Abstract
Members of the kinesin superfamily are force-generating ATPases that drive movement and influence cytoskeleton organization in cells. Often, more than one kinesin is implicated in a cellular process, and many kinesins are proposed to have overlapping functions. By using conventional kinesin as a model system, we have developed an approach to activate or inhibit a specific kinesin allele in the presence of other similar motor proteins. Modified ATP analogs are described that do not activate either conventional kinesin or another superfamily member, Eg5. However, a kinesin allele with Arg-14 in its nucleotide binding pocket mutated to alanine can use a subset of these nucleotide analogs to drive microtubule gliding. Cyclopentyl-ATP is one such analog. Cyclopentyl-adenylylimidodiphosphate, a nonhydrolyzable form of this analog, inhibits the mutant allele in microtubule-gliding assays, but not wild-type kinesin or Eg5. We anticipate that the incorporation of kinesin mutants and allele-specific activators and inhibitors in in vitro assays should clarify the role of individual motor proteins in complex cellular processes.
Enzymes in the kinesin superfamily use the free energy of ATP hydrolysis to drive intracellular movement and influence cytoskeleton organization (1). More than 90 members of this family are known. Historically, kinesins have been proposed to move cellular cargo along polar microtubule tracks. More recently it has been shown that these ATPases can modulate dynamics of the underlying microtubule network (2), couple movement of cargo to the microtubule polymerization or depolymerization (3), and crosslink microtubules in dynamic structures (4). Kinesins thus play central roles in mitotic and meiotic spindle formation, chromosome alignment and separation, axonal transport, endocytosis, secretion, and membrane trafficking. The cargo associated with these motor proteins includes intracellular vesicles, organelles, chromosomes, kinetochores, intermediate filaments, microtubules, and even other motors (reviewed in refs. 5 and 6).
For many of these processes, more than one kinesin is implicated, and the specific cargo associated with a given motor protein has been difficult to establish. For example, conventional kinesin (7) (the founding member of the family) is one of a subset of kinesins involved in organelle transport in mammalian cells. This group includes KIF1, KIF2, KIFC2/C3, and KIF4; and more recently, 18 new murine KIFs have been reported, many of which may functionally overlap with the transport kinesins (reviewed in ref. 6). It thus has been difficult to tie down the in vivo function(s) of conventional kinesin. Experiments using antisense techniques and microinjection of inhibitory antibodies have been further complicated by recent observations of efficient endoplasmic reticulum to Golgi transport in the absence of microtubules, albeit under restricted conditions (reviewed in ref. 8). Similar problems have been encountered in dissecting the function of kinesins in mitosis. Extensive genetic analysis of motors in Saccharomyces cerevisiae has linked all but one of the six kinesins to spindle function. None of these five motors are individually required for the viability of yeast, implying that more than one motor is associated with essential aspects of spindle movement (9, 10). Immunodepletion and add-back approaches in Xenopus extract spindle assembly assays have provided similarly ambiguous data (11).
Small molecules that conditionally activate or inactivate a protein are valuable tools for analyzing cellular functions of proteins (12). Their use provides an alternative to conventional biochemical and genetic approaches. However, to date there have been few reports of small molecules that can reversibly alter the function of motor proteins. Butanedione monoxime has been used to probe the role of myosin in cell movement (13), but its specificity has been questioned (14). A natural product inhibitor of kinesin has been reported (15), but is thought not to be selective for different kinesins and thus is not useful for probing the role of one specific kinesin in a complex process. Hyman et al. (16) have used ATP analogs to distinguish between microtubule motility at kinetochores driven by a kinesin and a dynein, but again, this approach is unlikely to distinguish between different kinesins. Thus we currently lack small molecule activators or inhibitors that are specific for one member of the kinesin family.
Adenylylimidodiphosphate (AMPPNP) is a nonhydrolyzable nucleotide analog that inhibits kinesins and generates a nonmotile rigor state such that the kinesin-nucleotide complex is locked onto microtubules (17). First observed in squid axoplasmic organelles and subsequently used to purify conventional kinesin (7), the rigor state has been found to be a general phenomenon for motors in the kinesin superfamily. AMPPNP has been used as an inhibitor for motor proteins in complex extract systems (18) and in live cells (19). Given the large number of ATP-utilizing enzymes in these systems, the assumption of inhibitor specificity is dubious.
