Abstract
Saccharomyces cerevisiae mother cells undergo an aging program that includes morphologic changes, sterility, redistribution of the Sir transcriptional silencing complex from HM loci and telomeres to the nucleolus, alterations in nucleolar architecture, and accumulation of extrachromosomal ribosomal DNA circles (ERCs). We report here that cells starved for nutrients during prolonged periods in stationary phase show a decrease in generational lifespan when they reenter the cell cycle. This shortened lifespan is not transmitted to progeny cells, indicating that it is not due to irreversible genetic damage. The decrease in the lifespan is accompanied by all of the changes of accelerated aging with the notable exception that ERC accumulation is not augmented compared with generation-matched, nonstarved cells. These results suggest a number of models, including one in which starvation reveals a component of aging that works in parallel with the accumulation of ERCs. Stationary-phase yeast cells may be a useful system for identifying factors that affect aging in other nondividing eukaryotic cells.
Keywords: recombination
Mother cells in the budding yeast Saccharomyces cerevisiae undergo a finite number of divisions before cessation of cell growth. This aging is accompanied by morphologic changes, including an increase in cell size, the onset of sterility, enlargement and fragmentation of the nucleolus, and redistribution of the Sir3 and Sir4 proteins from telomeres and HM loci to the nucleolus (1). Sterility in aging cells is caused by expression of the normally silent mating (HM) loci (2) and is a reliable marker of the aging process (1).
Genetic alterations have been identified that increase or decrease the number of divisions that a mother cell undergoes (3–5). For example, a gain-of-function mutation in Sir4p, a component of a transcriptional silencing complex, extends lifespan by 50% (6). In contrast, deletion of SGS1, encoding a RecQ-like DNA helicase concentrated in the nucleolus, diminishes lifespan by 60% and produces other manifestations of accelerated aging (1). Interestingly, SGS1 is the yeast homolog of the human WRN gene. Defects in WRN cause Werner’s Syndrome (7), a disease that exhibits many signs of premature aging (8).
The nucleolar changes in aging yeast cells are associated with accumulation of extrachromosomal ribosomal DNA (rDNA) circles (ERCs) generated by homologous recombination of tandemly arrayed copies of rDNA (9, 10). ERCs accumulate because of their replication at each cell cycle and preferential segregation to mother cells at each division (11, 12). Creating an ERC ectopically by using a site-specific recombinase can shorten the lifespan, indicating that ERCs are a cause of aging. ERC accumulation may arrest growth by sequestering critical proteins involved in transcription and/or DNA replication (12).
Yeast cells deprived of nutrients can survive for prolonged periods of time in stationary phase (13). This robust survival requires the activity of copper/zinc superoxide dismutase (14), illustrating the importance of detoxification of oxygen radicals during this period. Interestingly, the survival defect in sod1 mutants can be reversed by expression of human Bcl-2 (15), indicating a possible conserved mechanism of cell survival in many eukaryotes. It has been suggested that the survival of cells in stationary phase yeast cultures may be a model for aging in mammals, particularly for tissues composed of nondividing cell populations (15).
Here we set out to examine whether yeast cells held in stationary phase exhibit any of the phenotypes found in replicatively aging mother cells. Except for an aberrant nucleolar morphology in a small fraction of cells, stationary-phase cells resembled normal, young mother cells. However, when nutrients were returned to allow resumption of cell division, survivors of stationary phase displayed a much shorter replicative lifespan than nonstarved controls. Cells with a shortened lifespan exhibited most of the manifestations of accelerated aging, including sterility. The surprising exception was that their levels of ERC accumulation did not differ from age-matched, nonstarved controls. These findings raise the possibility that novel pathways of aging, in addition to accumulation of ERCs, may operate in mother yeast cells.
MATERIALS AND METHODS
Strains and Growth Conditions.
Strain YB332 (MATa ura3–52 his3Δ200 ade2–101 lys2–80 leu2–3, 112) is described in ref. 16. Cells were incubated in YPD (1% yeast extract/2% peptone/2% dextrose) at 24°C. Entry into stationary phase was monitored by assaying aliquots of the cultures at OD600.
Immunohistochemistry.
