Abstract
Background: Desulfovibrios produce sulphide, which is toxic to colonic epithelial cells. These bacteria have previously been linked to ulcerative colitis. Traditional methods of culturing these organisms are slow, and often unreliable, while molecular approaches are either non-quantitative or lack sensitivity.
Aims: To develop a sensitive method for quantitating desulfovibrios in stools and biopsy tissue, and to investigate the effects of age and disease on these bacteria.
Methods: Rectal biopsies were taken from 10 colitis patients and 10 healthy controls. Stool samples were obtained from 10 healthy infants (mean age 1.01 (0.18) years), 10 healthy young adults (26.7 (1.2) years), and 10 healthy elderly people (71.7 (1.2) years). Primers were designed and developed for analysing Desulfovibrio populations in the bowel using real time polymerase chain reaction (PCR).
Results: The PCR primers were highly specific for desulfovibrios. Large numbers (approximately 106–107/g) occurred in biopsies in colitis patients and healthy subjects, and no disease related differences were observed. Measurements of mucosal desulfovibrios over 12 months showed marked changes in some patients. Infants (106–107/g) and elderly people (107–108/g) had significantly higher numbers of desulfovibrios in stools compared with young adults (105/g).
Conclusions: Real time PCR analysis of desulfovibrios was an efficient and accurate method for studying these potentially harmful microorganisms. Desulfovibrios were ubiquitous in the bowel, irrespective of age. As rectal mucosae were heavily colonised in health and disease, if these bacteria play a role in colitis, some host defect, possibly in sulphide detoxication pathways or in bacterial antigen handling, is required for manifestations of pathogenicity.
Keywords: intestinal bacteria, sulphate reducing bacteria, desulfovibrios, ulcerative colitis, mucosal bacteria
Evidence from animal models and the study of patients with ulcerative colitis (UC) implicates the intestinal microbiota in disease aetiology. However, no specific pathogen has been identified as being causal, and UC is widely thought to result from a genetically determined but abnormal immune response to bacterial species in the normal gut microflora.1
Sulphate reducing bacteria (SRB) of the genus Desulfovibrio can occur in large numbers in faeces2 and have been linked to UC.3 It has been reported4 that healthy people have a lower prevalence of SRB in faeces (24%) than inflammatory bowel disease (IBD) patients (68%) while other investigations, using culturing methods, have shown that mucosa associated SRB were present in 92% of UC tissues and in 52% of non-IBD subjects.5 However, SRB were found to be present in all of the biopsies studied when they were analysed by conventional polymerase chain reaction (PCR).
SRB in the large bowel reduce sulphate to sulphide, which is toxic to colonic epithelial cells.6 Sulphide inhibits butyrate metabolism in colonocytes,7,8 inhibits phagocytosis and bacterial killing,9 and induces hyperproliferation and metabolic abnormalities in epithelial cells, similar to those observed in UC.10 In humans, untreated UC patients were observed to have significantly higher faecal sulphide excretion than healthy controls, while 5-ASA, the main drug used to treat UC, was reported to inhibit sulphide production.11
Conventional culture based methods for studying SRB are time consuming due to slow bacterial growth rates,12 and like other methods of culture, do not necessarily provide an accurate assessment of the types and numbers of bacteria present in a biological sample.5,13,14 However, it has been shown that variable regions of 16S ribosomal RNA (rRNA) gene sequences provide the basis for development of specific primers/probes that can be used for culture independent analyses of bacterial populations.15–20
The purpose of this study was to develop specific real time PCR primers for rapid, sensitive, high throughput determinations of intestinal desulfovibrios, based on analysis of 16S rRNA genes. These bacteria may play an important role in gut health, and because lumenal communities are known to change with age14 and distinct bacterial populations have been reported to occur on the gut epithelium,21 the occurrence of desulfovibrios in different age groups and on mucosal tissue was investigated.
MATERIALS AND METHODS
Bacterial strains
The following bacteria, obtained from the National Collection of Industrial and Marine Bacteria (Aberdeen, UK), the National Collection of Type Cultures (London, UK), Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunscheig, Germany), American Type Culture Collection (Manassas, Virginia, USA), and laboratory stock cultures at the University of Dundee (DUN-), were used to evaluate primer efficacy: Faecalibacterium prausnitzii ATCC 27768, F prausnitzii NCIMB 13872, Bifidobacterium catenulatum NCIMB 702246, Bif adolescentis NCIMB 702231, Bif bifidum NCIMB 702715, Bif breve NCTC 11815, Eubacterium limosum DUN-112, Eub rectale DUN-128, Eub aerofaciens DUN-207, Eub cylindroides DUN-119, Clostridium clostridioforme DUN-120, C malenominatum DUN-102, C perfringens NCTC 8346, C butyricum NCIMB 7423, C bifermentans DUN-156, C perfringens NCTC 8533, C sordellii DUN-113, C tetani NCTC 5404, C difficile NCTC 11223, C histolyticum DSM 2158, Desulfovibrio desulfuricans NCIMB 12833, Dsv vulgaris NCIMB 8303, Dsv desulfuricans NCIMB 8307, Desulfococcus multivorans NCIMB 12965, Desulfobacter vibrioformis NCIMB 13525, Dsb vibrioformis NCIMB 13525, Desulfotomaculum ruminis DSM 2154, Dsm ruminis NCIMB 8452, Dsm ruminis (Professor GR Gibson, University of Reading, UK), Bacteroides fragilis NCTC 9343, B ovatus DUN-203, B thetaiotaomicron NCTC 10582, B vulgatus DUN-116, Escherichia coli ATCC 11775, E coli NCTC 9001, Enterococcus faecalis ATCC 51299, Ent faecium NCTC 12202, Lactobacillus plantarum DUN-145, L. acidophilus DSM 20079, Peptostreptococcus anaerobius DUN-167, Shigella sonnei DUN-198, S flexneri DUN-200, Salmonella typhimurium DUN-217, and Veillonella parvula NCTC 12140.