By using conventional kinesin as a model system, we have developed an approach to control the function of individual kinesins. We have designed and synthesized ATP and AMPPNP analogs that activate and inhibit a kinesin allele mutated in the nucleotide binding pocket but do not affect wild-type kinesin. Altering inhibitor or substrate specificity for a protein with a complementary mutation has been used to regulate protein function in other systems. Cyclosporin analogs that are cell-specific calcineurin inhibitors have been developed (20, 21). The physiological substrates of kinases have been determined by using mutated enzymes and modified ATP derivatives (22), and the roles of small GTPs in protein translocation have been studied through the use of xanthosine triphosphate-utilizing enzymes (23). Here we report ATP analogs that cannot be used by the wild-type conventional kinesin to generate motion. A kinesin allele mutated in the nucleotide binding pocket is described that can use a subset of these nucleotides to drive motility. Nonhydrolyzable versions of these nucleotides can function as inhibitors for the mutant kinesin and do not inhibit the wild-type motor or a different kinesin superfamily member, Eg5. We plan to use these kinesin mutant-nucleotide analog pairs in assays to deconvolute the function of conventional kinesin in the context of other motor proteins. The high homology and structural similarity of kinesins also should allow this approach to be applicable to other members in this superfamily, including those involved in spindle assembly.
MATERIALS AND METHODS
Synthesis of ATP and AMPPNP Analogs.
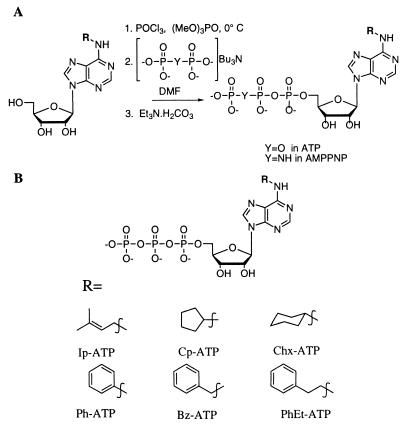
Compounds were synthesized from commercial N-6 substituted adenosines (Sigma and Research Biochemical, Natick, MA). Triphosphates were synthesized as described by Shah et al. (22). To synthesize AMPPNP derivatives, the tributylammonium pyrophosphate used in the triphosphate synthesis was replaced with an equivalent amount of tributylammonium imidodiphosphate (prepared as described by Yount et al., ref. 24). The triethyl ammonium salts of the analogs were purified by using anion exchange chromatography and characterized by mass spectral analysis and analytical chromatography. The cyclopentyl (Cp) ATP and AMPPNP analogs were converted to their potassium salts by mixing with excess KCl and desalting by gel filtration chromatography (G-10 Sephadex, Sigma).
Mutagenesis and Protein Expression.
Kinesin and kinesin mutants were expressed in bacteria [BL21(DE3) pLysS (Novagen)] as recombinant proteins comprising amino acid residues 1–560 and a C-terminal histidine tag. The plasmid encoding the wild-type kinesin has been described (25). The Eg5 motor domain, encoding residues 1–591, fused to a C-terminal histidine tag, was constructed by PCR amplification from a full-length Eg5 clone (26). The histidine tag and the restriction sites were included in the primers, and the digested PCR product was ligated into pRSET A (Invitrogen). Site-specific mutagenesis was performed by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Sequencing of plasmids from randomly selected clones confirmed the mutagenesis. Kinesins expressed in bacteria were purified by nickel affinity followed by Superose 6 gel filtration (Amersham Pharmacia), as described by Desai et al. (2).
Motility Assays.
To assess the nucleotide selectivity for each conventional kinesin allele, the motor-driven gliding velocity of microtubules over a glass coverslip in the presence of each different ATP analog was measured. A ≈10-μl flow chamber consisting of a glass slide and coverslip, separated by double-sided adhesive tape, first was filled with 0.5 mg/ml casein. After a 3-min incubation, the chamber was washed twice with 2 vol of buffer A [BRB80 (80 mM Pipes, 1 mM MgCl2, 1 mM EGTA, pH 6.8 with KOH) + ATP analog with equimolar MgCl2]. The motor protein (typically 50 μg/ml) diluted in buffer A, then is flowed into the chamber and incubated for 5 min. This step is followed by a wash consisting of 3 vol of buffer A with an oxygen-depletion system [71.5 mM β-mercaptoethanol, 22.5 mM glucose, 0.22 mg/ml glucose oxidase (Sigma G-2133), 0.036 mg/ml catalase (Sigma C-40)]. A solution of rhodamine-labeled microtubules (0.2 μM) (prepared by using standard procedures; ref. 2) in buffer A with the oxygen-depletion mix then is flowed in, and the gliding microtubules are visualized by time-lapse fluorescence microscopy. Velocities were measured by using the program trace (software developed by Aneil Mallavarapu, Millennium Pharmaceuticals, Cambridge, MA). Motility assays for the motor Eg5 were carried out with two modifications of the above protocol. Casein was replaced by BSA, and Eg5 was flowed into the chamber before the BSA solution.
