,6 A77D,7 and V162del.11–14 Heterozygosity for these mutations results in a form of iron overload associated with high serum ferritin levels and heavy deposition of iron in reticuloendothelial cells.
Iron overload in the Solomon Islands has been reported previously.15 It was described in a large Melanesian pedigree comprising 81 surviving relatives. A total of 31 members were shown to have iron overload by measurement of serum ferritin concentrations and transferrin saturation. Iron overload was confirmed in all subjects who underwent liver biopsy. Iron overload increased with age and some degree of fibrosis or cirrhosis was present in nearly all affected members. Genetic analysis suggested an autosomal dominant mode of inheritance. Linkage to the HLA-A and B loci was excluded, suggesting that this disorder is unrelated to HFE.15
We report the identification of a new nucleotide substitution in the ferroportin1 gene (431A>C; N144T) associated with severe iron overload in a patient from the Solomon Islands. We also describe a novel restriction endonuclease based detection assay which will be useful in screening for both N144T and N144H mutations.
PATIENTS AND METHODS
This study was approved by and performed in accordance with the ethical standards of the Queensland Institute of Medical Research Human Research Ethics Committee and with the Helsinki Declaration of 1975, as revised in 1983. Informed and written consent was obtained from the patient for all the studies described in this report.
Patient details
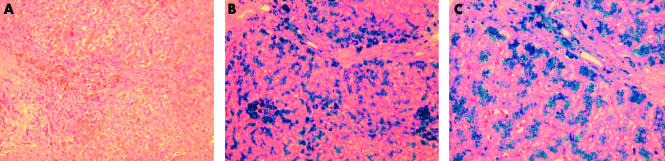
A 48 year old male from the Solomon Islands underwent a routine medical examination. A cardiac murmur and hepatomegaly were detected during physical examination. Subsequent biochemical evaluation showed an elevated alanine aminotransferase level of 82 U/l, serum iron concentration of 40 μmol/l, transferrin saturation of 80%, and serum ferritin concentration of 2937 μg/l. Serum copper and ceruloplasmin levels were normal. The patient was negative for the two common mutations of HFE, C282Y and H63D. Liver biopsy showed grade 4 iron stores predominantly in the parenchymal cells with significant iron deposits in the Kupffer cells and portal tract macrophages (fig 1 ▶). The hepatic iron concentration was 363 μmol/g and the hepatic iron index was 7.6. In addition, portal fibrosis was observed but no cirrhosis was evident.
Figure 1.
Liver biopsy sections from a patient showing portal fibrosis and iron overload. Liver biopsy showing (A) portal fibrosis and (B, C) Perls’ Prussian Blue staining showing heavy (grade 4) parenchymal iron accumulation with significant Kupffer cell and macrophage siderosis. (A and B, 100× magnification; C, 200× magnification.)
The patient was born in the Solomon Islands and both parents and grandparents were of Melanesian-Solomon Islands extraction. He is not directly related to the family studied by Eason and colleagues15 but a more distant ancestral connection cannot be ruled out.
Efforts were made to contact and obtain blood samples from family members and the previously described pedigree studied by Eason and colleagues.15 However, due to circumstances beyond our control none could be contacted.
Controls
A control group comprising 100 normal healthy Australian individuals was studied to determine the frequency of a novel ferroportin1 substitution and exclude it as a common polymorphism. A control group from the Solomon Islands population was not available for this study.
Molecular studies
Genomic DNA isolated from peripheral blood leucocytes was used as a template in polymerase chain reactions (PCRs). The entire coding sequence and splice sites of ferroportin1 were amplified and sequenced, as described previously.11
Analysis of the nucleotide sequence of exon 5 of ferroportin1 revealed that the novel nucleotide substitution 431A>C could be detected by a restriction endonuclease based assay. Amplification of part of exon 5 (primers IRG5F 5′ CTGCTATATCCTGATCATCACTAT3′ and IRG5R 5′ GAAAGCCAAATTACTTGCTAGTT3′) results in a product of 136 base pairs (bp). When digested with the enzyme Tsp509I (New England Biolabs, Massachusetts, USA) the wild-type sequence yields two bands of 37 and 99 bp. The 431A>C substitution (and 430A>C, N144H mutation) destroys the enzyme recognition site and results in no cleaved products. DNA fragments were run on 2% agarose-TBE gels and visualised using ethidium bromide.