Subjects and specimens
Rectal biopsies were taken from 10 UC patients and 10 normal controls who had not received any bowel preparation. Diagnosis of UC was confirmed in all cases at colonoscopy, and by histology. All UC patients had actively inflamed mucosae categorised as either mild or moderately severe. All UC biopsies were of inflamed tissue. Biopsies were collected aseptically and frozen immediately at −80°C in sterile Eppendorf tubes. Stools were obtained from 30 healthy subjects, comprising 10 infants, 10 young adults, and 10 elderly people. Clinical features, numbers, age, and gender of the subjects are shown in table 1 ▶. Informed consent was obtained for these investigations which were approved by the Tayside Committee on Medical Research Ethics, Dundee.
Table 1.
Characteristics of patients and subjects used in this study
| Biopsies | Stools | ||||
| UC patients (n = 10) | Controls (n = 10) | Infants (n = 10) | Adults (n = 10) | Elderly (n = 10) | |
| Age (y) | |||||
| Range | 28–65 | 19–76 | 22 days–23 mo | 20–35 | 64–83 |
| Mean (SEM) | 50.2 (2.8) | 50.8 (4.7) | 1.01 (0.18) | 26.7 (1.2) | 71.7 (1.2) |
| Sex (M/F) | 4/6 | 4/6 | 5/5 | 3/7 | 0/10 |
| UC duration (y)* | 9.2 (1.6) | – | – | – | – |
| Therapy for UC | – | – | – | ||
| None | 2 | – | – | – | – |
| Balsalazide | 2 | – | – | – | – |
| Salazopyrine | 2 | – | – | – | – |
| Minocycline | 1 | – | – | – | – |
| Asacol | 3 | – | – | – | – |
*Values are mean (SEM).
UC, ulcerative colitis.
Primer design
Primer Premier for Windows version 5.0 (Premier Biosoft International, Palo Alto, California, USA) was used for searching, aligning, editing, and handling primers for collected bacterial sequences. The Ribosomal Database Project was used for screening specific primer sets. Desulfovibrio specific primers were designed on the basis of 16S rRNA gene sequences, available at the National Center for Biotechnology Information databases. A wide range of bacterial species was tested to confirm the specificity of the developed primers to target bacteria using conventional PCR. The forward primer sequence was DSV691-F: 5′-CCGTAGATATCTGGAGGAACATCAG-3′, and the reverse primer sequence was DSV826-R: 5′-ACATCTAGCATCCATCGTTTACAGC-3′. Designed primers were purchased from Invitrogen Life Technologies (Paisley, UK).
DNA extraction from bacterial cultures
Intestinal isolates and a range of culture collection type strains were used as controls in testing specificities of the PCR primers. SRB were grown in Postgate’s medium B in Universal bottles,12 and other organisms were cultured on Wilkins-Chalgren agar plates in an anaerobic chamber (atmosphere N2 80%; CO2 10%; H2 10%) at 37°C. Cell pellets from liquid media or bacterial colonies from plate swabs were resuspended in 450 μl of sterile water and 50 μl of lysozyme (50 mg/ml), which was added to digest bacterial cell walls. Reaction mixtures were incubated in a water bath at 37°C for 30 minutes. Proteinase K solution l (25 μl of 20 mg/ml), 50 μl of 20% sodium dodecyl sulphate, 500 μl of H2O, and 350 mg glass beads (0.1 mm diameter) were then added, and the mixture bead beaten before and after incubation in a water bath at 70°C for 10 minutes. Total DNA was obtained after centrifugation at 5000 g (three minutes).
DNA extraction and purification from biopsy material and stools
Frozen tissue sections were extracted using QIAmp DNA spin columns (Qiagen Ltd, West Sussex, UK). Briefly, biopsy specimens (2–5 mg) were suspended in lysis buffer and lysozyme solutions before incubating at 55°C. Proteinase K, buffer ATL, and ethanol were sequentially added, and the mixture bead beaten before and after incubation at 70°C. Bacterial cell lysates were obtained by centrifugation at 5000 g (three minutes).
Stool samples were extracted for bacterial DNA using a Qiagen Stool Kit. Frozen stool samples were weighed in 200 mg quantities, and 1.4 ml of ASL buffer (preheated at 95°C) was added. Mixtures were bead beaten for two minutes before and after heating at 95°C (five minutes). Supernatants containing bacterial DNA were collected after centrifugation at 5000 g (two minutes). Inhibitex tablets were used to remove faecal debris, and bacterial cell lysis was completed by adding 25 μl of proteinase K solution and 600 μl of buffer AL, and incubating at 70°C for 10 minutes.
Biopsy and faecal DNA lysates were adsorbed onto minicolumn membranes, washed, and eluted according to their respective Qiagen kit procedures. The purified DNA was eluted in 200 μl of elution buffer.
Amplification in conventional PCR
For conventional PCR, a Techne Genius PCR machine (Techne Ltd, Duxford, UK) was used. PCR consisted of 35 cycles of initial DNA denaturation step at 95°C (one minute), followed by annealing at 62°C (one minute), and elongation at 72°C (45 seconds). The procedure was completed with a final elongation step at 72°C (five minutes). Determinations of optimum temperature were done using a Mastercycler Gradient PCR machine (Eppendorf, Hamburg, Germany).