Inhibition of the kinesin motility by AMPPNP analogs was assayed in the presence of the ATP analog that generated the maximum velocity for a given allele. In a typical assay, the microtubule-gliding velocity in the presence of the appropriate nucleotide was measured as described above. The same flow chamber then was subjected to two washes of buffer A (with the nucleotide), oxygen-depleting mixture, microtubules, and AMPPNP analog (with equimolar MgCl2). The microtubule-gliding velocity then was determined as described. All velocity data represent the results from two or more experiments, and the mean velocity is shown with the SE. Competitive inhibition (see Fig. 5C) is modeled on parameters for Dixon plots (27).
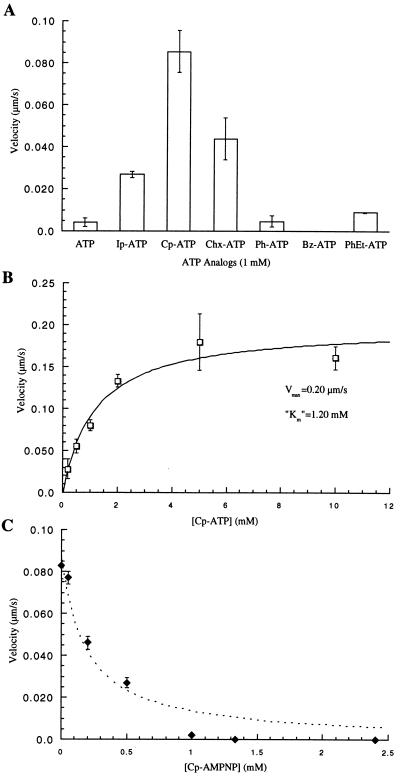
Figure 5.
Characterization of the R14A kinesin mutant. (A) Measurement of microtubule-gliding velocities in the presence of different nucleotide analogs reveals that the mutant kinesin can use Cp-ATP more efficiently than unmodified ATP. (B) R14A kinesin-driven gliding velocities depend on the Cp-ATP concentration, and the data fit well to the Michaelis Menten model [velocity = Vmax∗[Cp-ATP]/(Km + [Cp-ATP])]. (C) Inhibition of R14A kinesin-dependent microtubule gliding by Cp-AMPPNP, in the presence of 1 mM Cp-ATP, shows a deviation from competitive inhibition at higher concentrations (broken line) and concentrations greater than 1 mM Cp-AMPPNP completely inhibit the mutant motor. This aspect of kinesin inhibition by AMPPNP has been characterized previously (27).
RESULTS
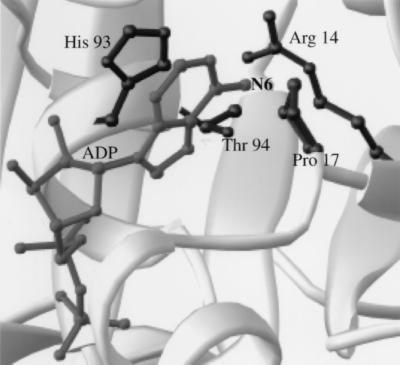
To design nucleotide analogs that are excluded from the ATP binding pocket of kinesins, we examined the x-ray crystal structures of kinesins (28–30). Fig. 1 shows the ADP-bound head domain of human conventional kinesin. This kinesin is a tetramer consisting of two heavy chains (110–130 kDa) and two light chains (60–80 kDa) with its heavy chain consisting of a head domain, attached by a neck region to the coiled-coil containing stalk domains, ending in a tail (1). The head domain (residues 1–349, for human conventional kinesin) is sufficient for ATP hydrolysis-driven motility. The purine ring of the bound ADP is buried in the pocket. The ribose hydroxyls and the phosphate groups project away from this pocket and are solvent-exposed. The residues that interact with the base are highlighted in Fig. 1. N-6 and N-7 of adenine are directed toward the protein with Pro-17 and His-93 packing on either side of the purine ring. Arg-14 from kinesin is directed toward the nucleotide’s N-6 and the side chain of Thr-94 is within 5 Å. We synthesized nucleotide analogs modified at this N-6 position, anticipating that they should be excluded from the wild-type motor’s binding pocket. Derivatization of the ribose hydroxyl groups was not expected to affect interactions with the protein directly, and modification of the phosphates was likely to interfere with the ATPase activity of the enzyme.
