Abstract
Background: Hereditary non-polyposis colorectal cancer (HNPCC) (Lynch cancer family syndrome I (LCFS1) and II (LCFS2)) is one of the most common hereditary cancer disorders. HNPCC results from dominantly inherited germline mutations in mismatch repair (MMR) genes, leading to genomic instability and cancer. No predictive physical signs of HNPCC are available to date.
Aims: Increased complexity in tumour associated vascular growth has been reported. Here, we tested the hypothesis that an increased vascular network complexity is a phenotypic marker for LCFS2.
Methods: Fourteen subjects from an LCFS2 kindred (gene carriers, n = 5; non-carriers, n = 9) and 30 controls were examined. Fractal dimension (D) at two scales (D (1–46), and D (1–15), tortuosity (minimum path dimension, Dmin), and relative Lempel-Ziev complexity (L-Z) of the vascular networks from the lower gingival and vestibular oral mucosa were measured.
Results: LCFS2 networks exhibited a significantly increased overall complexity at both larger (D (1–46): 1.82 (0.04) v 1.68 (0.08); p<0.0001) and smaller (D (1–15): 1.51 (0.11) v 1.20 (0.09); p<0.0001) scales, increased destructured randomness (L-Z: 0.77 (0.09) v 0.56 (0.03); p<0.0001), and decreased vessel tortuosity (Dmin: 1.02 (0.03) v 1.07 (0.04); p = 0.0005) compared with control patterns. The vascular networks of LCFS2 gene carriers showed higher complexity at the smaller scale (D (1–15): 1.59 (0.12) v 1.47 (0.07); p = 0.034), and higher destructured randomness (L-Z: 0.85 (0.11) v 0.73 (0.05); p = 0.013) than those of non-carriers.
Conclusions: Increased oral vascular network complexity is a previously unrecognised phenotypic marker for LCFS2, and is related to gene mutation carrier status.
Keywords: colorectal neoplasms, hereditary non-polyposis/diagnosis, blood vessels, population surveillance, colorectal cancer
Hereditary non-polyposis colorectal cancer (HNPCC)1,2 (Lynch cancer family syndrome I (LCFS1)3 and II (LCSF2)4), is a paradigmatic example of a dominantly inherited cancer syndrome. Germline mutations in five different mismatch repair (MMR) genes (MSH2, MSH6, MLH1, MLH3, and PMS2) are linked to HNPCC.1–4 Genomic deletions in MLH1 and MSH2 are believed to account for approximately 70–90% of causal mutations in HNPCC families,1,5 although over 400 different MMR gene mutations and approximately 100 intragenic polymorphic variations have been reported to date (International HNPCC mutation database, http://www.nfdht.nl).6 HNPCC gene carriers have a high risk (60–85%) of developing colorectal cancer (CRC), and other cancer types, primarily endometrial cancer (30–50%), at an early age.1–4,7,8 With the exception of the cutaneous signs of cancer risk relevant to the Muir-Torre syndrome in a subset of LCFS2 patients,9,10 no distinguishing phenotypic stigmata of cancer susceptibility in HNPCC are available to date.1,11 In the present study, we tested the hypothesis that an increased vascular network complexity is a phenotypic marker for LCFS2.
METHODS
Subjects
Fourteen surviving members from a large Southern Italy LCFS2 kindred (eight males, six females; aged 40.2 (11.7) years (range 26–71)) were examined. Detailed characteristic features of this extensively affected family have been reported previously, including early age of onset of tumours, full details of the pedigree, clinical parameters, increased frequency of CRCs (mainly with proximal location), as well as high occurrence of gastric, endometrial, and multiple primary malignancies.12–14 LCFS2 gene mutation carriers (n = 5) were differentiated from non-carriers (n = 9) using a previously established linkage analysis.15,16 Two of five gene carrier patients had a history of proven cancer at the time of examination. Given the specific end point of the present study, family members without genetic testing available were excluded (four males, one female; aged 16.2 (5.7) years (range 10–18)). Thirty genetically unrelated age and sex matched healthy controls (17 males, 13 females; aged 40.0 (10.5) years (range 26–71)) were also enrolled. None of the examined subjects had known inherited connective tissue or vascular diseases. No cases of CRC were present in the family history or pedigrees of controls. Approval by the research ethics committee and informed consent for examination and photographic documentation were obtained.