Purification and cloning of PCR products
Reaction products (amplicons) were purified using QIAquick spin PCR purification kit (Qiagen) according to the manufacturer’s instructions. PCR products were eluted in 30 μl. Purified PCR products were then used for sequencing and cloning. PCR products were ligated into the pGEMT Vector, as specified by Promega (Madison, USA). Competent E coli JM109 cells were transformed with ligation products by heat shock at 42°C. LB medium supplemented with ampicillin (200 μg/ml) was used for cloning and subculture. Cells containing the correct plasmid insert were confirmed with conventional PCR and sequencing.
Calculation of starting 16S rRNA gene copy numbers
Plasmid DNA was purified using the Wizard Plus SV Minipreps DNA Purification Kit (Promega). Miniprep concentrations were determined by electrophoresis and comparison of band strengths against molecular marker DNA (2 log DNA Ladder N3200L; New England Biolabs Ltd, Herts, UK). DNA concentrations were then converted into 16S rRNA gene copy numbers.
Real time PCR
Quantitation of bacterial DNA was performed using an iCycler Real-Time PCR Detection System (BioRad, Hercules, California, USA). DNA melting curves were used to monitor product specificities. Detection was based on fluorescence resonance energy transfer, with a SYBR Green 490 fluorophore. The cycle number at which the signal was first detected correlated with the original concentration of DNA template, and the starting copy number of amplicons was inversely proportional to the real time threshold cycle.
Analysis of PCR amplification and melting curves was made using iCycler Optical System Software, version 3.0 (BioRad). Conditions included one cycle of 95°C for three minutes followed by 38 cycles of denaturation (95°C) and annealing (30 seconds) at 62°C. The melt curve analysis was run for 43 repeats, increasing by 1°C. Plasmid standards and samples were simultaneously assayed in duplicate.
Bacterial recovery from tissue material
Bacterial recovery tests were conducted to validate DNA extraction and the analysis techniques. Serial dilutions of Dsv desulfuricans (103–107 bacterial cells/ml) were made in anaerobic phosphate buffered saline and added to colonic tissue cut into six subsections to resemble six similar samples of biopsy material. Total DNA was extracted using the Qiagen Tissue Kit procedure. Desulfovibrio DNA recoveries from treated and untreated tissue were then compared with five dilutions of the bacteria with no added tissue.
Statistical analysis
Data analysis was done using the SPSS v9.0 Statistics Package (SPSS, Chicago, USA). Copy numbers of 16S rRNA genes per mg of sample were transformed into logarithms and normally distributed data were subjected to statistical analysis. One way analysis (Tukey multiple comparison) of variance was used for comparison of bacterial densities between different specimens. The χ2 test was used for determining associations of bacteria detection and types of samples.
Chemicals
Unless stated otherwise, all chemicals were obtained from Sigma (Poole, Dorset, UK). Bacteriological culture media were purchased from Oxoid Ltd (Basingstoke, Hampshire, UK).
RESULTS
Development of real time PCR for enumerating intestinal desulfovibrios
A highly specific PCR primer set was developed for quantitating intestinal desulfovibrios in stools and biopsy tissues (table 2 ▶). Desulfovibrio desulfuricans, which had no nucleotide mismatch with the primers, and Dsv vulgaris, which had a mismatch of two nucleotides with the primer pair, showed strong PCR positivity.
Table 2.
Alignment of sequences of amplimers of desulfovibrios with corresponding regions of 16S rRNA genes of related species*†
| Species | Accession | Forward primer | Position | Reverse primer | Position | ||||||||||||||||||||||||||||||||||||||||||||||||
| C | C | G | T | A | G | A | T | A | T | C | T | G | G | A | G | G | A | A | C | A | T | C | A | G | A | C | A | T | C | T | A | G | C | A | T | C | C | A | T | C | G | T | T | T | A | C | A | G | C | ||||
| Desulfovibrios | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Desulfovibrio spp | AF0566089 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | A | G | A | C | 677 | T | G | T | A | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 812 |
| Desulfovibrio aminophilus | AF067964 | G | G | C | A | T | C | T | A | T | A | T | A | C | C | T | C | C | T | T | G | T | C | G | A | C | 688 | T | G | T | C | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 823 |
| Desulfovibrio oryzae | AF273083 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | A | G | A | C | 696 | T | G | T | C | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 831 |
| Desulfovibrio desulfuricans | AF354664 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | A | G | A | C | 691 | T | G | T | A | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 826 |
| Desulfovibrio gigantus | AF418170 | G | G | C | A | T | C | T | A | T | A | G | C | C | C | T | C | C | T | T | G | T | C | G | G | C | 698 | T | G | T | C | G | A | T | C | T | G | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 833 |
| Desulfovibrio vulgaris | AF418179 | G | G | C | A | T | C | T | G | T | A | G | A | C | C | T | C | C | T | T | G | T | A | G | T | C | 695 | T | G | T | C | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | G | C | G | 830 |
| Desulfovibrio intestinalis | AJ295680 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | A | G | T | C | 567 | T | G | T | A | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 702 |
| Desulfovibrio magneticus | D43944 | G | G | C | A | T | C | T | A | T | A | G | C | C | C | T | C | C | T | T | G | T | C | G | G | C | 698 | T | G | T | C | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 833 |
| Desulfovibrio gigas | M34400 | A | A | A | C | C | T | C | A | G | A | G | A | C | C | T | C | C | T | T | A | G | A | A | T | C | 435 | T | R | T | C | N | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 839 |