Cells (5 × 107) were washed twice in PBS and fixed for 20 min in 3.7% formaldehyde. Fixed cells were washed twice in PBS and resuspended in 1 ml of 0.1 M EDTA (pH 8)/10 mM DTT and incubated for 10 min at 30°C with gentle shaking. After two washes in PBS, cells were resuspended in 1 ml of YPD/20% (wt/vol) sorbitol containing 6 mg of Zymolyase (ICN, 100 units/mg) and incubated at 30°C for 15–60 min. Spheroplasts were washed once in YPD/sorbitol and resuspended in 200 μl of YPD, spotted onto Vectabond-coated glass slides (Vector Laboratories), air-dried at room temperature for 10–15 min, and then treated for 6 min with methanol (−20°C) followed by acetone (1 min; −20°C). The fixed spheroplasts were then preincubated at room temperature for 1 h in blocking buffer (1% BSA/0.1% Triton X-100, prepared in PBS). Multilabel immunohistochemistry was performed by using the following sequence of reagent additions: (i) rabbit anti-Sir3p (final dilution 1:100 in blocking buffer; 12-h incubation at 37°C); (ii) three washes in blocking buffer (15 min each; all at room temperature); (iii) FITC-tagged goat anti-rabbit IgG (1 h at 37°C); (iv) three more washes as in ii; (v) mouse mAbs to yeast fibrillarin (Nop1p; 1:100; 12-h incubation at 37°C); (vi) three more washes in blocking buffer; (vii) indocarbocyanine (Cy3)-tagged goat anti-mouse IgG (1 h at 37°C); (viii) three washes in blocking buffer; and (ix) 4′,6′-diamidino-2-phenylindole (DAPI, Sigma; 2 mg/ml in PBS; 90-sec incubation at room temperature to stain nuclear DNA).
Pulse-Field Gel Electrophoresis.
Cultures were incubated in YPD at 24°C. Cells (2 × 107) were harvested at various times during logarithmic, postdiauxic, and stationary phases, and spheroplasts were prepared and then lysed while embedded in agarose plugs. The total cellular lysate, containing chromosomal DNA, was subjected to pulse-field gel electrophoresis according to procedures described (17).
Preparation of Old Cell Populations by Sorting.
Biotin labeling and sorting of logarithmic-phase cultures. The sorting procedure exploits the fact that biotinylated surface proteins are retained in mothers but are not found in their daughters, because the daughters’ cell walls are newly synthesized. Cells that had, on average, undergone seven divisions were obtained as follows. Cells (2 × 108) from a mid-logarithmic-phase culture (OD600 = 0.5–1.0) were washed once in PBS, resuspended in 1 ml of PBS containing 7 mg of sulfosuccinimidyl-6′-(biotinamido)-6-hexanamido hexanoate (Pierce), and incubated for 15 min at 24°C with occasional shaking. Biotin that had not reacted with cell-surface proteins was removed by four washes in PBS (1 ml each). Cells were resuspended in 1 liter of YPD (2.5% dextrose), incubated at 24°C to OD600 = 0.8, harvested by centrifuging (8 min at 4,400 × g), and resuspended in 35 ml of PBS containing 70 mg/ml streptavidin-magnetic beads (PerSeptive Biosystems). The mixture was then incubated for 2 h at 4°C with brief swirling every 15 min. Biotin-labeled cells with bound streptavidin- magnetic beads were recovered with a magnetic sorter (PerSeptive Biosystems, Framingham, MA) and washed with ice-cold YPD. The cycle of sorting, followed by washing, was performed a total of eight times. An aliquot of the sorted, washed cells was stained with fluorescent brightener 28 (Sigma), and the number of bud scars per cell was defined with a fluorescence microscope (n = 50–75 cells analyzed per preparation). Generation 1 cells were obtained from the population that was left behind when the biotinylated cells were magnetically sorted away.
To isolate cells that had undergone an average of 12 divisions, the population obtained from the sort described above was resuspended in 1 liter of YPD (2.5% dextrose) and incubated for an additional 12–13 h at 24°C. At the end of this period, cell sorting was repeated. Average bud scar counts were defined as above.
Labeling and sorting of cells passaged through stationary phase.
The same procedure was used to sort cells that had been in stationary phase for 3 weeks and then placed in fresh YPD. The only difference was that number of cells that were biotinylated was 10-fold greater.
Sterility Assays.
At various generations, individual mothers, from cohorts of 70–80 cells, were moved from one area of a YPD/agar plate and placed next to a 0.5-cm Whatman filter paper disk soaked in 0.25 mM α-factor (Sigma). The response to pheromone (shmooing and division) was scored 4–6 h later. Cells were then moved to another region of the plate and allowed to complete their lifespan. All assays were performed at 24°C.
Assays for ERC Accumulation.