Figure 1.
The nucleotide binding pocket of human conventional kinesin bound to ADP. Side chains of residues within 6 Å of the N6 nitrogen of the nucleotide are highlighted. The figure was created with ribbons (41).
Shimizu et al. (31) have observed that the conventional kinesin nucleotide binding pocket is able to accommodate nucleotides with substituents as large as dimethyl at the N6 position. At 1 mM dimethyl-ATP, the kinesin-driven microtubule-gliding velocity is only 5-fold slower than that at equal ATP concentrations. Therefore, to exclude a synthetic nucleotide analog from the kinesin binding pocket larger substitutions at the exocyclic nitrogen were incorporated. Fig. 2A shows the synthesis of the ATP analogs by using published protocols (22). We report an extension of these methods for a one-pot synthesis of AMPPNP analogs where the tributylammonium pyrophosphate used for the ATP synthesis is replaced with tributylammonium imidodiphosphate. The nucleotide analogs we synthesized are shown in Fig. 2B.
Figure 2.
(A) Synthesis of the ATP and AMPPNP analogs. (B) Structures of the nucleotide analogs synthesized.
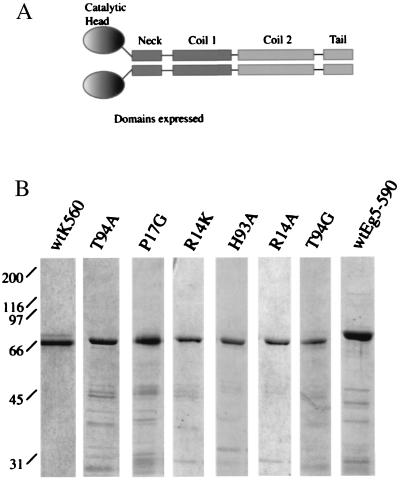
Analysis of these ATP analogs was performed by using a human kinesin construct (residues 1–560) (K560), containing the motor domain and half the stalk (Fig. 3A) followed by a C-terminal polyhistidine tag. This bacterially expressed kinesin construct has been reported to be primarily dimeric in solution and move processively along microtubules with velocities similar to those measured for native kinesin (25). All mutants were purified by using procedures similar to those used for the wild-type motor (Fig. 3B).
Figure 3.
(A) The domain structure of the kinesin dimer. Kinesin residues including the head, neck, and a portion of the coiled-coil regions were expressed in bacteria and purified. (B) Coomasie-stained gels of the wild-type and mutant kinesins used in our assays. As assessed by gel filtration chromatography, these proteins eluted as soluble dimers.