| Desulfovibrio fairfieldensis | U2221 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | A | G | T | C | 677 | T | G | T | A | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 812 |
| Desulfovibrio gabonensis | U31080 | G | G | C | A | T | C | T | A | T | A | G | G | C | C | T | C | C | T | T | G | T | C | G | T | C | 653 | T | G | T | C | G | A | T | A | G | G | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 788 |
| Desulfovibrio bastinii | U53462 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | C | G | T | C | 658 | T | G | T | C | G | A | T | C | T | T | A | G | G | T | A | G | C | N | N | N | N | G | T | C | G | 793 |
| Desulfovibrio gracilis | U53464 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | C | G | T | C | 658 | T | G | T | C | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 793 |
| Desulfovibrio caledonensis | U53465 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | C | G | G | C | 658 | T | G | T | C | G | A | T | A | T | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 793 |
| Desulfovibrio termitidis | X87409 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | A | G | T | C | 676 | T | G | T | C | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 811 |
| Desulfovibrio vietnamensis | X93994 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | C | G | G | C | 676 | T | G | T | C | G | A | T | C | T | T | A | G | G | T | A | G | C | A | A | A | T | G | G | C | G | 811 |
| Desulfovibrio aespoeensis | X95230 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | C | G | G | C | 682 | T | G | T | C | G | A | T | A | T | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 817 |
| Desulfovibrio africanus | X99236 | G | G | C | A | T | C | T | A | T | A | G | G | C | C | T | C | C | T | T | G | T | C | G | T | C | 676 | T | G | T | C | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | G | C | T | 811 |
| Desulfovibrio halophilus | X99237 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | C | G | G | C | 673 | T | G | T | C | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | G | C | G | 808 |
| Desulfovibrio indonensis | Y09504 | G | G | C | A | T | C | T | A | T | A | G | G | C | C | T | C | C | T | T | G | T | C | G | T | C | 698 | T | G | T | C | G | A | T | A | G | G | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 833 |
| Desulfovibrio alaskensis | Y11984 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | C | G | C | C | 677 | T | G | T | C | G | A | T | C | T | T | A | G | G | T | A | G | C | A | A | A | T | G | G | C | G | 812 |
| Desulfovibrio zosterae | Y18049 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | C | G | T | C | 682 | T | G | T | A | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 817 |
| Desulfovibrio longreachensis | Z24450 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | A | G | T | C | 673 | T | G | T | C | G | A | T | C | G | T | A | G | G | T | A | G | C | A | A | A | T | G | T | C | G | 808 |
| Other SRB genera | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Desulfotomaculum ruminis | M34418 | G | G | C | A | T | C | T | A | T | T | G | A | C | C | T | C | C | T | T | T | T | C | G | T | C | 956 | T | G | T | C | G | A | T | C | G | T | C | T | G | T | A | G | C | A | A | A | T | G | G | C | G | 1091 |
| Desulfobacter sp. | M34416 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | C | G | G | A | 712 | C | T | G | C | C | A | G | C | A | C | A | A | G | C | G | G | C | G | A | A | A | G | T | A | A | 826 |
| Desulfococcus biacutus | AJ277887 | G | G | C | A | T | C | T | A | T | A | G | A | C | C | T | C | C | T | T | G | T | C | G | G | C | 947 | T | G | T | A | G | A | G | C | C | T | C | C | G | A | A | C | G | C | T | G | G | A | G | C | G | 1518 |
| Desulfobulbus propionicus | M34410 | G | G | C | A | T | C | T | A | T | A | G | G | C | C | T | C | C | T | T | T | T | C | G | G | C | 708 | G | G | T | A | G | A | T | C | T | T | C | A | G | T | A | G | C | A | A | A | T | G | T | C | G | 843 |
| Desulfobacter vibrioformis | U12254 | G | G | C | A | T | C | T | A | T | A | G | T | C | C | T | C | C | T | T | G | T | G | G | C | T | 694 | T | C | T | G | G | T | C | G | A | T | T | G | G | T | A | G | C | A | A | C | G | G | A | A | C | 1284 |
| Desulfococcus multivorans | M34405 | C | G | C | A | T | C | T | A | T | A | G | T | C | C | T | C | C | T | T | G | T | G | G | C | C | 713 | C | C | T | C | T | A | G | C | G | A | N | G | G | G | A | N | N | C | A | T | G | T | C | G | None | |
*Nucleotides that are different from the target sequence for each organism are shown in underlined bold face. Primer alignment was done using Primer Premier Software.
†Sequence position according to 16S rRNA gene in E coli.
The two strains of Dsv desulfuricans and Dsv vulgaris yielded a single electrophoretic band (135 bp amplicon) when amplified by conventional PCR. Colonic bacteria such as bacteroides, clostridia, bifidobacteria, enterococcus, escherichia, faecalibacterium, eubacteria, shigella, salmonella, veillonella, peptostreptococcus, and lactobacillus gave no bands (fig 1 ▶). Other SRB known to be present in the human large intestine, in lower numbers than desulfovibrios (desulfotomaculum, desulfobacter, desulfococcus), were tested for specificity of the primers. After 35 cycles of PCR amplification (annealing temperature 54°C) only desulfovibrios were detected (fig 2A ▶) at about 20 ng of genomic DNA. Temperature gradient PCR showed an optimum annealing temperature of 62°C while no false positive amplifications were found over a wide range of annealing temperatures. As indicated by real time PCR, the negative derivative of fluorescence versus temperature (−d(RFU)/dT), over the course of PCR product melt (fig 2B ▶), generated a specific peak for desulfovibrios at 89 (1)°C. The average efficiency of real time PCR from several experiments was found to be 97% (fig 3 ▶).