Oral vascular network analysis
The lower gingival and vestibular oral mucosa was chosen as the study area due to its high vasculature pattern visibility and easy accessibility. A 704 mm2 size (32×22 mm) area of the lower gingival and vestibular oral mucosa was photographed for each subject (1:1 ratio, orthogonal projection). All photographs were taken by a single operator using a Yashica Dental Eye photocamera with an automated on-axis flashbulb and a 55 mm f 1:4 Yashica lens (Yashica-Kyocera Co., Kyoto, Japan). Kodak Elite Chrome 100 ISO/21 DIN films (Kodak-Eastman Kodak Co., Rochester, New York, USA) were used and developed according to the standard E-6 procedure. Images were digitised using a Canon Canoscan FS2710 (Canon Inc., Tokyo, Japan) scanner (colour resolution: 680 dpi, 6.67× magnification) with a Windows ’98 operating system (Microsoft Co., Redmond, Washington, USA).
The obtained images were converted into binary skeletonised form for geometric pattern analysis. Manual outline of the two dimensional trajectories of the vascular network was performed using Adobe Photoshop (Adobe Systems Inc., San Jose, California, USA) on a Sony 19′′ Trinitron Multiscan G420 screen (16 μm/pixel resolution) (Sony Co., Tokyo, Japan) by two operators who were unaware of the subject’s category. The two dimensional lattices were analysed using the Image Pro-Plus version 1.3 image analysis software (Image Pro-Plus-Media; Cybernetics Inc., Silver Spring, Maryland, USA). Non-readable areas were <5% of the total area. After enlargement to 1.4× magnification, images were processed to threshold the vessel network without background interference and the networks were subsequently converted into an outline of single pixels.
The oral vascular networks were characterised by analysing their complexity (fractal dimension(D), at two scales), tortuosity (minimum path fractal dimension (Dmin)), and randomness (relative Lempel-Ziv complexity (L-Z)) of the vascular loops. Fractal dimensions of the two dimensional skeletonised images were measured with the box counting algorithm, using the relation N(L)∼L−D, where L is the box size and N(L) is the number of squares.17 As natural fractals show upper and lower limits, the local fractal dimension (D) was determined for two regions of box lengths, <740 μm (pixels 1–46, D(1–46)) and <140 μm (pixels 1–15, D(1–15)), respectively. The measuring procedure was calibrated against shapes of known fractal dimension with an inaccuracy of ±2%. The fractal dimension of the minimum path, Dmin, was computed for each vascular cluster from the power law lc = rDmin, where Dmin is the exponent that governs the dependence of the minimum path length between two points (lc) on the Pythagorean distance (r) between them in a fractal random material.18 After enlarging the image to 2.4× magnification, thinning to 1 pixel, and discarding all areas with a diameter <3 pixels, the half perimeter (xi) and the maximum diameter (yi) of either the vessel loops or vessel free areas in the single pixel two dimensional lattice were measured using an automated procedure (Image Pro-Plus version 1.3 image analysis software; examined areas for each sample: 500–1000). The slope of the log/log plot xi/yi represented Dmin.
The method was validated with the original one by Herrmann and Stanley,18 with a maximum shift of ±3%. To determine the algorithmic complexity of the vascular patterns, relative Lempel-Ziv (L-Z)19 values were calculated according to the Kaspar and Schuster algorithm20 using the Chaos Data Analyzer version 2.1 (1995) software package (CDA: Pro; The Academic Software Library, North Carolina State University, Raleigh, North Carolina, USA). Vascular network lattices from a 251×251 pixels window of the original image were transformed into 16 732 points containing one dimensional vectors, and each datum point was converted into a single binary digit according to whether the design is touched ( = 1) or not ( = 0). Relative L-Z values may range from near 0 for a deterministic equation to approximately 1.0 for totally destructured random phenomena. Using the described procedure, the relative L-Z values for the sinus function and white noise were 0.0044 and 1.047, respectively.