Total cellular DNA was prepared from stationary phase and from sorted cells (12) and fractionated by electrophoresis through 0.7% agarose in TAE buffer (40 mM Tris⋅acetate/1 mM EDTA, pH 7.5) (1 V/cm gel length with buffer recirculation for 30 h). After transfer to GeneScreen Plus (NEN), blots were probed with a 32P-labeled, 2.8-kilobase EcoRI fragment containing rDNA sequences derived from pNL47 (12). After hybridization (12 h at 65°C in 1% SDS/1M NaCl/10% dextran sulfate/500 μg/ml salmon sperm DNA), blots were washed, and ERC levels defined with storage PhosphorImaging system (Molecular Dynamics).
RESULTS
Stationary Phase Cells Do Not Exhibit Aging Phenotypes.
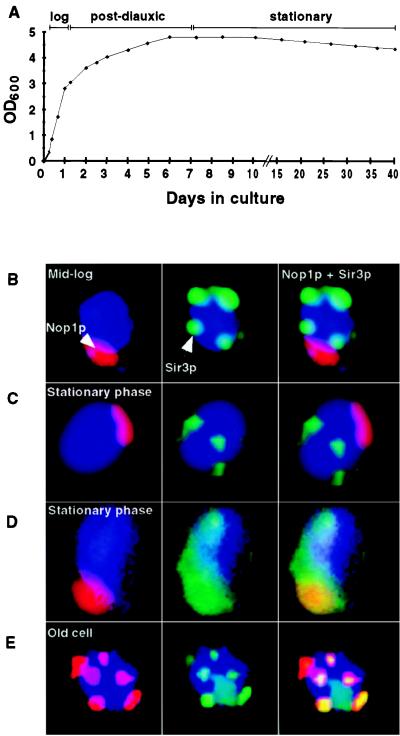
We examined whether any of the phenotypes of replicative aging in nonstarved mother cells were evident in cells held for prolonged periods in stationary phase. Typically, a wild-type strain reaches stationary phase after a 7-day incubation at 24°C in standard rich medium (Fig. 1A). The size and integrity of the nucleolus and the intracellular location of Sir3p were examined during logarithmic phase, during the shift from logarithmic to stationary phase (postdiauxic transition), and at various times during stationary phase. Analyses were performed by using a strain with a standard genetic background (S288C; ref. 16). At each time point, 5 × 107 cells in duplicate aliquots were stained with previously characterized antibodies to fibrillarin (Nop1p), a nucleolar marker, and Sir3p (1). Eleven different time points were surveyed (spanning 1–40 days in YPD).
Figure 1.
Stationary-phase cells do not resemble replicatively aging mother cells. (A) Growth curve in YPD at 24°C. (B) Multilabel immunohistochemical study of cells harvested at mid-logarithmic phase. Nop1p (fibrillarin) appears red and marks the nucleolus. Sir3p is stained green and is located outside the nucleolus and is telomeric. DNA is stained blue with DAPI. (C) Typical cells (80–85% of the population) present in 3-week-old stationary-phase cultures. Sir3p is present at telomeres. The nucleolus is crescent shaped and intact. (D) Representative of a subpopulation of cells present in a 3-week-old stationary-phase culture. Sir3p is redistributed throughout the nucleus, including the nucleolus (yellow). Staining patterns shown in B–D were scored in several hundred cells per aliquot per time point per experiment (n = 2 independent experiments). (E) Old cell recovered from a nonstarved culture by sorting (see Materials and Methods). The average bud scar count of the preparation was 15.5 ± 3.3. There is nucleolar fragmentation and redistribution of Sir3p to these fragments.
Fig. 1B shows the pattern observed in mid-logarithmic phase or young cells. Sir3p is located at discrete telomeric sites and is absent from the intact nucleolus. This pattern persisted through the postdiauxic phase and as cells entered stationary phase, although during the transitional postdiauxic period, nucleolar size was generally reduced (1). Fig. 1C shows typical cells after 21 days in stationary phase. In 80–85% of these cells, nucleoli remained intact and Sir3p was telomeric, i.e., a staining pattern typical of wild-type young cells.
In a small fraction of stationary-phase cells (15–20%), Sir3p was lost from telomeres and redistributed to an enlarged but intact nucleolus (Fig. 1D). The fractional representation of this subpopulation did not increase over a 3-week period in stationary phase, even though proliferative potential was reduced by ≈50% during this interval [as judged by the ability of cells to form colonies when plated on rich media (YPD/agar); data not shown]. Sir3p redistribution and nucleolar enlargement in this subpopulation of stationary-phase cells was superficially reminiscent of replicative aging with two notable exceptions: in replicative aging, there is nucleolar fragmentation (Fig. 1E), and the fraction of the population exhibiting Sir complex redistribution and nucleolar changes increases with successive generations (1).