RESULTS AND DISCUSSION
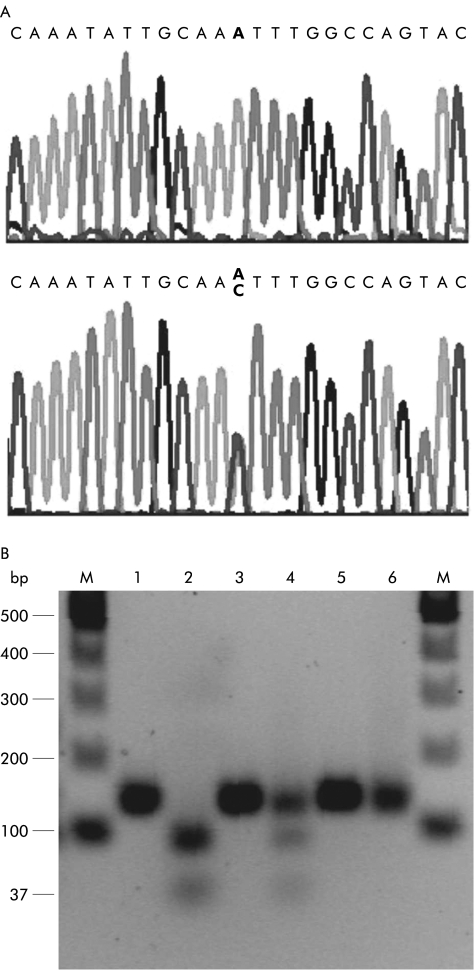
Analysis of the DNA sequence of ferroportin1 in a patient with a severe form of haemochromatosis from the Solomon Islands revealed a novel single nucleotide substitution (431A>C) in exon 5 (fig 2A ▶). The substitution results in a change of residue 144 from an asparagine to a threonine (N144T).
Figure 2.
Detection of mutations by DNA sequencing and restriction endonuclease polymorphism. Exon 5 of ferroportin1 was amplified using primers IRG5F and IRG5R. (A) DNA sequence of the wild-type (top) and mutant ferroportin1 (bottom). (B) Lanes 1, 3, and 5 are uncut DNA; lanes 2, 4, and 6 are DNA digested with Tsp 509I. Lanes 1 and 2 are wild- type DNA, lanes 3 and 4 are DNA isolated from the Solomon Islands patient, and lanes 5 and 6 are cloned exon 5 DNA in a plasmid with the 430A>C mutation. The polymerase chain reaction product obtained is 136 base pairs (bp) in length. When cut with Tsp 509I, the wild- type sequence gives two bands of 37 and 99 bp (lane 2). The patient (lane 4) is heterozygous for the 431A>C mutation; this results in three bands, one uncut of 136 bp, and the two cut bands of 37 and 99 bp. The mutant control is a synthetic plasmid control (430A>C) and is thus homozygous (lane 6).
The presence of the 431A>C substitution was confirmed using a Tsp509I restriction endonuclease digestion assay (fig 2B ▶). Interestingly, this assay can also be used to detect the 430A>C (N144H) mutation which was identified in a Dutch family with type 4 haemochromatosis.6
A control group comprising 100 normal healthy individuals was analysed using the Tsp509I restriction endonuclease assay to determine the frequency of the 431A>C substitution in the general population. It was not detected in the control group, suggesting that 431A>C is likely to be a disease causing mutation and not a common polymorphism.
It has been known for many years that a form of hereditary iron overload unrelated to HFE existed in the Solomon Islands but the molecular basis of the disease had not previously been examined.15 Whether the mutation described here is the cause of haemochromatosis in the family studied by Eason and colleagues15 remains to be determined.
The mutation we have identified in a Solomon Islands patient affects the same amino acid as the mutation described by Njajou and colleagues6 (430A>C;N144H), however, asparagine is substituted by threonine instead of histidine. This asparagine residue is highly conserved across species and is likely to be important in the correct functioning of ferroportin1. Njajou and colleagues6 suggested that N144H is a gain of function mutation. However, in agreement with Montosi and colleagues,7 we propose that heterozygosity for N144T, N144H, and all other previously reported mutations of ferroportin1 causes loss of function leading to impaired iron export, abnormal body iron homeostasis, and iron overload.