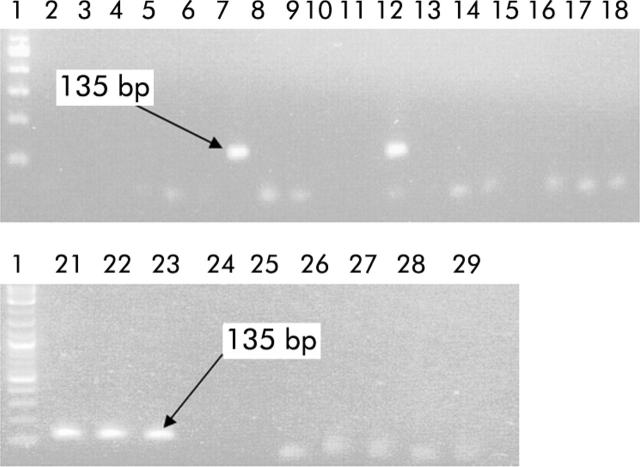
Figure 1.
Specificity of the desulfovibrio polymerase chain reaction (PCR) assay. Electrophoresis was done on an agarose (2% w/v) gel with PCR products obtained with primer pair DSV691-F and DSV826-R, and genomic DNA from various bacterial templates. DNA ladder (100 bp) (lane 1), Bacteroides fragilis NCTC 9343 (lane 2), Faecalibacterium prausnitzii ATCC 27768 (lane 3), Clostridium difficile NTCC 11223 (lane 4), C histolyticum DSM 2158 (lane 5), Lactobacillus acidophilus DSM 20079 (lane 6), E coli ATCC 11775 (lane 7), Desulfovibrio desulfuricans NCIMB 12833 (lane 8), Bifidobacterium catenulatum NCIMB 702246 (lane 9), Bif bifidum NCIMB 702715 (lane 10), Bif breve NCTC 11815 (lane 11), Faecalibacterium prausnitzii NCIMB 13872 (lane 12), Desulfovibrio vulgaris NCIMB 8303 (lane 13), Eubacterium limosum DUN-112 (lane 14), C clostridioforme DUN-120 (lane 15), C malenominatum DUN-102 (lane 16), Eubacterium aerofaciens DUN-207 (lane 17), Clostridium perfringens NCTC 8346 (lane 18). Desulfovibrio desulfuricans NCIMB 12833 (lane 21), Dsv vulgaris NCIMB 8303 (lane 22), Dsv desulfuricans NCIMB 8307 (lane 23), Desulfotomaculum ruminis NCIMB 8452 (lane 24), Desulfococcus multivorans NCIMB 12965 (lane 25), Desulfobacter vibrioformis NCIMB 13525 (lane 26), Dsb vibrioformis NCIMB 13525 (obtained as DNA) (lane 27), Desulfotomaculum ruminis DSM 2154 (lane 28), Dsm ruminis-Reading (lane 29), negative control (lane 30). PCR positivity is indicated by the presence of a 135 bp PCR product.
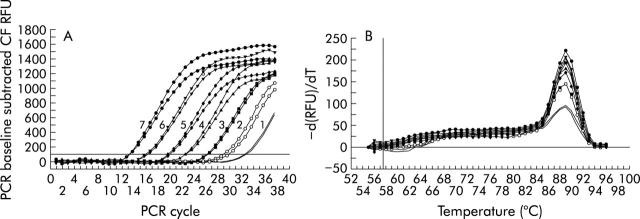
Figure 2.
Post-amplification melt curve analysis of desulfovibrios. Fluorescence of SYBR Green with the progress of amplicon cycles is shown in (A). Values are duplicates of different dilutions of the plasmid used as standard. 16S rRNA gene copy numbers ranging from log10 1.0 to 7.0 are shown in duplicate. The rate of change of fluorescence against temperature is shown in (B). The peak (T = 89 (1)) is specific to the genus Desulfovibrio. Symbols and lines correspond to amplifications shown in (A). PCR, polymerase chain reaction; CF, curve fit; RFU, relative fluorescent unit.
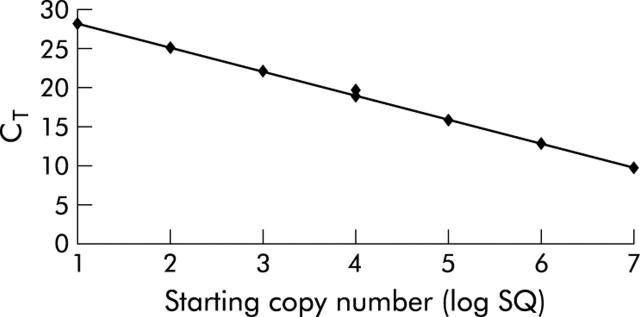
Figure 3.
Quantitation of desulfovibrios. A standard curve for desulfovibrios was constructed with 10-fold dilution of plasmid from 101–107 copies. Threshold cycle (CT) is plotted against the starting copy number. Values on the line represent the result of duplicates. The equation for the curve is: CT = −3.425logSQ+33.16945 (r = 0.997). The efficiency of the determination was calculated to be 97%.
