Abstract
Background: Although it has been reported that different hepatitis B virus (HBV) genotypes induce different clinical characteristics in patients with chronic liver diseases (CLD), there have been few reports that have detailed the distribution of HBV genotypes in acute forms of liver disease.
Methods: HBV genotypes were determined in 61 patients who had acute forms of liver disease (45 had acute self limited hepatitis (AH) and 16 had fulminant hepatitis (FH)) and in 531 patients with CLD, including 19 patients with severe acute exacerbation of CLD. We also analysed the enhancer II, core promoter, and precore region sequences for the presence of mutations.
Results: Expression of genotype B in patients with acute forms of liver disease was significantly greater than in those with CLD (39.3% v 11.7%, respectively; p<0.001). Furthermore, expression of genotype B was significantly greater in patients with FH than in those with AH (62.5% v 31.1%, respectively; p=0.027). The precore mutation A1896 and the core promoter mutation at nt 1753 and 1754 were found more frequently in FH than in AH, and genotype B was predominant in FH regardless of the presence of these mutations.
Conclusions: HBV genotype B was found more frequently in patients with acute forms of liver disease than in patients with CLD, and more frequently in patients with FH than in those with AH. These results suggest that this HBV genotype may induce more severe liver damage than other viral genotypes, at least in patients from Chiba, Japan.
Keywords: acute self limited hepatitis, fulminant hepatitis, hepatitis B virus, genotype
Hepatitis B virus (HBV) is one of the major aetiological agents of acute and chronic liver diseases worldwide. HBV is classified into seven genotypes according to the phylogenetic analysis of its genomic sequences. The first four genotypes (genotypes A–D) were first described by Okamoto and colleagues.1 Six years later, two additional genotypes2(genotypes E, F), and most recently genotype G, were described.3 The geographical distributions of these genotypes are different. Genotypes A and D were shown to be prevalent in Europe and the USA4,5 while genotypes E and F were found to be restricted to Africa6 and to be prevalent in Central and South America, respectively.7,8 Genotypes B and C were reported to be common in East Asia.9 In Japan, genotypes B and C were reported in 12% and 85%, respectively, of patients with chronic liver diseases (CLD).10 Recently, differences in the clinical features of CLD between individuals harbouring viruses of different genotypes were reported, with genotype A being shown to cause milder liver disease than genotype D.11 Orito et al also reported that patients with HBV of genotype C might undergo seroconversion at a later age than those with genotype B.12 Genotype C was reported to associate with more severe histological inflammation than genotype B.13,14 It has also been reported that in patients with chronic hepatitis, the response to interferon was lower in patients infected with genotype C than in those infected with genotype B.15 Thus the clinical course of acute liver diseases may also be different in patients with different viral genotypes. We therefore examined the distribution of HBV genotypes in patients who had an acute form of liver disease and compared it with the distribution seen in patients with CLD and acute exacerbation of CLD.
We previously showed that a G to A mutation at nt 1896 in the precore region (A1896), and mutations in the core region of HBV, were frequently found in fatal acute forms of liver disease.16,17 There have been a few reports that have described a relationship between precore mutations and fulminant hepatitis (FH).18–23 Similarly, Sato et al reported the presence of an A to T mutation at nt 1762 and a G to A mutation at nt 1764 in the core promoter region (T1762 and A1764) in Japanese patients with FH who did not manifest precore stop codon mutations.24 However, a study of patients from the USA and Europe failed to reveal such an association.25–30 We have also reported that mutations in the enhancer II/core promoter region of the virus were observed more frequently in patients with FH than in those with acute self limited hepatitis (AH) although the mutations in nucleotides 1762 and 1764 were not often seen.31 Thus the involvement of mutations in the core promoter and precore regions of HBV in the development of FH is still controversial and the relationship between these mutations and the different viral genotypes is not known. To examine the relation of these mutations to the different viral genotypes, we compared the HBV sequences in the core promoter and precore regions in each genotype.
MATERIALS AND METHODS
Patients
Sixty one patients with HBV related acute forms of liver disease that were consecutively admitted to Chiba University Hospital between 1980 and 2001 were examined in this study. They comprised 45 patients with AH and 16 with FH (table 1 ▶). HBV related acute forms of liver disease were diagnosed based on either the appearance of hepatitis B surface antigen (HBsAg) or the presence of >1 titre of immunoglobulin M (IgM) antibody to hepatitis B core antigen. Patients with a prolonged prothrombin time that was less than 40% of control values and who developed encephalopathy during the clinical course of their AH were diagnosed with FH. Patients who tested positive for HBsAg were considered to be HBV carriers, and those who had an abnormal alanine aminotransferase (ALT) level at some point during their observation period were diagnosed with CLD. CLD patients who had an elevated ALT of more than 600 IU/l, a total bilirubin of more than 3.0 mg/dl, or a prolonged prothrombin time of less than 50% of control values at any time during the clinical course of their disease were diagnosed with severe acute exacerbation of CLD. None of the patients with HBV related acute forms of liver disease or severe acute exacerbation of CLD tested positive for either anti-hepatitis C antibody (anti-HCV) or anti-hepatitis D antibody (anti-HDV). From the 590 patients with CLD who visited Chiba University Hospital between 1980 and 2001, 531 control patients with CLD were selected who: (1) were inhabitants of the Chiba region and (2) were negative for both anti-HCV and anti-HDV. The HBV genotypes of the above CLD patients were determined. The characteristics of the CLD patients examined in this study are also included in table 1 ▶.