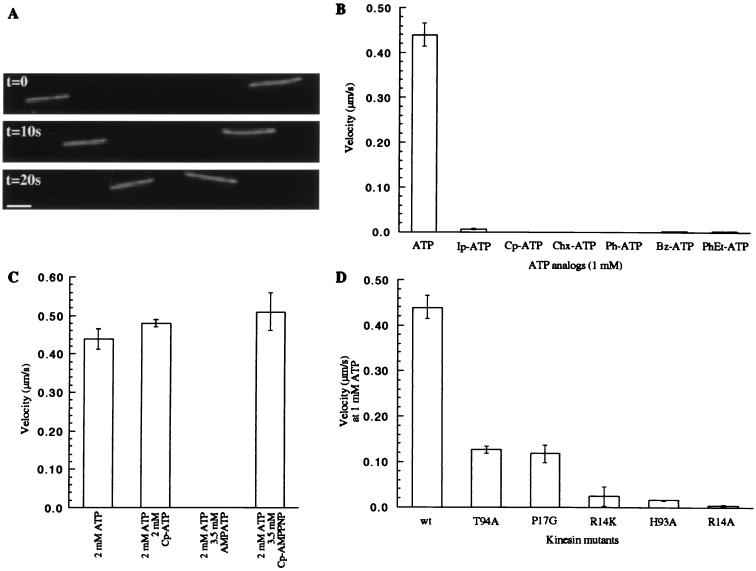
We selected the kinesin-driven microtubule-gliding velocity as a measure of the protein-nucleotide interaction and nucleotide turnover. Fig. 4A shows three images from such a motility assay with the expressed wild-type kinesin (K560). The linear distance traversed by a microtubule in fixed time intervals yielded the gliding velocity. As anticipated, ATP analogs with large substitutions at the N6 position were not efficiently used by the wild-type motor to drive movement (Fig. 4B). An approximately 63-fold reduction in velocity was measured for the isopentenyl analog (Ip-ATP), whereas no gliding was observed for the wild-type motor in the presence of Cp-ATP and ATP analogs with larger substitutions. These modified ATP analogs did not inhibit the microtubule gliding driven by the wild-type motor in the presence of ATP (Fig. 4C shows data for Cp-ATP). Furthermore, nonhydrolyzable AMPPNP analogs with large substituents did not inhibit kinesin motility in the presence of ATP, implying that the modifications on the purine ring prevented the nucleotide analogs from interacting in the binding pocket (data not shown). Specifically, the kinesin-dependent microtubule-gliding velocities in 2 mM ATP were unchanged after the addition of Cp-AMPPNP at concentrations as high as 3.5 mM (Fig. 4C). In the presence of 2 mM ATP, unsubstituted AMPPNP at 3.5 mM completely inhibits kinesin motility.
Figure 4.
(A) Three frames from a time-lapse fluorescence microscopy video of a kinesin-driven microtubule-gliding assay are shown. The distance rhodamine-labeled microtubules move in fixed time intervals determines the velocity. The images were acquired with a 60×, 1.4 NA Nikon objective and a Princeton Instruments cooled charge-coupled device camera. Bar = 2 μm. (B) The microtubule-gliding velocity of the wild-type kinesin in the presence of different ATP analogs (1 mM) is shown. Modification of the nucleotide with a Cp group yields an ATP analog that is incapable of driving kinesin-dependent movement. (C) Cp-ATP does not inhibit wild-type kinesin motility in the presence of ATP. AMPPNP at 3.5 mM completely inhibits kinesin motility in the presence of 2 mM ATP. The nonhydrolyzable Cp-AMPPNP does not reduce ATP-dependent kinesin motility. (D) A comparison of the gliding velocities of the different mutant kinesins in the presence of 1 mM ATP.
To expand the kinesin nucleotide binding pocket, we decided to mutate residues proximal to the N6 on the purine ring. Although the kinesin structure (Fig. 1) is a snapshot of a dynamic motor that cycles between different conformations, we do not expect significant reorganization of the residues closest to the N6 nitrogen. Alanine-scanning mutagenesis has been used to study the microtubule interaction site of the motor (25), but kinesins with mutations in residues that interact with the ATP purine ring have not been characterized. Mutations in the nucleotide triphosphate interacting P-loop (residues 84–92 in human kinesin) are known where the steric bulk of the side chains of residues is increased (32–34). These mutations probably disrupt ATP binding, and some of these mutants have been characterized as dominant rigor motors with no motile function and constitutive microtubule association.
We introduced mutations in kinesin that increased the size of its nucleotide binding pocket in small increments. The Arg-14 to lysine (R14K), the Pro-17 to glycine (P17G), and the Thr-94 to alanine (T94A) were conservative relative to the Arg-14 to alanine (R14A), His-93 to alanine (H93A), and Thr-94 to glycine (T94G) mutations. Sequence alignment of plus- and minus-end directed kinesin motor domains shows that Arg-14, Pro-17, and Thr-94 are highly conserved, whereas His-93 is less conserved and may be replaced by aromatic amino acids (Phe in Xenopus Eg5 and Tyr in Drosophila Ncd) (29). Fig. 3B shows SDS/PAGE analysis of the mutant kinesins expressed in bacteria and purified. All expressed mutants gave equivalent yields of soluble protein that was dimeric by the criterion of gel filtration chromatography.