Recovery of desulfovibrio DNA from tissue sections
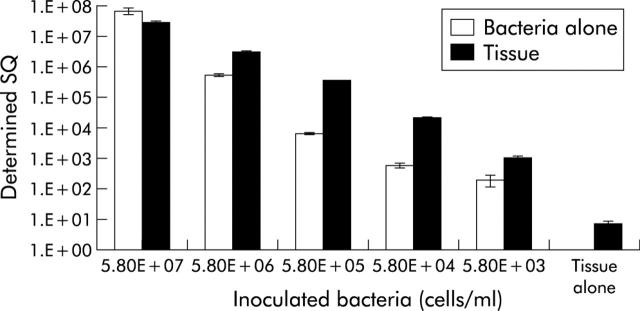
The effect of tissue material on recovery of desulfovibrio DNA is shown in fig 4 ▶. Untreated tissue sections harboured desulfovibrios and consequently, with the exception of the 107 inoculum, bacterial DNA was extracted from tissue in higher amounts than in controls. Results showed a good correlation between numbers of added bacteria and DNA recoveries from tissue, demonstrating high efficiencies in the extraction methods and real time PCR assay procedures. Irrespective of the amount of added bacteria, different tissue sections had variable bacterial densities due to differences in tissue size.
Figure 4.
Recovery of desulfovibrio DNA added to tissue samples by real time polymerase chain reaction (PCR). The target bacterium (Desulfovibrio desulfuricans) was added in 10-fold dilutions from a culture grown in Postgate’s medium B to colonic tissue. The tissue was cut into six subsamples and each was treated with the bacterial preparation before extraction of genomic DNA. Means of triplicate copy number of 16S rRNA genes were plotted against bacterial cell numbers (mean (SEM)). A sample without added sulphate reducing bacteria was used to determine whether the original tissue samples contained desulfovibrios.
Measurements of desulfovibrios in stools and biopsies
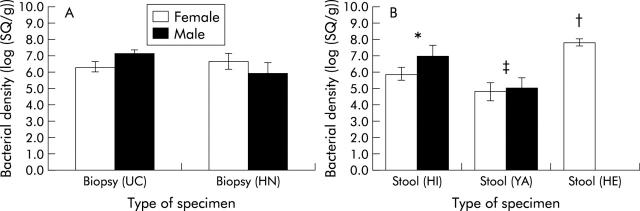
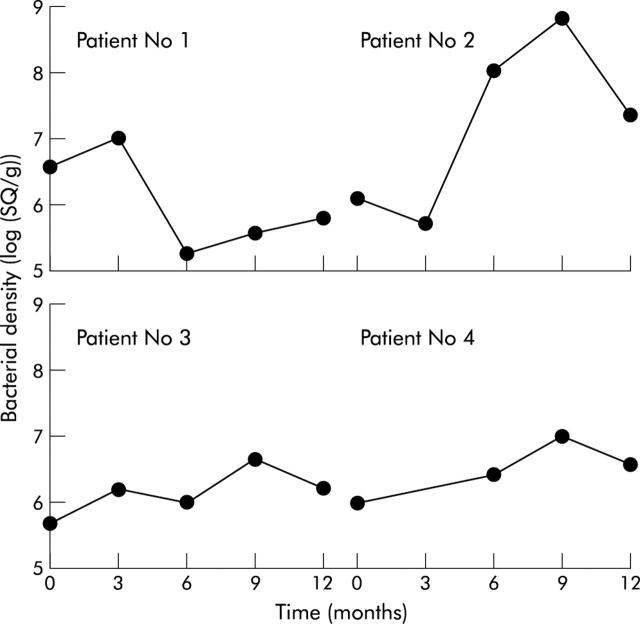
Quantitation of intestinal desulfovibrios in UC and healthy rectal biopsies, and stools from three different population age groups is shown in fig 5A and B ▶. In principle, the sensitivity of the method was about 10 bacteria, but median values were between 105 per gram of sample in healthy young adults to 108 in the elderly. The lowest prevalence of desulfovibrios was observed in faecal samples from healthy young adults (7/10) whose stools also had the lowest numbers of desulfovibrios. These organisms were found in 8/10 normal biopsy samples and 9/10 UC biopsies. Eight of the 10 UC patients were receiving therapy, and no real differences were seen in SRB counts (no therapy, n = 2, 6.9 (0.08)/g biopsy; balsalazide, n = 2, 7.1 (0.22); salazopyrine, n = 2, 6.9 (0.35); minocycline, n = 1, 6.59; Asacol, n = 3, 6.1 (0.67)). There were also no significant differences in SRB counts between UC and healthy rectal biopsies. To assess the stability of mucosal desulfovibrios, bacteria were studied over a 12 month period in four UC patients (fig 6 ▶). Results showed great variations in numbers in two of the patients, over several orders of magnitude, whereas populations were stable in the other two individuals.
Figure 5.
Quantitation of desulfovibrios from biopsies (A) and different stool specimens (B). Bacterial density is expressed as copy number of 16S rRNA genes for desulfovibrios (SQ) per g of biopsy or stool. Biopsies from patients with ulcerative colitis (UC) and biopsies from healthy normal adults (HN), and faeces from healthy infants (HI), healthy young adults (YA), and healthy elderly people (HE) are shown. Values are mean (SEM) for each group. Significant difference, irrespective of sex: *p = 0.008 compared with stool YA; †p = 0.021 compared with stool HI; and ‡p = 0.000 compared with stool HE.
Figure 6.
Stability of desulfovibrio populations on the rectal mucosa in four patients with ulcerative colitis.
All stools from healthy elderly people and infants contained desulfovibrios, and individuals in these groups also had the highest numbers of SRB in their faeces. The χ2 test showed that these associations were statistically significant (Pearson χ2 = 16; p = 0.003). Comparisons made between the mean values of bacterial cell population density and one way analysis of variance showed significant differences. The main difference related to the high levels of desulfovibrios in stool samples from healthy elderly people. However, there were no significant differences between males and females in the groups containing both sexes, with the exception of slightly higher male counts than female values in UC biopsy tissue which were just below statistical significance (p = 0.068).