Table 1.
Patient profiles
| Case | No studied | Sex (M:F) | Age (y)* | ALT (U/l)* | HBeAg positivity (%) | HBV DNA (log copies/ml)* |
| Acute forms of liver disease | 61 | 41:20 | 34.1 (11.1) | 3143 (3251) | 54.1a | 4.58 (1.31)d |
| (AH) | 45 | 29:16 | 37.1 (12.5) | 3092 (2870) | 65.2b | 4.48 (1.26)e |
| (FH) | 16 | 12:4 | 43.0 (9.20) | 2959 (4068) | 25.0b | 4.71 (1.40)e |
| CLD | 531 | 360:171 | 38.5 (13.9) | 113.4 (127.3) | 58.7a c | 6.19 (1.65)df |
| (Acute exacerbation of CLD) | 19 | 14:5 | 46.4 (12.3) | 1271 (1875) | 36.8c | 5.91 (1.74)f |
| Total | 592 | 401:191 | 38.3 (13.7) | 370.8 (1238.1) | 57.9 | 5.97 (1.51) |
*Values are mean (SD).
aNS; b p=0.007; cNS; d p<0.001; eNS; fNS.
AH, acute self limited hepatitis; ALT, alanine aminotransferase; CLD, chronic liver diseases; FH, fulminant hepatitis; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus.
(AH and FH are included in acute forms of liver dissase.)
(Acute exacerbation of CLD is included in CLD.)
HBV genotypes and the nucleotide sequence of the core promoter and precore regions of the virus were examined in 69 patients (37 of 45 patients with AH, 15 of 16 with FH, and 17 of 18 with acute exacerbation of CLD) whose serum samples were available.
Serological markers
HBsAg, hepatitis B e antigen (HBeAg), and anti-HDV were determined using an enzyme linked immunosorbant assay (ELISA) (Abbot Laboratory, Chicago, Illinois, USA). The presence of anti-HCV was also determined by ELISA (Ortho Diagnostics, Tokyo, Japan). Serum HBV DNA was quantified using the polymerase chain reaction (PCR) assay (Amplicor HBV Monitor; Roche Diagnostics, Tokyo, Japan). The linear range of this assay was 2.6–7.6 log copies /ml.
Determination of HBV genotypes
HBV genotype was determined in patient sera by ELISA (HBV Genotype EIA; Tokushu-Meneki Laboratory, Tokyo, Japan) based on the methodology described by Usuda et al.32 Genotypes were also examined by the method of Mizokami et al in cases where the ELISA results were inconclusive.33 Briefly, S-gene sequences that were amplified by semi nested PCR were digested with restriction endonucleases, and the genotypes were determined by the restriction fragment length polymorphisms of the digested PCR products. Our results from the use of either of the above techniques were comparable.
Determination of HBV sequences
Sequences of HBV genomes were determined by the direct sequencing method after PCR amplification.34 To amplify the enhancer II (nt 1688 to 1773), core promoter (nt 1742 to 1849), and precore regions (nt 1814 to 1900), several primers were prepared. The primers for the first PCR were 5 -GTCTGTGCCTTCTCATCTGCC-3 (sense, nt 1553–1573) and 5 -AGAATAGCTTGCCTGAGTGC-3 (antisense, nt 2060–2079), and the primers for the second PCR were 5 -ACGTCGCATGGAGACCACCG-3 (sense, nt1603–1622) and 5 -GAAAGAAGTCAGAAGGCAAA-3 (antisense, nt 1954–1973). Briefly, 10 µl of DNA, extracted from the serum, were added to 5 µl of dNTP, 1 µl of Ex-Taq polymerase (Takara Syuzou, Kyoto, Japan), 5 µl of 10×Taq polymerase buffer, and 100 pmol of outer sense and antisense primers. The amplification profile was two minutes at 96 C, followed by 25 cycles at 96 C for one minute (denaturation), 55 C for one minute (annealing), and 72 C for one minute (extension). For the second PCR, 1 µl of the first PCR product was added to the same reaction buffer with inner sense and antisense primers except for the outer sense and antisense primers. The second PCR products were purified and sequenced using a Dye Terminator Kit (Applied Biosystems, Foster City, California, USA). Sequences were compared with the reported sequences of each genotype (genotype A, X70185; genotype B, X97851; genotype C, X04615).