Each kinesin mutant was tested in an in vitro microtubule-gliding assay. Remarkably, all but one of the mutant motors retained motility. The T94G kinesin showed no microtubule binding and hence, no motility. This mutant kinesin may be useful as a dominant negative allele in cellular assays. Data for the motile kinesin mutants are shown in Fig. 4D. The velocities range from 29% to 0.9% of that measured for the wild-type motor at equal ATP concentrations. An 18-fold decrease in velocity was observed for the R14K mutation, where the guanidyl functionality in the arginine side chain is replaced with an aminomethyl. Loss of the imidazole of the histidine in the H93A mutant resulted in a similar decrease in velocity. Here the hydrophobic interaction of the alanine side-chain methyl with the base may replace the aromatic stacking interactions between the purine ring of the nucleotide and the histidine imidazole.
Each mutant was tested for motility in the presence of the ATP analogs we synthesized. The nucleotide selectivity for kinesins with conservative mutations approximated that of the wild-type kinesin (K560), with ATP being the preferred substrate for motility (data not shown). However, the R14A mutant showed a rather different preference for N6 substitutions (Fig. 5A), whereby an increase in hydrophobic bulk at the N6 position compensated for the loss in electrostatic interactions between purine and guanidine in the unmodified binding pocket. The isopentenyl group on ATP results in an almost 7-fold increase in the motor’s velocity relative to that in ATP, whereas the Cp group allows the R14A mutant to drive microtubule gliding to within 20% of the wild-type motor. Further increasing the size of the substituent on ATP to cyclohexyl, phenyl, benzyl, and phenethyl, leads to a decrease in measured velocity, indicating that the hole in the nucleotide binding pocket may be smaller than these modifications on the purine ring.
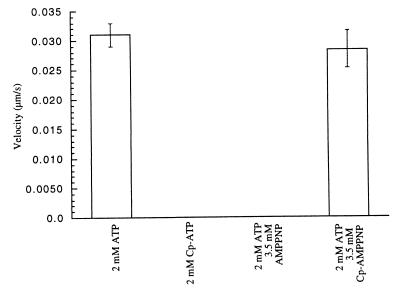
Fig. 5B shows the dependence of the R14A kinesin velocity on the Cp-ATP concentration. The data fit well to a Michaelis Menten model (27). The maximum observed velocity of this motor protein is within 35% of the wild-type kinesin velocity in ATP. The Km, however, is 20-fold higher than that of the wild-type kinesin in the presence of ATP (27). This R14A kinesin shows essentially no activity (22-fold lower) at 1 mM unmodified ATP. Furthermore, a nonhydrolyzable analog of Cp-ATP, Cp-AMPPNP, completely inhibits this kinesin allele in the presence of Cp-ATP (Fig. 5C). However, 3.5 mM Cp-AMPPNP does not inhibit wild-type kinesin in the presence of ATP (Fig. 4C), and the wild-type motor is unable to use Cp-ATP to drive microtububle gliding (Fig. 4B).
To characterize the specificity of the activator (Cp-ATP) and inhibitor (Cp-AMPPNP) further, we tested a homologous kinesin, Eg5. This enzyme has an essential function in mitosis and like kinesin, has a motor domain at its N terminus (11, 35). We expressed and purified the region corresponding to residues 1–560 of human kinesin (amino acid identity between expressed regions of human conventional kinesin and Xenopus Eg5 is 32%) (Fig. 3B). The microtubule-gliding velocity of this motor has been reported to be 10-fold lower than that of kinesin (35). As observed with wild-type kinesin, wild-type Eg5 showed no detectable microtubule gliding in the presence of Cp-ATP. Similarly, its gliding velocity stimulated by ATP was not inhibited by Cp-AMPPNP, at concentrations as high as 3.5 mM (Fig. 6).
Figure 6.
The specificity of the activator and the inhibitor for the R14A kinesin mutant. The kinesin superfamily member, Eg5, shows no detectable microtubule gliding in the presence of 2 mM Cp-ATP. Cp-AMPPNP also is unable to inhibit the enzyme at concentrations that unmodified AMPPNP completely inhibits its activity.
CONCLUSION
We report a mutant allele of conventional kinesin (R14A) that has a greatly reduced microtubule-gliding velocity in unmodified ATP, but is specifically activated by a modified ATP analog, Cp-ATP. Cp-ATP cannot be used efficiently by either the wild-type motor or by Eg5, another kinesin superfamily member. Similarly, a nonhydrolyzable version of Cp-ATP, Cp-AMPPNP, does not inhibit the wild-type kinesins, but does inhibit the R14A kinesin allele in a nonmotile, microtubule-bound rigor state. We also have characterized kinesin alleles that have reduced microtubule-gliding velocities in the presence of unmodified ATP.
