Abstract
Background: Chronic ethanol consumption is associated with an increased risk of upper aerodigestive tract cancer. As acetaldehyde seems to be a carcinogenic factor associated with chronic alcohol consumption, alcoholics with the alcohol dehydrogenase (ADH) 1C*1 allele seem to be particularly at risk as this allele encodes for a rapidly ethanol metabolising enzyme leading to increased acetaldehyde levels. Recent epidemiological studies resulted in contradictory results and therefore we have investigated ADH1C genotypes in heavy alcohol consumers only.
Methods: We analysed the ADH1C genotype in 107 heavy drinkers with upper aerodigestive tract cancer and in 103 age matched alcoholic controls without cancer who consumed similar amounts of alcohol. Genotyping of the ADH1C locus was performed using polymerase chain reaction based on restriction fragment length polymorphism methods on leucocyte DNA. In addition, ethanol was administered orally (0.3 g/kg body weight) to 21 healthy volunteers with the ADH1C*1,1, ADH1C*1,2, and ADH1C*2,2 genotypes, and 12 volunteers with various ADH genotypes consumed ethanol ad libitum (mean 211 (29) g). Subsequently, salivary acetaldehyde concentrations were measured by gas chromatography or high performance liquid chromatography.
Results: The allele frequency of the ADH1C*1 allele was found to be significantly increased in heavy drinkers with upper aerodigestive tract cancer compared with age matched alcoholic controls without cancer (61.7% v 49.0%; p = 0.011). The unadjusted and adjusted odds ratios for all cancer cases versus all alcoholic controls were 1.67 and 1.69, respectively. Healthy volunteers homozygous for the ADH1C*1 allele had higher salivary acetaldehyde concentrations following alcohol ingestion than volunteers heterozygous for ADH1C (p = 0.056) or homozygous for ADH1C*2 (p = 0.011).
Conclusions: These data demonstrate that heavy drinkers homozygous for the ADH1C*1 allele have a predisposition to develop upper aerodigestive tract cancer, possibly due to elevated salivary acetaldehyde levels following alcohol consumption.
Keywords: alcohol dehydrogenase, upper aerodigestive tract cancer, acetaldehyde, saliva
Excessive chronic alcohol consumption is a major risk factor for the development of upper aerodigestive tract cancer (UADTC).1–9 As the majority of heavy drinkers smoke and only a minority (10–20%) develop UADTC,10 constitutional factors, predisposing some alcoholics to develop these tumours, may be of importance.
Although the mechanisms of ethanol associated carcinogenesis are not known, there is increasing evidence that acetaldehyde (AA) rather than ethanol itself is responsible for the cocarcinogenic effect of alcohol.11 AA interferes with DNA synthesis and repair at many sites, and consequently promotes tumour development.11–18 It causes point mutations in certain genes, induces sister chromatide exchanges, and gross chromosomal aberrations.19–25 AA also induces inflammation and metaplasia of the tracheal epithelium,18,26,27 a delay in cell cycle progression,28 stimulation of apoptosis,28 and enhanced cell injury associated with hyperregeneration.29–31 Moreover, when inhaled, it causes nasopharyngeal and laryngeal carcinoma.17,18 According to the International Agency for Research on Cancer, there is sufficient evidence to identify AA as a carcinogen in animals.32
AA is predominantly produced from ethanol by alcohol dehydrogenase (ADH) and is further metabolised to AA by aldehyde dehydrogenase (ALDH).33 Both enzymes exhibit a genetic polymorphism which influences the rate of conversion of ethanol to AA and of AA to acetate, thereby affecting AA levels in the body.33,34 A high percentage of Orientals have the inactive form of ALDH2 in which the mutant allele ALDH2*2 encodes an inactive subunit. When the enzyme is inactive, the body fails to metabolise AA rapidly, thus leading to excessive accumulation of AA. In individuals with inactive heterozygous ALDH2*1,2, blood35 and salivary36 AA concentrations are significantly higher than in those with active ALDH2*1,1. Indeed, it was demonstrated that the frequency of the inactive ALDH2*2 allele is significantly increased in alcoholics with cancer of the oral cavity, oropharynx, hypopharynx, larynx, and oesophagus.37,38
As the ALDH2 mutation does not exist in Caucasians, the effect of the ADH1C polymorphism is not overshadowed. The ADH enzyme encoded by the ADH1C*1 allele metabolises ethanol to AA 2.5 times faster than that encoded by the ADH1C*2 allele.33 Although ethanol oxidation largely occurs in the liver, ADH activity is also present in the mucosa of the alimentary tract.39,40 Contradictory results have been reported with respect to the ADH1C genotype and UADTC. While one study from France41 with a relatively small number of patients and another one performed in Puerto Rico,42 a country known for its worldwide highest incidence of oral cancer, reported a positive correlation between ADH1C*1 allele frequency and the risk of developing UADTC, a larger French study43 and a case control study from North Carolina44 and Texas45 did not confirm these findings. It has to be emphasised that in all of these epidemiological studies, individuals with a wide range of alcohol intake were analysed using predominantly interview techniques to quantitate alcohol consumption.