Statistics
Results are expressed as mean (SD). Frequencies between groups were compared using the 2×2 χ2 test, and group means were compared using the Student’s t test. A p value of less than 0.05 was considered to be statistically significant.
RESULTS
Characteristics of patients whose HBV genotypes were investigated
Table 1 ▶ shows the clinical and laboratory data of the patients whose HBV genotypes were investigated. There were no statistically significant differences in mean age or sex between patients with AH, FH, CLD, and acute exacerbation of CLD. The percentage of FH patients who were positive for HBeAg was significantly lower than that seen in patients with AH (25.0% v 65.2%; p<0.001). Mean HBV DNA levels were significantly lower in patients who had an acute form of liver disease than in those with CLD (4.58 (1.31) v 6.19 (1.65) log copies/ml; p<0.001), although they were comparable in patients with AH and FH (4.48 (1.26) v 4.71 (1.40) log copies/ml; NS) and in patients with CLD and acute exacerbation of CLD (6.19 (1.65) v 5.91 (1.74) log copies/ml; NS).
Distribution of HBV genotypes in patients with an acute form of liver disease or CLD
The distribution of HBV genotypes in the various forms of liver disease is shown in table 2 ▶. Among the 61 patients who had an acute form of liver disease, six (9.8%), 24 (39.3%), and 31 (50.8%) patients were positive for viral genotype A, genotype B, and genotype C, respectively. Genotype B was found significantly more often in patients with an acute form of liver disease than in patients with CLD (24/61 (39.3%) v 62/531 (11.7%); p<0.001). There was no difference in the frequency of genotype B between patients with CLD and those with acute exacerbation of CLD (62/531 (11.7%) v 4/19 (21.1%); NS). Genotype B tended to be found more often in patients with FH than in patients with AH (10/16 (62.5%) v 14/45 (31.3%); p=0.027).
Table 2.
Distribution of HBV genotypes in various forms of hepatitis B
| Diagnosis | No studied | Genotype A | Genotype B | Genotype C |
| Acute forms of liver disease | 61 | 6 (9.8%) | 24 (39.3%)a | 31 (50.8%) |
| (AH) | 45 | 5 (11.1%) | 14 (31.1%)b | 26 (57.8%) |
| (FH) | 16 | 1 (6.2%) | 10 (62.5%)b | 5 (31.3%) |
| CLD | 531 | 10 (1.9%) | 62 (11.7%)ac | 459 (86.4%) |
| (Acute exacerbation of CLD) | 19 | 2 (10.5%) | 4 (21.1%)c | 13 (68.4%) |
| Total | 592 | 16 | 86 | 490 |
a p<0.001; b p=0.027; cNS.
a b c The number of patients with and without HBV genotype B was compared between disease groups.
HBV, hepatitis B virus; AH, acute self limited hepatitis; CLD, chronic liver diseases; FH, fulminant hepatitis.
(AH and FH are included in acute forms of liver disease.)
(Acute exacerbation of CLD is included in CLD.)
Precore region (nt1896) mutation in HBV genotypes
Of the 69 patients whose HBV sequences were investigated, five contained virus with genotype A, 27 contained virus with genotype B, and 37 contained virus with genotype C. The nucleotide sequences of the enhancer II, core promoter, and precore regions in each genotype are shown in fig 1 ▶. Regarding mutations in the precore region, the number of patients with A1896 is shown in table 3 ▶. A1896 was found more frequently in patients with FH than in patients with AH (p<0.001). The percentages of patients with A1896 in genotypes B and C were comparable (11/27 (40.7%) v 10/37 (32.3%); NS). In patients with FH, the distributions of HBV genotypes A, B, and C with and without A1896 were 0, 7, 4 and 1, 3, 0, respectively; hence the percentages of genotype B patients who either had or did not have this mutation were comparable (7/11 (63.6%) v 3/4 (75%); NS). Thus genotype B tended to be predominant in FH, independent of the presence or absence of A1896. No patient carrying genotype A had A1896 although the number of patients with genotype A was very small. For this reason, comparisons were only made between genotypes B or C.