Our kinesin alleles and cognate nucleotide analogs should be useful to probe both cell biological and biophysical aspects of kinesin function. To illustrate this point we outline two types of experiments. An outstanding question in kinesin cell biology is the identity of the vesicles that kinesin transports in vivo and the nature of the kinesin receptor on those vesicles (36). The kinesin targeting problem has been addressed in a variety of cell-free systems, by using AMPPNP and antibodies to implicate kinesin in, for example, Golgi membrane transport (37, 38). It should now be possible to transfect cells with our mutant alleles, prepare cell-free extracts, add Cp-ATP, observe vesicle motility, and ask whether the movement of a particular vesicle type is retarded by Cp-AMPPNP in a dose-dependent manner. A positive result would confirm that kinesin is moving the vesicle type in question. Because rigor inhibition should be dominant to unperturbed motor (32–34), this experiment should work even while wild-type kinesin is present on the vesicles along with the mutant allele. A key biophysical question for kinesin has been how stepping activity of the two heads is coordinated so as to obtain processive movement along the microtubule (1). If kinesin heterodimers were made adapting known technology (39) so that one head was wild type and the other a R14A mutant, some interesting experiments are possible. For example, with the mutant head tethered with Cp-AMPPNP to a microtubule, the rate of ATP hydrolysis by the wild-type head could be measured. This experiment would probe the functional flexibility of the neck region connecting the two heads.
Our results imply the possibility of new experimental approaches to other kinesin superfamily proteins. Structures of the head domains of kinesin superfamily members from human (28), Drosophila (29), and yeast (30) show very high homology in the nucleotide binding pocket. Thus it should be possible to create equivalent mutant alleles of other kinesins and to use these in conjunction with Cp-ATP and Cp-AMPPNP to dissect their overlapping functions in cell biological processes. We plan to take this approach to probe the role of kinesin superfamily members in mitosis in the Xenopus extract system (11). In this system the wild-type motor can be immunodepleted and replaced by a mutant allele (40), which should allow us to use both Cp-ATP as a specific activator and Cp-AMPPNP as a specific inhibitor in analyzing function. We envisage, for example, using this technology to examine the role of Eg5 in establishing spindle bipolarity (11).
We find that ATP analogs with very similar modifications to those used as substrates for mutated tyrosine kinases (22) can be used to create allele-specific activators and inhibitors of kinesin. Tyrosine kinases and kinesin are no more homologous in their ATP binding sites than almost any pair of ATP-hydrolyzing enzymes. Thus our work implies that this strategy should be generalizeable to other ATPases, including those for which high-resolution structural data will be difficult to obtain, such as ATPase membrane pumps. Allele-specific activators and inhibitors so far have been very useful for probing the function of cyclophilins (20), small GTPases (23), and kinases (22). Our work should lead directly to equivalent analysis of kinesins in vesicle trafficking and mitosis, and in principle, it opens up the whole panoply of ATPases for such analysis.
Acknowledgments
We are grateful to Arshad Desai and Aaron Straight for their encouragement and technical guidance; Michelle Shirazu, Jack Taunton, and Thomas Mayer for helpful comments on the manuscript; and Mykol Larvie for assistance with ribbons. This work was supported by National Institutes of Health Grant GM39565. T.M.K. is a Runyon-Winchell Fellow.