The vascular network analysis was reproducible, with mean intra and interobserver coefficients of variation of ⩽5.0% and ⩽10%, respectively. Differences among group means were analysed by the t test. Differences between multiple groups were evaluated by one way ANOVA, and post-hoc pairwise differences were tested using the Student-Newman-Keuls statistics. A two sided p<0.05 was considered to indicate statistical significance, and the Bonferroni corrected significance levels were used for multiple t tests. The MedCalc version 7.0 statistical software package (MedCalc Software, Mariakerke, Belgium) was used.
RESULTS
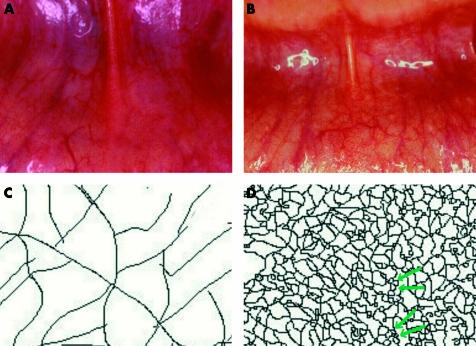
Oral vascular networks in LCFS2 patients showed geometric pattern abnormalities (that is, increased vessel density and presence of multiple vascular loops features) compared with the typical dichotomic branching patterns of control networks (fig 1 ▶). LCFS2 associated vascular networks exhibited a significantly increased overall complexity (table 1 ▶) at both larger (D(1–46) difference: 0.14 (95% confidence interval (CI) 0.09–0.19); t = 6.39, df = 42,p<0.0001)) and smaller (D(1–15) difference: 0.31 (95% CI 0.24–0.37); t = 9.96, df = 42, p<0.0001)) scales, increased destructured randomness (L-Z difference: 0.21 (95% CI 0.17–0.24); t = 11.7, df = 42, p<0.0001), and decreased vessel tortuosity (Dmin difference: 0.047 (95% CI 0.022–0.072); t = 3.75, df = 42, p = 0.0005) compared with control vascular networks. The vascular networks of the LCFS2 gene mutation carriers showed higher complexity at the smaller scale (D(1–15) difference: 0.12 (95% CI 0.011–0.23); t = 2.39, df = 12, p = 0.034), and a more marked destructured randomness (L-Z difference: 0.12 (95% CI 0.03–0.21); t = 2.9, df = 12, p = 0.013) than those of the non-carrier LCFS2 individuals.
Figure 1.
Lynch cancer family syndrome II (LCFS2) associated vascular network abnormalities. (A) Control oral vascular network; (B) abnormal oral vascular density of the vascular network in an LCFS2 gene carrier patient, with multiple loops features; (C) skeletonised control vascular network, with typical dichotomic branching pattern (D (1–46) = 1.57; D (1–15) = 1.07; Dmin = 1.08; L-Z = 0.49); (D) skeletonised vascular network pattern of an LCFS2 gene carrier patients showing increased density of trajectories and multiple loops aspects (greenarrows) (D (1–46) = 1.79; D (1–15) = 1.68; Dmin = 0.99; L-Z = 0.92). Note (C) and (D) represent ∼60× enlarged details of (A) and (B), respectively. D, fractal dimension; Dmin, minimum path fractal dimension; L-Z, relative Lempel-Ziv complexity.
Table 1.
Lynch cancer family syndrome II (LCFS2) associated vascular networks (LCFS2−) display significantly increased complexity, together with increased destructured randomness, and decreased vessel tortuosity compared with control (LCFS2, group C) networks. LCFS2 gene mutation carriers (group A) show higher complexity at smaller scales and a more marked destructured randomness than non-carriers (group B)
| Vascular network variable | LCFS2+ | LCFS2− controls (group C, n = 30) | F2,41 values | p Value | Significant pairwise differences | |
| Gene carriers (group A, n = 5) | Non-carriers (group B, n = 9) | |||||
| D (1–46) | 1.84 (0.07) | 1.82 (0.02) | 1.68 (0.08) | 17.97 | 2.5×10−5 | A v C; B v C |
| D (1–15) | 1.59 (0.12) | 1.47 (0.07) | 1.20 (0.09) | 71.83 | <10−6 | A v B; A v C; B v C |
| Dmin | 0.99 (0.02) | 1.03 (0.04) | 1.07 (0.04) | 9.07 | 0.00054 | A v C; B v C |
| L-Z | 0.85 (0.106) | 0.73 (0.05) | 0.56 (0.03) | 20.78 | 6×10−6 | A v B; A v C; B v C |
Data are means (SD).
p values refer to one way ANOVA results.