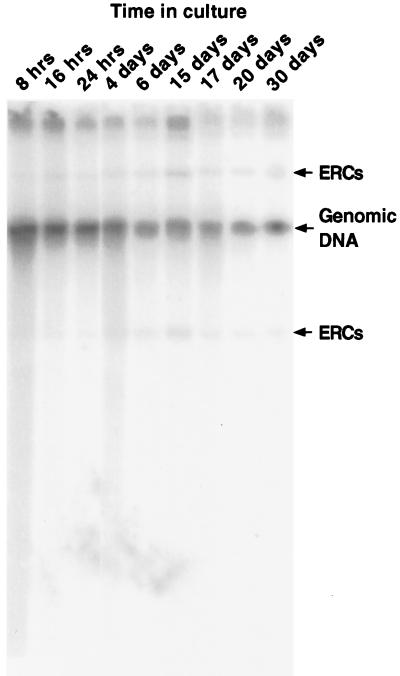
Because dividing cells age on rich media, nucleolar enlargement and redistribution of the Sir complex are thought to be mediated by ERC accumulation (12). Because of the subpopulation of stationary-phase cells noted above, we examined ERC levels as a function of time spent in stationary phase. ERC levels did not change appreciably from mid-logarithmic phase through at least 3 weeks in stationary phase (Fig. 2). Thus, the redistribution of Sir3p and the presence of abnormal nucleoli in this minority population of stationary-phase cells cannot be attributed to ERC proliferation. Furthermore, this population of cells did not produce buds when reexposed to nutrients (data not shown), indicating that they were unable to reenter the cell cycle.
Figure 2.
ERCs do not accumulate with increasing time spent in stationary phase. Total cellular DNA was prepared from cultures harvested at various times after incubation at 24°C in YPD, and probed with [32P]rDNA. The arrows indicate the positions of migration of ERCs and genomic DNA.
In summary, the vast majority of stationary-phase cells do not exhibit any of the manifestations of aging: their nucleolar morphology, Sir3p localization, and ERC levels are indistinguishable from young logarithmic-phase cells.
Passage Through Stationary Phase Reduces Mean Lifespan.
We next determined whether cells that survive passage through stationary phase and resume mitotic growth have a lifespan equivalent to cells that have never experienced starvation. The mean lifespan of nonstarved wild-type cells is 27.5 generations (data not shown). Most cells entering stationary phase are young: 50% of cells will have never divided, 25% are mothers that have divided once, 12.5% have divided twice, 6.25% have divided three times, etc.
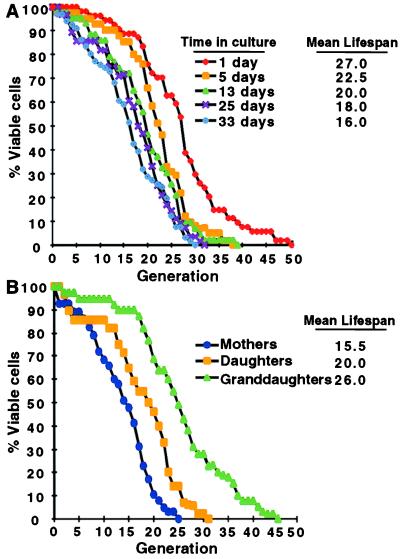
Cells were exposed to fresh YPD medium at various times before and during stationary phase (Fig. 3A). At each time point surveyed, lifespan analysis was begun on a cohort of randomly selected cells immediately after plating on YPD/agar. Cells that did not divide were discarded (up to 50% for cells that had spent the longest period in stationary phase). After each cell division, the smaller daughters were removed from their mothers by micromanipulation, and the total number of divisions that each mother underwent was scored. Mean lifespan was defined as the generation where 50% of mothers in the cohort were still dividing.
Figure 3.
Passage through stationary phase results in a shortened lifespan when cells are reexposed to nutrients. (A) Lifespans of cells recovered from cultures after the indicated incubation times in YPD (n = 37–51 mothers scored per group). The mean lifespans for each group (defined as the generation where 50% of mothers are still dividing) are noted. (B) Mothers refer to viable cells recovered after 3 weeks in stationary phase. Daughters and granddaughters were obtained from a cohort of 44 mothers and 39 daughters, respectively. The difference in mean lifespans between mothers, daughters, and granddaughters is statistically significant as judged by the nonparametric Wilcoxon signed rank test.