DISCUSSION
Desulfovibrios are the predominant SRB in the human large intestine.4,22 In addition to their links with IBD,3–7 these bacteria are involved in a number of important processes in the bowel, including hydrogen disposal and other nutrient cross feeding reactions.2,23–25 In view of their metabolic significance in the gut, it is important that reliable technologies are available for their detection in faeces and mucosal tissue.
Investigating the human colonic microbiota using culturing techniques has certain limitations, such as the requirement for samples to be processed quickly, the need for specialised anaerobic techniques, and low cell recoveries, while bacterial isolations and identifications are time consuming and prohibitive for large scale studies. However, current developments in molecular analysis have overcome many of these restrictions, and the design and employment of carefully evaluated primers is proving to be of value in identifying bacteria in complex microbial communities.26 Nevertheless, probes developed for a particular group of bacteria may also detect unrelated organisms.20,27,28
The PCR primers developed here specifically detected and quantitated desulfovibrios in biopsy material and faeces. The method provided a highly sensitive means of identifying and quantitating these organisms, and facilitated high throughput bacterial analysis of biological samples. Tests using a wide range of gut microorganisms showed that the desulfovibrio primers did not target other bacteria (table 2 ▶, fig 1 ▶), including other intestinal SRB.29 Real time PCR analysis had no secondary peaks or peaks corresponding to primer dimers (fig 2A ▶) while the plasmid derived PCR amplicon was successfully diluted to copy numbers in the order of units, without loss of linearity (fig 3 ▶). Lysis of the cells by heat treatment,30,31 mechanical disruption, and enzymic and chemical extraction favoured recovery of bacterial DNA.
Real time PCR showed that desulfovibrios were ubiquitous in faeces, irrespective of age (fig 5B ▶). However, there was an age related association with respect to numbers of these bacteria in stools. In particular, faeces from older people (64–83 years) showed significantly higher numbers of desulfovibrios compared with their younger counterparts (age range 20–35 years). This may reflect differences in sulphate availability through diet or endogenous sources, such as chondroitin sulphate and mucins,32 as well as being a reflection of slower intestinal transit times in older people because SRB are relatively slow growing bacteria and this would facilitate proliferation of these organisms.3 The occurrence of desulfovibrios in infants was very surprising because young children have immature developing microbiota33 while SRB have a specialised mode of energy generation that makes them dependent on other intestinal microorganisms for their growth requirements.34
As well as occurring in high numbers in faeces, desulfovibrios heavily colonised mucosal tissue (fig 5A ▶, fig 6 ▶). Although rectal biopsies from UC patients had generally higher numbers of desulfovibrios than healthy people, the differences were not statistically significant. This would suggest that if these bacteria play a role in UC, some host defect, possibly in sulphide detoxication pathways or in bacterial antigen handling, is required for disease to occur. Once inflammatory processes have been initiated, or the mucosal antibacterial response compromised, desulfovibrios may produce antigens that could damage the integrity of epithelial cells,5 in addition to sulphide. However, sulphide is thought to be the single most important cytotoxic bacterial metabolite where inhibition of butyrate oxidation in epithelial cells has been demonstrated in UC patients.35,36
Interestingly, although the patient numbers involved in the present investigation were low, anti-inflammatory drugs, which have previously been linked to inhibition of faecal SRB in UC,37 appeared to have little effect on mucosal populations, which may reflect the inherent resistance of bacteria growing in biofilms to antimicrobial substances.38 The potential link between mucosal SRB and UC is intriguing. Studies are now needed to determine how these biofilm communities interact with host immunity and metabolic processes in the mucosa.
Acknowledgments
This work was supported by the Medical Research Council and the Scottish Executive Government Chief Scientist’s Office.
Abbreviations
IBD, inflammatory bowel disease
rRNA, ribosomal RNA
PCR, polymerase chain reaction
SRB, sulphate reducing bacteria
UC, ulcerative colitis
REFERENCES
- 1.Cummings JH, Macfarlane GT, Macfarlane S. Intestinal bacteria and ulcerative colitis. Curr Iss Intest Microbiol 2003;4:9–20. [PubMed] [Google Scholar]
- 2.Gibson GR, Macfarlane S, Macfarlane GT. Metabolic interactions involving sulphate-reducing and methanogenic bacteria in the human large intestine. FEMS Microbiol Ecol 1993;12:117–25. [Google Scholar]
- 3.Gibson GR, Cummings JH, Macfarlane GT. Growth and activities of sulphate-reducing bacteria in gut contents from healthy subjects and patients with ulcerative colitis. FEMS Microbiol Ecol 1991;86:103–12. [Google Scholar]
- 4.Loubinoux J, Bronowicji J-P, Pereira IAC, et al. Sulfate-reducing bacteria in human feces and their association with inflammatory diseases. FEMS Microbiol Ecol 2002;40:107–12. [DOI] [PubMed] [Google Scholar]
- 5.Zinkevich V, Beech IB. Screening of sulfate-reducing bacteria in colonoscopy samples from healthy and colitic gut mucosa. FEMS Microbiol Ecol 2000;34:147–55. [DOI] [PubMed] [Google Scholar]
- 6.Pitcher MCL, Beatty ER, Harris RM, et al. Sulfur metabolism in ulcerative colitis. Investigation of detoxification enzymes in peripheral blood. Dig Dis Sci 1998;43:2080–5. [DOI] [PubMed] [Google Scholar]
- 7.Roediger WEW, Duncan A, Kapaniris O, et al. Sulphide impairment of substrate oxidation in rat colonocytes: A biochemical basis for ulcerative colitis? Clin Sci 1993;85:1–5. [DOI] [PubMed] [Google Scholar]
- 8.Roediger WEW, Duncan A, Kapaniris O, et al. Reducing sulfur compounds of the colon impair colonocyte nutrition: implications for ulcerative colitis. Gastroenterology 1993;104:802–9. [DOI] [PubMed] [Google Scholar]
- 9.Gardiner KR, Halliday MI, Barclay GR, et al. Significance of systemic endotoxaemia in inflammatory bowel disease. Gut 1996;36:897–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christl S, Eisner HD, Kasper H, et al. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa: a potential role for these agents in the pathogenesis of ulcerative colitis. Dig Dis Sci 1996;41:2477–81. [DOI] [PubMed] [Google Scholar]
- 11.Pitcher MCL, Beatty ER, Cummings JH. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 2000;46:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macfarlane GT, Gibson GR. Sulphate-reducing bacteria. In: Levett PN, ed. Anaerobic microbiology—a practical approach. Oxford: Oxford University Press, 1991:201–22.