DISCUSSION
HNPCC accounts for approximately 3–6% of all colorectal cancers.21,22 Early identification of HNPCC gene carriers is essential as surveillance has been reported to reduce CRC incidence and overall mortality.23,24 However, in the absence of predictive physical signs, no universal consensus exists on the most cost effective strategy for detecting potential gene carriers,25 with population screening by mutation analysis not being a feasible option.26 The main findings of the present study indicate the presence of a previously unrecognised abnormal vascular network complexity in the oral mucosa of LCFS2 patients, which is related to gene mutation carrier status. This observation may provide a useful phenotypic linkage of LCFS2 in both the presymptomatic detection of DNA genetic testing candidates in HNPCC families and CRC case selection for mutational analysis. The mechanisms underlying the increased vascular complexity observed in the oral cavity of the LCFS2 subjects remain unclear. However, it appears unlikely that an MMR defect may directly affect normal vasculogenesis. On the other hand, a close relationship between extracellular matrix and blood vessel geometry is known,27,28 and an abnormal complexity of tumour vascular networks has been previously related to changes in the local properties of the extracellular matrix.29 As a consequence, the presence of a vascular network geometry abnormality in LCFS2 patients generates the hypothesis of the coexistence of a hitherto unrecognised subclinical systemic extracellular matrix abnormality in this hereditary cancer disorder.
Acknowledgments
We thank Professor Alessandro Barducci (Faculty of Engineering, University of Siena, and Institute for Applied Physics IFAC-CNR, National Research Council of Italy, Florence, Italy), Mr Roberto Casini (Orintex srl, Prato, Italy), and Professor Stefano Marmi (Scuola Normale Superiore, Pisa, Italy) for helpful discussions, inspiration, and criticism; Ombretta Bugiani and Roberto Faleri (Central Library, University Medical School, Siena, Italy) for online research assistance; and especially all the HNPCC kindred who kindly accepted to participate in the study, and to whom this paper is dedicated.
Abbreviations
HNPCC, hereditary non-polyposis colorectal cancer
LCFS1, Lynch cancer family syndrome I
LCFS2, Lynch cancer family syndrome II
MMR, mismatch repair
CRC, colorectal carcinoma
D, fractal dimension
Dmin, minimum path fractal dimension
L-Z, relative Lempel-Ziv complexity
REFERENCES
- 1.Lynch HT, Lynch JF. Hereditary cancer: family history, diagnosis, molecular genetics, ecogenetics, and management strategies. Biochimie 2002;84:3–17. [DOI] [PubMed] [Google Scholar]
- 2.Online Mendelian Inheritance in Man, OMIM (TM). Johns Hopkins University: Baltimore, MD MIM Number: No 120435 (date last edited: 01/08/2003). World Wide Web URL http://www.ncbi.nlm.nih.gov/omim/.
- 3.Online Mendelian Inheritance in Man, OMIM (TM). Johns Hopkins University: Baltimore, MD MIM Number: No 114500. (Date last edited: 9/30/2002). World Wide Web URL http://www.ncbi.nlm.nih.gov/omim/.
- 4.Online Mendelian Inheritance in Man, OMIM (TM). Johns Hopkins University: Baltimore, MD MIM Number: No 114400. (Date last edited: 10/20/2002). World Wide Web URL http://www.ncbi.nlm.nih.gov/omim/.