ABBREVIATIONS
- AMPPNP
adenylylimidodiphosphate
- Cp
cyclopentyl
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Vale R D, Fletterick R J. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- 2.Desai A, Verma S, Mitchison T J, Walczak C E. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 3.Wood K W, Sakowicz R, Goldstein L S, Cleveland D W. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- 4.Sharp D J, McDonald K L, Brown H M, Matthies H J, Walczak C, Vale R D, Mitchison T J, Scholey J M. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walczak C E, Mitchison T J. Cell. 1996;85:943–946. doi: 10.1016/s0092-8674(00)81295-7. [DOI] [PubMed] [Google Scholar]
- 6.Hirokawa N. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 7.Vale R D, Reese T S, Sheetz M P. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom G S, Goldstein L S. J Cell Biol. 1998;140:1277–1280. doi: 10.1083/jcb.140.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saunders W S, Hoyt M A. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- 10.Hoyt M A, Hyman A A, Bahler M. Proc Natl Acad Sci USA. 1997;94:12747–12748. doi: 10.1073/pnas.94.24.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walczak C E, Vernos I, Mitchison T J, Karsenti E, Heald R. Curr Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- 12.Hung D T, Jamison T F, Schreiber S L. Chem Biol. 1996;3:623–639. doi: 10.1016/s1074-5521(96)90129-5. [DOI] [PubMed] [Google Scholar]
- 13.Cramer L P, Mitchison T J. J Cell Biol. 1995;131:179–189. doi: 10.1083/jcb.131.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinberg G, McIntosh J R. Eur J Cell Biol. 1998;77:284–293. doi: 10.1016/S0171-9335(98)80087-3. [DOI] [PubMed] [Google Scholar]
- 15.Sakowicz R, Berdelis M S, Ray K, Blackburn C L, Hopmann C, Faulkner D J, Goldstein L S. Science. 1998;280:292–295. doi: 10.1126/science.280.5361.292. [DOI] [PubMed] [Google Scholar]
- 16.Hyman A A, Middleton K, Centola M, Mitchison T J, Carbon J. Nature (London) 1992;359:533–536. doi: 10.1038/359533a0. [DOI] [PubMed] [Google Scholar]
- 17.Saxton W M. Methods Cell Biol. 1994;44:279–288. doi: 10.1016/s0091-679x(08)60919-x. [DOI] [PubMed] [Google Scholar]
- 18.Desai A, Maddox P S, Mitchison T J, Salmon E D. J Cell Biol. 1998;141:703–713. doi: 10.1083/jcb.141.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee G M. J Cell Sci. 1989;94:425–441. doi: 10.1242/jcs.94.3.425. [DOI] [PubMed] [Google Scholar]
- 20.Belshaw P J, Schreiber S L. J Am Chem Soc. 1997;119:1805–1806. [Google Scholar]
- 21.Belshaw P J, Schoepfer J G, Liu K-Q, Morrison K L, Schreiber S L. Angew Chem Int Ed Engl. 1995;34:2129–2132. [Google Scholar]
- 22.Shah K, Liu Y, Deirmengian C, Shokat K M. Proc Natl Acad Sci USA. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers T, Walter P. Science. 1995;269:1422–1424. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- 24.Yount R G, Babcock D, Ballantyne W, Ojala D. Biochemistry. 1971;10:2484–2489. doi: 10.1021/bi00789a009. [DOI] [PubMed] [Google Scholar]
- 25.Woehlke G, Ruby A K, Hart C L, Ly B, Hom-Booher N, Vale R D. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- 26.Sawin K E, Mitchison T J. Proc Natl Acad Sci USA. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohn S A, Ingold A L, Scholey J M. J Biol Chem. 1989;264:4290–4297. [PubMed] [Google Scholar]
- 28.Kull F J, Sablin E P, Lau R, Fletterick R J, Vale R D. Nature (London) 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sablin E P, Kull F J, Cooke R, Vale R D, Fletterick R J. Nature (London) 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- 30.Gulick A M, Song H, Endow S A, Rayment I. Biochemistry. 1998;37:1769–1776. doi: 10.1021/bi972504o. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu T, Furusawa K, Ohashi S, Toyoshima Y Y, Okuno M, Malik F, Vale R D. J Cell Biol. 1991;112:1189–1197. doi: 10.1083/jcb.112.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meluh P B, Rose M D. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- 33.Rasooly R S, New C M, Zhang P, Hawley R S, Baker B S. Genetics. 1991;129:409–422. doi: 10.1093/genetics/129.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakata T, Hirokawa N. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawin K E, LeGuellec K, Philippe M, Mitchison T J. Nature (London) 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- 36.Sheetz M P. Eur J Biochem. 1999;262:19–25. doi: 10.1046/j.1432-1327.1999.00340.x. [DOI] [PubMed] [Google Scholar]
- 37.Verhey K J, Lizotte D L, Abramson T, Barenboim L, Schnapp B J, Rapoport T A. J Cell Biol. 1998;143:1053–1066. doi: 10.1083/jcb.143.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fullerton A T, Bau M Y, Conrad P A, Bloom G S. Mol Biol Cell. 1998;9:2699–2714. doi: 10.1091/mbc.9.10.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hancock W O, Howard J. J Cell Biol. 1998;140:1395–1405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walczak C E, Mitchison T J, Desai A. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- 41.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]