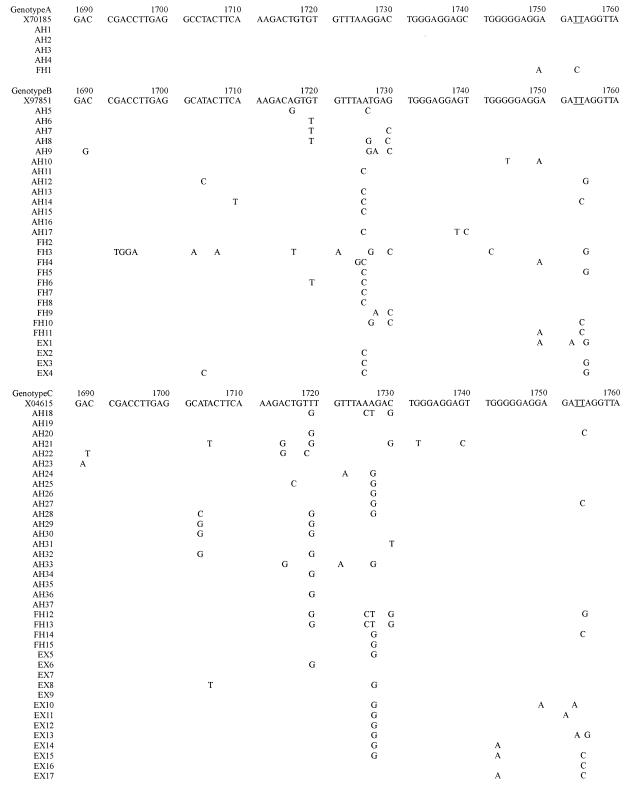
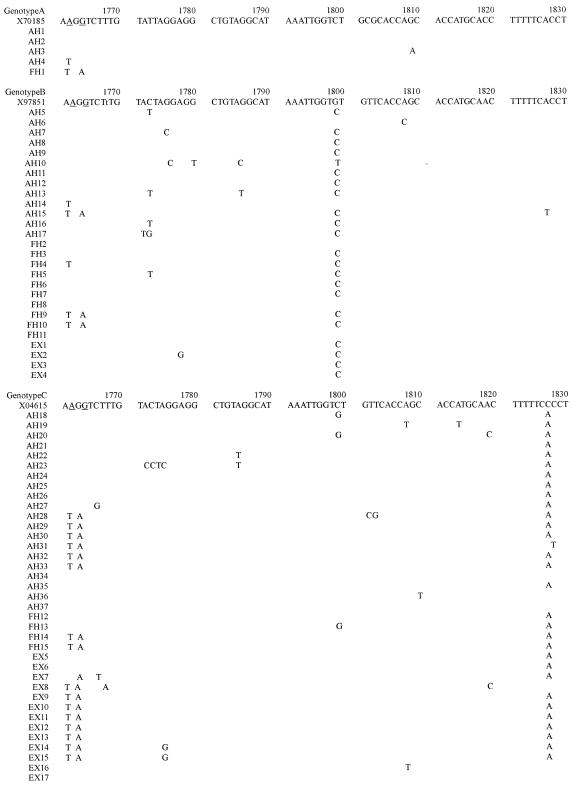
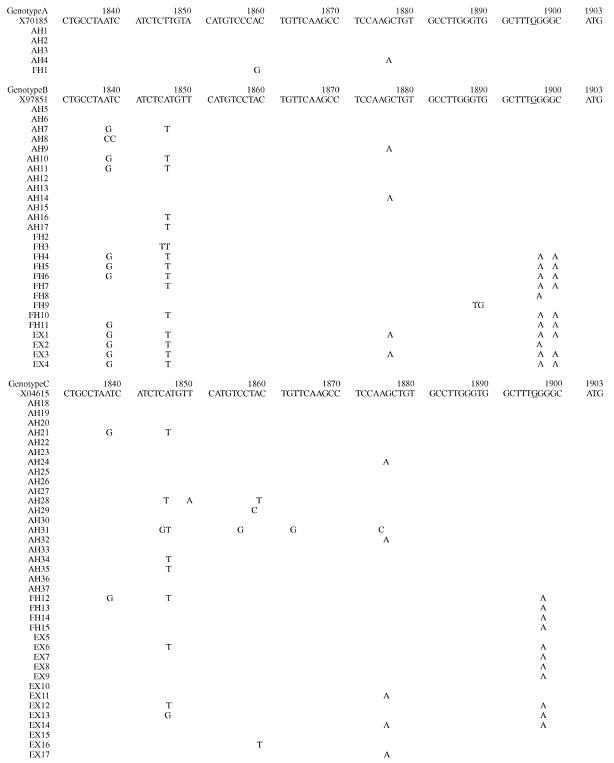
Figure 1.
The nucleotide sequence from nt 1688 to nt 1903 (see above, and on the following two pages), which included the enhancer II (nt 1688 to 1773), core promoter (nt 1742 to 1849), and precore regions (nt 1814 to 1900) is shown for each genotype, and is compared with the prototype sequence (genotype A, X70185; genotype B, X97851; genotype C, X04615). The nucleotide sequence of the HBV genome shown will appear in the DDBJ/EMBL/Genbank nucleotide sequence databases, and have accession numbers AB090170-AB090230 and AB099497-AB099504.