Increasing time spent in stationary phase led to a progressive shortening of the subsequent lifespan of surviving cells: dividing cells harvested from a mid-logarithmic phase culture had a mean lifespan of 27 generations, whereas cells recovered after 1 and 3 weeks in stationary phase had mean lifespans of 20 and 16 generations, respectively (Fig. 3A). There is a limit to the reduction in lifespan produced by the experience of stationary phase. Cells harvested after 5 weeks had a lifespan equivalent to those harvested after 3 weeks (data not shown).
The shortened lifespan of stationary-phase cells could result from cellular damage incurred in stationary phase. If damage were in the form of DNA mutations, the shortened lifespan should be heritable. To test this possibility, mothers were harvested after a 3-week period in stationary phase, and their lifespans were compared with the lifespans of their initial offspring (daughters) and, in turn, the daughters’ daughters (granddaughters). A partial restoration of lifespan returned in the daughters, and a full restoration was observed in the granddaughters (Fig. 3B).
Two other observations indicate that stationary phase is not associated with extensive double-stranded DNA damage. The first comes from an analysis of the stationary-phase survival of isogenic wild-type, rad51Δ, rad52Δ, and rad57Δ strains. The products of these three genes are normally involved in double-stranded break repair via two principal pathways of homologous recombination: RAD51, RAD52, and RAD57 and are required for strand invasion; RAD52 is also required for single-stranded annealing (reviewed in ref. 18). Removal of Rad51p, Rad52p, or Rad57p has no appreciable affect on the percentage of cells that reentered the cell cycle after up to 5 weeks of stationary-phase existence (data not shown). Second, pulse-field gels did not show any alteration in the integrity of chromosomes from stationary-phase cells (data not shown).
Phenotypes of Aging in Old Mother Cells Derived from Cells That Experienced Stationary Phase.
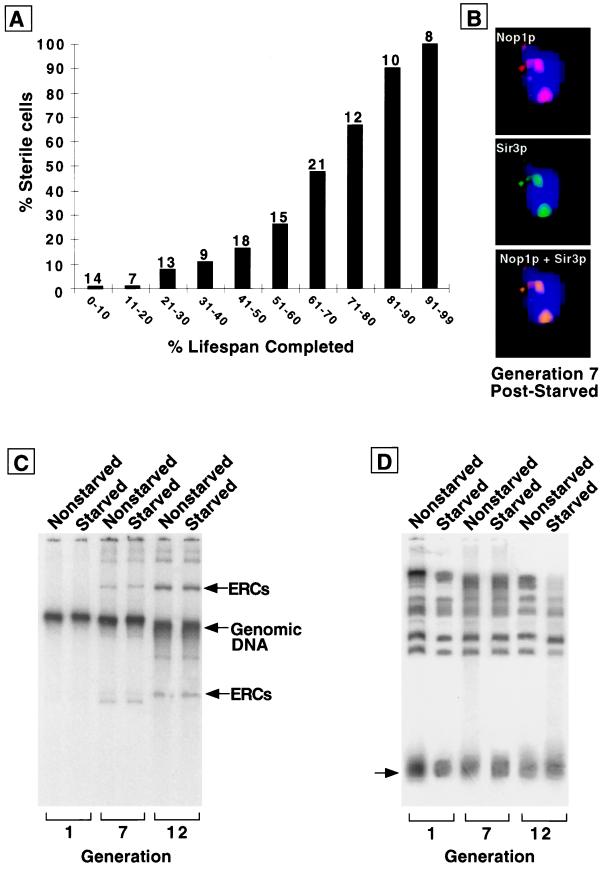
To determine whether the reduced lifespan resulting from time spent in stationary phase was a replica of accelerated aging, we examined whether these cells exhibit the known manifestations of aging. Cells that have a shortened lifespan because of lack of fitness die early and do not become sterile. In contrast, progressive sterility accompanies normal and accelerated aging (1). Acquisition of sterility can be assessed by exposing individual mothers to mating pheromone at various stages of their lifespan. Mating-competent mothers stop dividing and assume a characteristic morphologic state (shmooing), whereas sterile cells continue to divide. Cells that reentered the cell cycle after 3 weeks in stationary phase exhibited progressive increases in sterility during their shortened lifespan (Fig. 4A). The pattern of sterility observed in this population of survivors of stationary phase starvation resembles the pattern of sterility found in aging nonstarved wild-type cells and prematurely aging nonstarved sgs1 mutants (1).