In conclusion, we have identified a novel nucleotide substitution in ferroportin1 which results in an amino acid change, N144T. This change was associated with a severe form of iron overload in a patient from the Solomon Islands. Although an autosomal dominant form of iron overload has previously been described in this population,15 the molecular basis had not been established. This is the first identification of a mutation associated with haemochromatosis in this population. Whether this mutation is responsible for the previously reported cases of iron overload remains to be determined. The description of a novel and reliable assay to detect both N144T and N144H mutations will enable the rapid screening and diagnosis of type 4 haemochromatosis in the Solomon Islands and other populations. This will enable the early identification of affected individuals and allow treatment to prevent iron associated tissue damage.
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council of Australia (953219) and National Institutes of Health, USA (5R01DK057648-02).
Abbreviations
PCR, polymerase chain reaction
bp, base pairs
K E Arden and D F Wallace contributed equally to this work.
REFERENCES
- 1.Bothwell TH, Charlton RW, Motulsky AG. Hemochromatosis. In: Scriver CR, Beaudet AL, Sly WS, et al, eds. The Metabolic and Molecular Bases of Inherited Disease, 7th edn. New York: McGraw-Hill; 1995:2237–69.
- 2.Feder JN, Gnirke A, Thomas W, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 1996;13:399–408. [DOI] [PubMed] [Google Scholar]
- 3.Roetto A, Totaro A, Cazzola M, et al. The juvenile hemochromatosis locus maps to chromosome 1q. Am J Hum Genet 1999;64:1388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roetto A, Papanikolaou G, Politou M, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet 2003;33:21–2. [DOI] [PubMed] [Google Scholar]
- 5.Camaschella C, Roetto A, Cali A, et al. The gene TfR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet 2000;25:14–5. [DOI] [PubMed] [Google Scholar]
- 6.Njajou OT, Vaessen N, Joosse M, et al. A mutation in SLC11A3 is associated with autosomal dominant hemochromatosis. Nat Genet 2001;28:213–14. [DOI] [PubMed] [Google Scholar]
- 7.Montosi G, Donovan A, Totaro A, et al. Autosomal dominant hemochromatosis is associated with a mutation in the ferroportin (SLC11A3) gene. J Clin Invest 2001;108:619–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donovan A, Brownlie A, Zhou Y, et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 2000;403:778–81. [DOI] [PubMed] [Google Scholar]
- 9.McKie AT, Marciani P, Rolfs A, et al. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell 2000;5:299–309. [DOI] [PubMed] [Google Scholar]
- 10.Abboud S, Haile DJ. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem 2000;275:19906–12. [DOI] [PubMed] [Google Scholar]
- 11.Wallace DF, Pedersen P, Dixon JL, et al. Novel mutation in ferroportin1 is associated with autosomal dominant hemochromatosis. Blood 2002;100:692–4. [DOI] [PubMed] [Google Scholar]
- 12.Devalia V, Carter K, Walker AP, et al. Autosomal dominant reticuloendothelial iron overload associated with a 3-base pair deletion in the ferroportin 1 gene (SLC11A3). Blood 2002;100:695–7. [DOI] [PubMed] [Google Scholar]
- 13.Roetto A, Merryweather-Clarke AT, Daraio F, et al. A valine deletion of ferroportin 1: a common mutation in hemochromastosis type 4. Blood 2002;100:733–4. [DOI] [PubMed] [Google Scholar]
- 14.Cazzola M, Cremonesi L, Papaioannou M, et al. Genetic hyperferritinaemia and reticuloendothelial iron overload associated with a three base pair deletion in the coding region of the ferroportin gene (SLC11A3). Br J Haematol 2002;119:539–46 [DOI] [PubMed] [Google Scholar]
- 15.Eason RJ, Adams PC, Aston CE, et al. Familial iron overload with possible autosomal dominant inheritance. Aust NZ J Med 1990;20:226–30. [DOI] [PubMed] [Google Scholar]