Table 3.
Mutations in the precore region (nt 1896) in each genotype
| Diagnosis | Genotype A | Genotype B | Genotype C | Total |
| AH | 0/4 (0%) | 0/13 (0%) | 0/20 (0%) | 0/37 (0%)* |
| FH | 0/1 (0%) | 7/10 (70.0%) | 4/4 (100.0%) | 11/15 (73.3%)* |
| Acute exacerbation of CLD | 0/0 (0%) | 4/4 (100%) | 6/13 (46.2%) | 10/17 (55.6%) |
| Total | 0/5 (0%) | 11/27 (40.7%)* | 10/37 (42.3%)† | 21/69 (32.8%) |
*p<0.001 †NS.
AH, acute self limited hepatitis; FH, fulminant hepatitis; CLD, chronic liver disease.
Mutations in the core promoter region (nt 1762 and 1764/nt 1753 and 1754) of the different HBV genotypes
We compared our core promoter mutation data with previously reported HBV sequences. Mutations at nt 1762 and 1764 and nt 1753 and 1754 were found when they were compared with each respective prototype sequence. These were all located in AT rich regions of the prototype sequences in the core promoter region. The numbers of patients with T1762 and A1764 are shown in table 4 ▶. There was no difference in the frequency of these mutations between patients with FH and those with AH (5/15 (33.3%) v 7/37 (18.9%); NS). T1762 and A1764 tended to be seen more frequently in patients with genotype C than in those with genotype B (16/37 (43.2%) v 3/27 (11.1%); p=0.005).
Table 4.
Mutations in the core promoter region (nt 1762 and 1764) in each genotype
| Diagnosis | Genotype A | Genotype B | Genotype C | Total |
| AH | 0/4 (0%) | 1/13 (8.3%) | 6/20 (30.0%) | 7/37 (18.9%)a |
| FH | 1/1 (100%) | 2/10 (20.0%) | 2/4 (50.0%) | 5/15 (33.3%)a |
| Acute exacerbation of CLD | 0/0 (0%) | 0/4 (0%) | 8/13 (61.5%) | 8/17 (47.0%) |
| Total | 1/5 (20.0%) | 3/27 (11.1%)b | 16/37 (43.2%)b | 20/69 (29.0%) |
aNS; b p=0.005.
AH, acute self limited hepatitis; FH, fulminant hepatitis; CLD, chronic liver diseases.
The numbers of patients with C/A/G 1753 and/or C/G1754 are shown in table 5 ▶. These mutations were found more frequently in patients with FH than in patients with AH (7/15 (46.7%) v 4/37 (10.8%); p=0.004). The percentages of patients with these mutations in genotypes B and C were comparable (9/27 (33.3%) v 9/37 (18.9%); NS). In patients with FH, the distributions of HBV genotypes A, B, and C with and without these mutations were 1, 4, 2 and 0, 6, 2 respectively; hence the percentages of genotype B patients who either had or did not have these mutations were comparable (4/7 (57.1%) v 6/8 (75.0%); NS). Thus genotype B tended to predominate in FH independent of the presence or absence of these mutations.
Table 5.
Mutations in the core promoter region (nt1753 and 1754) in each genotype
| Diagnosis | Genotype A | Genotype B | Genotype C | Total |
| AH | 0/4 (0%) | 2/13 (15.4%) | 2/20 (10.0%) | 4/37 (10.8%)a |
| FH | 1/1 (100%) | 4/10 (40.0%) | 2/4 (50.0%) | 7/15 (46.7%)a |
| Acute exacerbation of CLD | 0/0 (0%) | 3/4 (75.0%) | 5/13 (38.5%) | 8/17 (47.1%) |
| Total | 1/5 (20.0%) | 9/27 (33.3%)b | 9/37 (18.9%)(b) | 19/69 (27.5%) |
a p=0.004 b p=NS.
DISCUSSION
Our results showed that genotypes B and C of HBV were present in about 40% and 50%, respectively, of patients with an acute form of liver disease. In contrast, genotype C was found in 86% of patients with CLD and 68.4% of patients with acute exacerbation of CLD, suggesting that the distribution of HBV genotype B in acute forms of liver disease was higher than that seen in chronic forms of the disease. Interestingly, the distribution of genotype B was higher in patients with FH than in patients with AH, suggesting a stronger association of genotype B with more severe acute forms of liver disease.