- 13.Schultsz C, van den Berg FM, Ten Kate FW, et al. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology 1999;117:1089–97. [DOI] [PubMed] [Google Scholar]
- 14.Hopkins MJ, Sharp R, Macfarlane GT. Age and disease-related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance and community cellular fatty acid profiles. Gut 2001;48:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto T, Morotomi M, Tanaka R. Species-specific oligonucleotide probe for five Bifidobacterium species detected in human intestinal microflora. Appl Environ Microbiol 1992;58:4076–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreader CA. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl Environ Microbiol 1995;61:1171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kok RG, de Waal A, Schut F, et al. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl Environ Microbiol 1996;62:3668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R, Cao W, Cerniglia CE. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol 1996;62:1242–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin C, Raskin L, Stahl DA. Microbial community structure in gastrointestinal tracts of domestic animals: comparative analyses using rRNA-targeted oligonucleotide probes. FEMS Microbiol Ecol 1997;22:281–94. [Google Scholar]
- 20.Kaufmann P, Pfefferkorn A, Teuber M, et al. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA targeted probes by colony hybridization and PCR. Appl Environ Microbiol 1997;63:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furrie E, Macfarlane S, Cummings JH, et al. Systemic antibodies towards mucosal bacteria in ulcerative colitis and Crohn’s disease differentially activate the innate immune response. Gut 2004;53:92–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson GR, Macfarlane GT, Cummings JH. Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate-reduction to methanogenesis in the large gut. J Appl Bacteriol 1988;65:103–11. [DOI] [PubMed] [Google Scholar]
- 23.Gibson GR, Cummings JH, Macfarlane GT. Competition for hydrogen between sulphate-reducing bacteria and methanogenic bacteria from the human large intestine. J Appl Bacteriol 1988;65:241–7. [DOI] [PubMed] [Google Scholar]
- 24.Gibson GR, Cummings JH, Macfarlane GT, et al. Alternative pathways for hydrogen disposal during fermentation in the human colon. Gut 1990;31:679–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson GR, Macfarlane GT, Cummings JH. Sulphate-reducing bacteria and hydrogen metabolism in the human large intestine. Gut 1993;34:437–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alm EW, Oerther DB, Larsen N, et al. The oligonucleotide probe database. Appl Environ Microbiol 1996;62:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manz W, Amann R, Ludwig W, et al. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacterium-bacteroides in the natural environment. Microbiology 1996;142:1097–2006. [DOI] [PubMed] [Google Scholar]
- 28.Langendijk PS, Schut F, Jansen GJ, et al. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol 1995;61:3069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibson GR, Macfarlane GT. Chemostat enrichment of sulphate-reducing bacteria from the large gut. Lett Appl Microbiol 1988;7:127–33. [Google Scholar]
- 30.Ashimoto A, Chen C, Bakker I, et al. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol Immunol 1996;11:266–73. [DOI] [PubMed] [Google Scholar]
- 31.Papapanou PN, Madianos PN, Dahlen G, et al. “Checkerboard” versus culture: a comparison between two methods for identification of subgingival microbiota. Eur J Oral Sci 1997;105:389–96. [DOI] [PubMed] [Google Scholar]
- 32.Macfarlane GT, Gibson GR. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine. In: Mackie RI, White BA, eds. Ecology and physiology of gastrointestinal microbes, vol 1: Gastrointestinal fermentations and ecosystems. New York: Chapman and Hall, 1996:269–318.
- 33.Bullen CL, Tearle PV, Willis AT. Bifidobacteria in the intestinal tract of infants: an in vivo study. J Med Microbiol 1976;9:325–27. [DOI] [PubMed] [Google Scholar]
- 34.Newton DF, Cummings JH, Macfarlane S, et al. Growth of a human intestinal Desulfovibrio desulfuricans in continuous cultures containing defined populations of saccharolytic and amino acid fermenting bacteria. J Appl Microbiol 1998;85:372–80. [DOI] [PubMed] [Google Scholar]
- 35.Roediger WEW, Moore J, Babidge W. Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig Dis Sci 1997;42:1571–9. [DOI] [PubMed] [Google Scholar]
- 36.Babidge W, Millard S, Roediger WEW. Sulfides impair short chain fatty acid beta oxidation at acyl-CoA dehydrogenase level in colonocytes: implications for ulcerative colitis. Mol Cell Biochem 1998;181:117–24. [DOI] [PubMed] [Google Scholar]
- 37.Pitcher MC, Beatty ER, Cummings JH. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 2000;46:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anwar H, Dasgupta MK, Costerton JW. Testing the susceptibility of bacteria in biofilms to antibacterial agents. Antimicrob Agents Chemother 1990;34:2043–6. [DOI] [PMC free article] [PubMed] [Google Scholar]