Figure 4.
The reduced lifespan of cells with prior stationary phase experience is accompanied by sterility and nucleolar changes but not more rapid ERC accumulation. (A) Sterility assay performed on cells maintained for 3 weeks in stationary phase before plating on YPD. The numbers above each bar represent the number of cells whose response to pheromone was scored at this point of their lifespan. (B) Multilabel immunohistochemical study of a typical cell from a population analyzed in A. Cells with an average generation of seven were obtained by sorting (average bud scar count = 7.2 ± 3.1). Sir3p colocalizes to nucleolar fragments. (C) ERC levels measured in nonstarved and starved cells. Starved cells are cells that have experienced 3 weeks in stationary phase before reexposure to fresh YPD. Nonstarved cells are cells obtained from logarithmic-phase cultures that have never been through stationary phase. Average bud scar count in the sorted starved populations labeled generations 1, 7, and 12 = 0.9 ± 1.1, 6.8 ± 2.6, and 12.4 ± 4.1, respectively; average bud scar count in the corresponding nonstarved populations = 0.9 ± 1.1, 7.4 ± 3.1, and 11.7 ± 3.8. The fractional representation of ERCs in each of the total cellular DNA preparations shown was defined by comparing signals from the ERC bands with signals from all bands by using a PhosphorImaging system. Fractional representation of ERCs for both nonstarved cells and starved cells are <1% at generation 1 and 23% at generation 7. At generation 12, the values are 32 and 33%, respectively. (D) Telomere length does not change appreciably as nonstarved and starved cells age. The same DNA preparations used in C were digested with XhoI. Southern blots were prepared and probed with a 600-bp fragment corresponding to the conserved Y′ region of yeast telomeres. The arrow points to a broad band, migrating at ≈1 kilobase, consisting of Y′ telomeres.
We also monitored other aging phenotypes: loss of Sir3p from telomeres and its redistribution to an enlarged, fragmented nucleolus. To perform these experiments, cells were sorted by the biotinylation method described previously (2). Twenty-five percent of cells in a population that had undergone seven divisions (n = 78) exhibited these changes (Fig. 4B). By generation 12, the percentage increased to 45% (n = 112). In a nonstarved, age-matched control, the fractions of cells with these changes were 8% at generation 7 and 19% at generation 12 (n = 223 and 135 cells surveyed, respectively; data not shown).
Finally, we measured ERC levels in cells that had been recovered after 3 weeks in stationary phase, reexposed to nutrients, and sorted after dividing an average of 1, 7, or 12 times. The results were compared with results from cells that had never been starved. ERC levels increased in both types of cells with increasing generations. Strikingly, the increase was equivalent (Fig. 4C), even though the generational time points represent different percentages of total lifespan for the cell types: cells without the experience of stationary phase that are allowed to undergo a mean total of 12 divisions have completed 44% of their mean lifespan (12 of 27.5 generations), whereas cells that have survived 3 weeks of stationary phase, been reexposed to nutrients, and allowed to divide an average total of 12 times, have achieved 75% of their lifespan (12 of 16 generations). Thus, the accumulation of ERCs in mothers that experienced stationary phase did not scale to their shortened lifespan.
Together, our findings suggest that ERCs are not the sole determinant of nucleolar enlargement and fragmentation: despite their equivalent levels of ERCs, nucleolar changes are more prominent in starved cells compared with nonstarved, generation-matched cells. Moreover, ERC accumulation may not be the only determinant of aging (see Discussion).
Telomere shortening correlates with aging in many eukaryotes, but not with generational aging in nonstarved yeast cells (19). We tested the possibility that the experience of starvation may “induce” an aging program that includes telomere shortening, by measuring telomere length in the same total genomic DNA preparations employed for the ERC analysis. Southern blots of XhoI-digested DNA were probed with a labeled 600-bp sequence derived from the conserved Y′ region of yeast telomeres (20). The results, presented in Fig. 4D, indicate that generational aging in rich media, or generational aging after a period of starvation, is not accompanied by appreciable changes in telomere length.
DISCUSSION
Aging in yeast is typically defined by the finite number of divisions that individual mother cells undergo, rather than the “chronologic time” that this process encompasses. When cell division is not limited by nutrient availability, generational and chronologic definitions of aging can coincide. The present study emphasizes the difficulty in viewing aging in yeast solely from the perspective of time: when nutrients are limiting and cells enter a dormant period (stationary phase), lifespan is increased in terms of time but is shortened in terms of generations.