Acute hepatitis B is thought to be transmitted horizontally. As such, our results might reflect the distribution of HBV genotypes in the general population. The distribution of HBV genotypes in patients with an acute form of liver disease suggests that individuals with genotype B are more prevalent in the general population—that is, the prevalence of HBV genotype B might be higher in the general population than in patients with CLD. Fujie et al reported a higher incidence of genotype B in asymptomatic carrier patients who were positive for antibody to hepatitis B e antigen (43.2%) than in patients with hepatocellular carcinoma who were positive for antibody to hepatitis B e antigen (5.9%), although these authors did not compare the prevalence of genotypes B and C in chronic liver diseases.35 Another reason for the higher distribution of genotype B in acute forms of liver disease might be that the percentage of clinical HBV infection compared with subclinical infection might be higher in patients infected with genotype B than in those with genotype C. Kozik et al reported that the ratio of clinical and subclinical HBV infection was 1:30 in Thai children, although they did not compare the ratio between HBV genotypes.36 From our results that showed that genotype B was more prevalent in patients with FH than AH, we speculate that genotype B of HBV may induce more liver damage than genotype C.
Currently, we do not know why genotype B viruses might cause more liver damage than genotype C viruses. As HBV is not a cytotoxic virus, hepatocyte death is caused by the immune response of the host to the viral infection. Therefore, the ability of these viruses to replicate themselves, and/or differences in their amino acid sequences that bind to hepatocyte HLA class I molecules, the target of peptide specific CTL responses probably play a role.37–43 Genotype B virus may have the motifs that strongly bind to HLA class I molecules, thereby resulting in activation of a stronger immune response. Full genomic sequence analysis of HBV in FH will be required to more directly address this question.
In our current study, the majority of patients with FH were found to have an A1896 precore stop codon mutation, which confirmed our previous report that most Japanese patients with fatal FH or fatal acute exacerbation of CLD had HBV with A1896.16 Interestingly, the precore stop codon mutation was not found in any of the five patients with genotype A, including the patient with FH. It has been speculated that A1896 could not occur in HBV genotype A (with T at nt 1858) because of its role in stabilising the stem loop structure of the encapsidation sequence.44 In contrast with the higher prevalence of genotypes B and C in the Japanese population, the predominant HBV genotypes in Europe and the US are genotypes A and D. Therefore, the frequency of the A1896 mutation in FH patients in Japan and the USA needs to be examined based on HBV genotypes.
Orito et al reported that T1762 and A1764 were found more frequently in Japanese patients with CLD who had genotype C virus than in those who had genotype B.12 Our results suggest that this tendency also exists in patients with an acute form of liver disease. Why T1762 and A1764 are found less frequently in viruses with genotype B remains unclear.
Our data also showed that viral mutations tended to be found more frequently in FH patients with nt 1753 and 1754 than in patients with AH. T to C/A/G mutations in nt 1753 were reported to be closely associated with progression of CLD.45 These mutations were found in AT rich regions of the core promoter and as such may change the binding efficiency of transcription factors to the core promoter region, as well as the production of precore and core messages.
In conclusion, we found that genotype B of the HBV occurred more frequently in patients with an acute form of liver disease than in those with CLD, and more frequently in patients with FH than in those with AH, in our cohort of patients in the Chiba region of Japan. Viral genotype as well as the presence of mutations at the core promoter and precore regions may be associated with more severe liver damage in these patients. The molecular mechanisms that account for these associations remain to be investigated.
Acknowledgments
We thank Dr M Shima of the Department of Public Health at the Graduate School of Medicine at Chiba University for his kind advice concerning the statistical analyses that were carried out in this study.