- 5.Peltomäki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 1997;113:1146–58. [DOI] [PubMed] [Google Scholar]
- 6.Kariola R, Otway R, Lonnqvist KE, et al. Two mismatch repair gene mutations found in a colon cancer patient—which one is pathogenic? Hum Genet 2003;112:105–9. [DOI] [PubMed] [Google Scholar]
- 7.Lynch HT, Lynch JF. The Lynch syndrome: melding natural history and molecular genetics to genetic counseling and cancer control. Cancer Control 1996;3:13–19. [DOI] [PubMed] [Google Scholar]
- 8.Watson P, Lynch HT. Extracolonic cancer in hereditary nonpolyposis colorectal cancer. Cancer 1993;71:677–85. [DOI] [PubMed] [Google Scholar]
- 9.Lynch HT, Lynch PM, Pester J, et al. The cancer family syndrome: rare cutaneous phenotypic linkage of Torre’s syndrome. Arch Intern Med 1981;141:607–11. [PubMed] [Google Scholar]
- 10.Lynch HT, Fusaro RM, Roberts L, et al. Muir-Torre syndrome in several members of a family with a variant of the cancer family syndrome. Br J Dermatol 1985;113:295–301. [DOI] [PubMed] [Google Scholar]
- 11.Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet 1999;36:801–18. [PMC free article] [PubMed] [Google Scholar]
- 12.Cristofaro G, Lynch HT, Caruso ML, et al. New phenotypic aspects in a family with Lynch syndrome II. Cancer 1987;60:51–8. [DOI] [PubMed] [Google Scholar]
- 13.Guanti G, Susca F, Cristofaro G, et al. Cancer family syndrome: cytogenetic investigations, in vitro tetraploidy, and biomarker studies in a large family. J Med Genet 1990;27:441–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch HT, Smyrk TC, Lynch PM, et al. Adenocarcinoma of the small bowel in Lynch syndrome II. Cancer 1989;64:2178–83. [DOI] [PubMed] [Google Scholar]
- 15.Lindblom A, Tannergard P, Werelius B, et al. Genetic mapping of a second locus predisposing to hereditary non-polyposis colon cancer. Nat Genet 1993;5:279–82. [DOI] [PubMed] [Google Scholar]
- 16.Piepoli A, Santoro R, Cristofaro G, et al. Linkage analysis identifies gene carriers among members of families with hereditary nonpolyposis colorectal cancer. Gastroenterology 1996;110:1404–9. [DOI] [PubMed] [Google Scholar]
- 17.Peitgen H- O, Jurgens H, Saupe D. Chaos and fractals. New frontiers of science. NY: Springer-Verlag, 1992.
- 18.Herrmann HJ, Stanley HE. The fractal dimension of the minimum path in two- and three-dimensional percolation. J Phys A Math Gen 1988;21:L829–33. [Google Scholar]
- 19.Ziv J, Lempel A. A universal algorithm for sequencial data compression. IEEE Trans Inf Technol Biomed 1997;23:337–43. [Google Scholar]
- 20.Kaspar F, Schuster HG. An easily calculable measure for the complexity of spatio-temporal patterns. Phys Rev A 1987;36:842–8. [DOI] [PubMed] [Google Scholar]
- 21.Lynch HT, Smyrk TC, Watson P, et al. Genetics, natural history, tumor spectrum, and pathology of hereditary nonpolyposis colorectal cancer: an updated review. Gastroenterology 1993;104:1535–49. [DOI] [PubMed] [Google Scholar]
- 22.Houlston RS, Collins A, Slack J, et al. Dominant genes for colorectal cancer are not rare. Ann Hum Genet 1992;56:99–103. [DOI] [PubMed] [Google Scholar]
- 23.Jarvinen HJ, Mecklin JP, Sistonen P. Screening reduces colorectal cancer rate in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 1995;108:1405–11. [DOI] [PubMed] [Google Scholar]
- 24.Jarvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 2000;118:829–34. [DOI] [PubMed] [Google Scholar]
- 25.Reyes CM, Allen BA, Terdiman JP, et al. Comparison of selection strategies for genetic testing of patients with hereditary nonpolyposis colorectal carcinoma: effectiveness and cost-effectiveness. Cancer 2002;95:1848–56. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell RJ, Farrington SM, Dunlop MG, et al. Mismatch repair genes hMLH1 and hMSH2 and colorectal cancer: a HuGE review. Am J Epidemiol 2002;156:885–902. [DOI] [PubMed] [Google Scholar]
- 27.Suwa N. Supracellular structural principle and geometry of blood vessels. Virchows Arch A 1981;390:161–79. [DOI] [PubMed] [Google Scholar]
- 28.Ingber DE, Folkman J. Mechanochemical switching between growth and differentiation during fibroblast growth factor-stimulated angiogenesis in vitro: role of extracellular matrix. J Cell Biol 1984;109:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain RK. Transport of molecules in the tumor interstitium: a review. Cancer Res 1987;47:3039–51. [PubMed] [Google Scholar]