The difficulty in defining the best parameter for measuring aging is not limited to S. cerevisiae. For many species, chronologic measurements of lifespan only account for the adult phase and exclude earlier prematurational stages (21). However, both prematurational and adult phases can exhibit remarkable variations in length (time) depending on environmental factors. Dauer larvae of Caenorhabditis elegans and microbial spores represent examples where the organismal response to nutrient deprivation involves entry into a nonreplicative “dormant” state (diaupause) that can vastly exceed the time period which encompasses the “life” of the organism during its growing or adult phase (21).
The relevance of these states to the study of aging is that organisms in diapause or dormancy still maintain metabolic activities, albeit at lower rates, and remain subject to the time-dependent decay of molecules and various forms of damage (oxidative, UV radiation) that may contribute to senescence (21). A central question is whether factors that determine survival during nonreplicative dormant states are related to the factors that affect longevity under nonstarved conditions. Because there are no distinct developmental (e.g., larval) or adult phases in S. cerevisiae, this organism provides an opportunity to examine whether time spent in this starvation-induced dormant state is associated with any of the known manifestations of aging, and/or whether time spent in stationary phase affects the subsequent aging program of dividing cells on reexposure to nutrients.
We monitored the phenotypes of stationary-phase cells, such as the morphology of the nucleolus, the localization of the Sir protein complex in nuclei, and the production of ERCs, and concluded that stationary-phase cells do not resemble replicatively aged nonstarved mother cells. Most interestingly, by following the replicative lifespan of mother cells that had survived stationary phase and reentered the cell cycle after reexposure to fresh nutrients, we discovered that lifespan was shortened. This shortening was proportional to time spent in stationary phase but approached a limit of about 50%. The shortening of lifespan cannot be caused by permanent damage, i.e., genetic changes, because daughters and granddaughters of these cells recover normal lifespans.
Why do these cells have a shortened lifespan? We monitored the aging phenotypes over the lifespans of these mother cells and found that most of the characteristic phenotypes scaled to their shortened lifespan. These include the progressive acquisition of sterility, the enlargement and fragmentation of the nucleolus, and the redistribution of the Sir protein complex from telomeres to the nucleolus. In these respects, these cells display accelerated aging. The one exceptional phenotype that did not scale in these cells is the accumulation of ERCs. Mother cells recovered from stationary-phase cultures show very similar kinetics of ERC accumulation as generation-matched nonstarved controls. This means that the phenotype of nucleolar fragmentation does not strictly correlate with ERC levels, and, in fact, may not represent the embodiment of ERCs. More importantly, these cells are growth-arrested at a level of ERCs that does not arrest control cells.
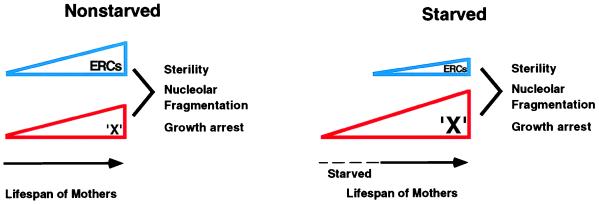
Several models can be invoked to explain the shortened lifespan of cells that have experienced stationary phase. In the first, we propose a cause of aging that is separate from ERCs and functions in parallel with ERCs, i.e., the accumulation of a substance termed “X” (see Fig. 5). In control cells, ERCs and X both accumulate with age and are summed to elicit the characteristic responses of sterility, nucleolar fragmentation, and growth arrest. We propose that X accumulates during stationary phase so that cells that reenter the cell cycle begin with an unusually high level of X, which advances their aging program. We do not know what X is. Nonetheless, this model suggests the possible existence of a second pathway of aging in yeast. The limit in the shortening of lifespan that can be elicited by time spent in stationary phase may represent a saturation of this second pathway. Thus, growth arrest of mother cells that reenter the cell cycle is not provoked by X alone, but only occurs when ERCs also reach a critical level.
Figure 5.
Model for diminished replicative lifespan of mother cells that have reentered the cell cycle after stationary phase. In control (nonstarved) cells, replicative aging of mother cells (solid time line) is caused by the sum of two processes, the accumulation of ERCs and the parallel accumulation of something else, termed X. These signals are summed and at some threshold elicit the aging phenotypes of sterility, nucleolar fragmentation, and growth arrest. Stationary-phase cells (starved) accumulate X while in stationary phase (dotted time line), so that their aging is advanced when they commence replicative growth (solid time line).