Abbreviations
AH, acute self limited hepatitis
ALT, alanine aminotransferase
CLD, chronic liver diseases
ELISA, enzyme linked immunosorbant assay
FH, fulminant hepatitis
HBV, hepatitis B virus
HBeAg, hepatitis B e antigen
HBsAg, hepatitis B surface antigen
anti-HCV, hepatitis C virus antibody
anti-HDV, hepatitis D virus antibody
PCR, polymerase chain reaction
A1896, G to A mutation at nt 1896 in the precore region
T1762 and A1764, A to T mutation at nt 1762 and G to A mutation at nt 1764 in the core promoter region
C/A/G 1753 and/or C/G1754, C/A/G to T mutation at nt 1753 and/or C/G to T mutation at nt 1754 in the core promoter region
REFERENCES
- 1.Okamoto H, Tsuda F, Sakugawa H, et al. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol 1988;69:2575–83. [DOI] [PubMed] [Google Scholar]
- 2.Norder H, Courouce AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 1994;198:489–503. [DOI] [PubMed] [Google Scholar]
- 3.Stuyver L, De Gendt S, Van Geyt C, et al. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol 2000;81(Pt 1):67–74. [DOI] [PubMed] [Google Scholar]
- 4.Lindh M, Andersson AS, Gusdal A. Genotypes, nt 1858 variants, and geographic origin of hepatitis B virus—large-scale analysis using a new genotyping method. J Infect Dis 1997;175:1285–93. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Frias F, Buti M, Jardi R, et al. Hepatitis B virus infection: precore mutants and its relation to viral genotypes and core mutations. Hepatology 1995;22:1641–7. [DOI] [PubMed] [Google Scholar]
- 6.Odemuyiwa SO, Mulders MN, Oyedele OI, et al. Phylogenetic analysis of new hepatitis B virus isolates from Nigeria supports endemicity of genotype E in West Africa. J Med Virol 2001;65:463–9. [PubMed] [Google Scholar]
- 7.Blitz L, Pujol FH, Swenson PD, et al. Antigenic diversity of hepatitis B virus strains of genotype F in Amerindians and other population groups from Venezuela. J Clin Microbiol 1998;36:648–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arauz-Ruiz P, Norder H, Visona KA, et al. Genotype F prevails in HBV infected patients of hispanic origin in Central America and may carry the precore stop mutant. J Med Virol 1997;51:305–12. [PubMed] [Google Scholar]
- 9.Ding X, Mizokami M, Yao G, et al. Hepatitis B virus genotype distribution among chronic hepatitis B virus carriers in Shanghai, China. Intervirology 2001;44:43–7. [DOI] [PubMed] [Google Scholar]
- 10.Orito E, Ichida T, Sakugawa H, et al. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology 2001;34:590–4. [DOI] [PubMed] [Google Scholar]
- 11.Grandjacques C, Pradat P, Stuyver L, et al. Rapid detection of genotypes and mutations in the pre-core promoter and the pre-core region of hepatitis B virus genome: correlation with viral persistence and disease severity. J Hepatol 2000;33:430–9. [DOI] [PubMed] [Google Scholar]
- 12.Orito E, Mizokami M, Sakugawa H, et al. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology 2001;33:218–23. [DOI] [PubMed] [Google Scholar]
- 13.Kao JH, Chen PJ, Lai MY, et al. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 2000;118:554–9. [DOI] [PubMed] [Google Scholar]
- 14.Lindh M, Hannoun C, Dhillon AP, et al. Core promoter mutations and genotypes in relation to viral replication and liver damage in East Asian hepatitis B virus carriers. J Infect Dis 1999;179:775–82. [DOI] [PubMed] [Google Scholar]
- 15.Kao JH, Wu NH, Chen PJ, et al. Hepatitis B genotypes and the response to interferon therapy. J Hepatol 2000;33:998–1002. [DOI] [PubMed] [Google Scholar]
- 16.Omata M, Ehata T, Yokosuka O, et al. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N Engl J Med 1991;324:1699–704. [DOI] [PubMed] [Google Scholar]
- 17.Ehata T, Omata M, Chuang WL, et al. Mutations in core nucleotide sequence of hepatitis B virus correlate with fulminant and severe hepatitis. J Clin Invest 1993;91:1206–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima M, Shimizu M, Tsuchimochi T, et al. Posttransfusion fulminant hepatitis B associated with precore-defective HBV mutants. Vox Sang 1991;60:34–9. [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa K, Huang JK, Wands JR, et al. Association of hepatitis B viral precore mutations with fulminant hepatitis B in Japan. Virology 1991;185:460–3. [DOI] [PubMed] [Google Scholar]
- 20.Aye TT, Uchida T, Becker SO, et al. Variations of hepatitis B virus precore/core gene sequence in acute and fulminant hepatitis B. Dig Dis Sci 1994;39:1281–7. [DOI] [PubMed] [Google Scholar]
- 21.Chu CM, Yeh CT, Chiu CT, et al. Precore mutant of hepatitis B virus prevails in acute and chronic infections in an area in which hepatitis B is endemic. J Clin Microbiol 1996;34:1815–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terazawa S, Kojima M, Yamanaka T, et al. Hepatitis B virus mutants with precore-region defects in two babies with fulminant hepatitis and their mothers positive for antibody to hepatitis B e antigen. Pediatr Res 1991;29:5–9. [DOI] [PubMed] [Google Scholar]