Another possible model for the shortened lifespan of cells that have experienced stationary phase is that they are more sensitive to ERCs. We do not yet understand the mechanism by which ERCs arrest the growth of cells, although they may titrate replication and/or transcription factors (12). Perhaps stationary phase depletes the relevant factors and cells that reenter the cell cycle are thereby sensitized to the toxic effect of ERCs.
Finally, we cannot rule out the possibility that stationary phase induces a novel mechanism of aging that does not involve ERCs at all. We do not favor this model because of the profound similarities in the premature aging of stationary-phase cells that reenter the cell cycle and control nonstarved cells.
In summary, these findings provide a clue of a second mechanism of aging in yeast that may operate in addition to the accumulation of ERCs. It is tempting to speculate that this mechanism, which appears to involve progressive changes in nondividing yeast cells, may be germane to the aging of nondividing tissues in higher eukaryotes. The molecular description of this mechanism becomes a high priority for a more complete understanding of aging in yeast and, perhaps, other organisms.
Acknowledgments
We thank Pierre Defossez and Kevin Mills for helpful discussions, reagents, and technical assistance. We are also grateful to Rajiv Bhatnagar, Lora Hooper, and Melissa Wong for their comments during the course of these studies. This work was supported by grants from the National Institutes of Health (AG11119 to L.G. and AI38200 to J.I.G.). K.A. was supported, in part, by a Glenn/American Federation of Aging Research scholarship. D.A.S. was supported by a Helen Hay Whitney Postdoctoral Fellowship.
ABBREVIATIONS
- rDNA
ribosomal DNA
- ERC
extrachromosomal rDNA circles
References
- 1.Sinclair D A, Mills K D, Guarente L. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 2.Smeal T, Claus J, Kennedy B, Cole F, Guarente L. Cell. 1996;84:633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- 3.D’mello N P, Childress A M, Franklin D S, Kale S P, Pinswasdi C, Jazwinski S M. J Biol Chem. 1994;269:15451–15459. [PubMed] [Google Scholar]
- 4.Sinclair D A, Mills K D, Guarente L. Annu Rev Microbiol. 1998;52:533–560. doi: 10.1146/annurev.micro.52.1.533. [DOI] [PubMed] [Google Scholar]
- 5.Sun J, Kale S P, Childress A M, Pinswasdi C, Jazwinski S M. J Biol Chem. 1994;269:18638–18645. [PubMed] [Google Scholar]
- 6.Kennedy B K, Austriaco N R, Zhang J, Guarente L. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- 7.Yu C E, Oshima J, Fu Y H, Wijsman E M, Hisama F, Alisch R, Matthew S, Nakura J, Miki T, Ouais S, Martin G M, Schellenberg G D. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 8.Martin G M. Birth Defects Orig Artic Ser. 1978;14:5–39. [PubMed] [Google Scholar]
- 9.Clark-Walker G D, Azad A A. Nucleic Acids Res. 1980;8:1009–1022. doi: 10.1093/nar/8.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larionov V L, Grishin A V, Smirnov M N. Gene. 1980;12:41–49. doi: 10.1016/0378-1119(80)90014-1. [DOI] [PubMed] [Google Scholar]
- 11.Larionov V L, Kouprina N, Karpova T. Gene. 1984;28:229–235. doi: 10.1016/0378-1119(84)90260-9. [DOI] [PubMed] [Google Scholar]
- 12.Sinclair D A, Guarente L. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 13.Werner-Washburne M, Braun E L, Crawford M E, Peck V M. Mol Microbiol. 1996;19:1159–1166. doi: 10.1111/j.1365-2958.1996.tb02461.x. [DOI] [PubMed] [Google Scholar]
- 14.Longo V D, Gralla E B, Valentine J S. J Biol Chem. 1996;271:12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 15.Longo V D, Ellerby L, Bredesen D E, Valentine J S, Gralla E B. J Cell Biol. 1997;137:1581–1588. doi: 10.1083/jcb.137.7.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson D R, Cok S J, Feldman H, Gordon J I. Proc Natl Acad Sci USA. 1994;91:10158–10162. doi: 10.1073/pnas.91.21.10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1994. pp. 2.5.9–2.5.17. [Google Scholar]
- 18.Baumann P, West S C. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 19.D’mello N P, Jazwinski S M. J Bacteriol. 1991;173:6709–6713. doi: 10.1128/jb.173.21.6709-6713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 21.Finch C E. Longevity, Senescence, and the Genome. Chicago: Univ. of Chicago Press; 1990. pp. 465–496. [Google Scholar]