- 23.Yotsumoto S, Kojima M, Shoji I, et al. Fulminant hepatitis related to transmission of hepatitis B variants with precore mutations between spouses. Hepatology 1992;16:31–5. [DOI] [PubMed] [Google Scholar]
- 24.Sato S, Suzuki K, Akahane Y, et al. Hepatitis B virus strains with mutations in the core promoter in patients with fulminant hepatitis. Ann Intern Med 1995;122:241–8. [DOI] [PubMed] [Google Scholar]
- 25.Laskus T, Persing DH, Nowicki MJ, et al. Nucleotide sequence analysis of the precore region in patients with fulminant hepatitis B in the United States. Gastroenterology 1993;105:1173–8. [DOI] [PubMed] [Google Scholar]
- 26.Liang TJ, Hasegawa K, Munoz SJ, et al. Hepatitis B virus precore mutation and fulminant hepatitis in the United States. A polymerase chain reaction-based assay for the detection of specific mutation. J Clin Invest 1994;93:550–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feray C, Gigou M, Samuel D, et al. Low prevalence of precore mutations in hepatitis B virus DNA in fulminant hepatitis type B in France. J Hepatol 1993;18:119–22. [DOI] [PubMed] [Google Scholar]
- 28.Laskus T, Rakela J, Nowicki MJ, et al. Hepatitis B virus core promoter sequence analysis in fulminant and chronic hepatitis B. Gastroenterology 1995;109:1618–23. [DOI] [PubMed] [Google Scholar]
- 29.Karayiannis P, Alexopoulou A, Hadziyannis S, et al. Fulminant hepatitis associated with hepatitis B virus e antigen- negative infection: importance of host factors. Hepatology 1995;22:1628–34. [PubMed] [Google Scholar]
- 30.Sterneck M, Kalinina T, Gunther S, et al. Functional analysis of HBV genomes from patients with fulminant hepatitis. Hepatology 1998;28:1390–7. [DOI] [PubMed] [Google Scholar]
- 31.Honda A, Yokosuka O, Suzuki K, et al. Detection of mutations in hepatitis B virus enhancer 2/core promoter and X protein regions in patients with fatal hepatitis B virus infection. J Med Virol 2000;62:167–76. [DOI] [PubMed] [Google Scholar]
- 32.Usuda S, Okamoto H, Iwanari H, et al. Serological detection of hepatitis B virus genotypes by ELISA with monoclonal antibodies to type-specific epitopes in the preS2-region product. J Virol Methods 1999;80:97–112. [DOI] [PubMed] [Google Scholar]
- 33.Mizokami M, Nakano T, Orito E, et al. Hepatitis B virus genotype assignment using restriction fragment length polymorphism patterns. FEBS Lett 1999;450:66–71. [DOI] [PubMed] [Google Scholar]
- 34.Yokosuka O, Omata M, Hosoda K, et al. Detection and direct sequencing of hepatitis B virus genome by DNA amplification method. Gastroenterology 1991;100:175–81. [DOI] [PubMed] [Google Scholar]
- 35.Fujie H, Moriya K, Shintani Y, et al. Hepatitis B virus genotypes and hepatocellular carcinoma in Japan. Gastroenterology 2001;120:1564–5. [DOI] [PubMed] [Google Scholar]
- 36.Kozik CA, Vaughn DW, Snitbhan R, et al. Hepatitis B virus infection in Thai children. Trop Med Int Health 2000;5:633–9. [DOI] [PubMed] [Google Scholar]
- 37.Penna A, Chisari FV, Bertoletti A, et al. Cytotoxic T lymphocytes recognize an HLA-A2-restricted epitope within the hepatitis B virus nucleocapsid antigen. J Exp Med 1991;174:1565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertoletti A, Ferrari C, Fiaccadori F, et al. HLA class I-restricted human cytotoxic T cells recognize endogenously synthesized hepatitis B virus nucleocapsid antigen. Proc Natl Acad Sci U S A 1991;88:10445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nayersina R, Fowler P, Guilhot S, et al. HLA A2 restricted cytotoxic T lymphocyte responses to multiple hepatitis B surface antigen epitopes during hepatitis B virus infection. J Immunol 1993;150:4659–71. [PubMed] [Google Scholar]
- 40.Rehermann B, Fowler P, Sidney J, et al. The cytotoxic T lymphocyte response to multiple hepatitis B virus polymerase epitopes during and after acute viral hepatitis. J Exp Med 1995;181:1047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertoletti A, Chisari FV, Penna A, et al. Definition of a minimal optimal cytotoxic T-cell epitope within the hepatitis B virus nucleocapsid protein. J Virol 1993;67:2376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Missale G, Redeker A, Person J, et al. HLA-A31- and HLA-Aw68-restricted cytotoxic T cell responses to a single hepatitis B virus nucleocapsid epitope during acute viral hepatitis. J Exp Med 1993;177:751–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin Y, Shih WK, Berkower I. Human T cell response to the surface antigen of hepatitis B virus (HBsAg). Endosomal and nonendosomal processing pathways are accessible to both endogenous and exogenous antigen. J Exp Med 1988;168:293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li JS, Tong SP, Wen YM, et al. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J Virol 1993;67:5402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi K, Ohta Y, Kanai K, et al. Clinical implications of mutations C-to-T1653 and T-to-C/A/G1753 of hepatitis B virus genotype C genome in chronic liver disease. Arch Virol 1999;144:1299–308. [DOI] [PubMed] [Google Scholar]