Hence we investigated for the first time the ADH1C genotype and allele frequency in 107 patients with heavy alcohol consumption and various types of UADTC. The data were compared with those of age matched patients consuming similar amounts of ethanol and suffering from other alcohol associated organ damage, such as cirrhosis of the liver or pancreatitis. In addition, we studied the effect of the ADH1C genotype on salivary AA levels as it has been claimed that high salivary AA concentrations in individuals with inactive ALDH 2 following alcohol intake36 is responsible for the increased risk of UADTC observed in these patients.
METHODS
Patients and controls
The study protocol was approved by the ethics committee of the University Hospitals in Mannheim and Heidelberg and all patients provided written informed consent.
Genotyping of ADH1C was performed on the serum of 107 Caucasians (89 males, mean age 58 (10) years; 18 females, mean age of 60 (12) years) with UADTC treated at the ENT Hospital Mannheim, University of Heidelberg, Germany. Tumour sites, number of patients, sex, age, alcohol consumption, and smoking habits are listed in table 1 ▶. History of alcohol consumption was obtained by personal interview. Questions also included quantity and duration of alcohol abuse. Verification and completion of the data was done in the pre-anaesthesia questionnaire on a multiple choice basis. For open questions, family members were interviewed.
Table 1.
Characteristics of alcoholic patients with upper aerodigestive tract cancer and alcoholic control subjects
| Group | n | Sex | Age (y) | Alcohol (g/day) | Smoker | ||||
| M | F | >80 | 20–80 | <20 | Yes | No | |||
| Alcoholic cirrhosis | 39 | 23 | 16 | 61 (8) | 39 | 0 | 0 | 35 | 4 |
| Alcoholic pancreatitis | 38 | 33 | 5 | 56 (10) | 38 | 0 | 0 | 37 | 1 |
| Alcoholics without organ injury | 26 | 11 | 15 | 56 (9) | 26 | 0 | 0 | 23 | 3 |
| All alcoholic controls | 103 | 67 | 36 | 58 (9) | 103 | 0 | 0 | 95 | 8 |
| Larynx cancer | 41 | 37 | 4 | 58 (11) | 19 | 22 | 0 | 38 | 3 |
| Oral cancer | 16 | 13 | 3 | 62 (12) | 6 | 10 | 0 | 14 | 2 |
| Hypopharynx cancer | 22 | 18 | 4 | 55 (8) | 11 | 11 | 0 | 20 | 2 |
| Oropharynx cancer | 8 | 5 | 3 | 59 (10) | 3 | 5 | 0 | 8 | 0 |
| Oesophageal cancer | 20 | 16 | 4 | 61 (9) | 14 | 6 | 0 | 20 | 0 |
| All cancers | 107 | 89 | 18 | 59 (11) | 53 | 54 | 0 | 99 | 8 |
An age matched hospital based control group consisted of 39 patients with biopsy proven alcoholic cirrhosis of the liver and a daily alcohol intake of more than 100 g for more than 10 years, 38 patients with alcoholic pancreatitis, and 26 alcohol dependent patients without organ injury who met MS-III-R criteria for alcohol dependence (table 1 ▶).46 All control patients were recruited from the Department of Medicine, Salem Medical Centre (University of Heidelberg), Heidelberg.
In Caucasians, the ADH1C genotype does not have any effect on the development of alcoholic liver cirrhosis.47–49
To prove that the alcohol control group has a similar ADH1C allele frequency as the general population, 48 healthy non-smoking volunteers who consumed less than 70 g of alcohol per week were additionally genotyped.
Genotype determination of ADH1C
Blood samples from 107 patients with cancer were used for analysis of restriction fragment length polymorphism. In four cases, DNA was isolated from tumorous cells, in 56 samples DNA was purified from whole blood leucocytes treated with EDTA or citrate, and 47 DNA samples were obtained from blotted venous blood. DNA isolated from venous blood blotted onto filter paper was also used in all controls. All determinations were performed in duplicate.
For DNA extraction from whole blood, FTA cards were used as blood carriers (Life Technologies, Gibco BRL, Paisley, UK). On these, white blood cells are lysed and nuclear DNA is immobilised within the matrix of the paper. Haeme and other inhibitors of polymerase chain reaction (PCR) amplification are removed by washing. Three pcs 2 mm discs of each FTA paper with dried blood samples were punched out by Harris Micro-Punch (Life Technologies, Gibco BRL). The discs were washed three times with 300 μl FTA purification reagent (Life Technologies, Gibco BRL) and twice with 300 μl of buffer (10 mM Tris HCl, pH 8.0, 0.1 mM EDTA) before being dried at 60°C for 30 minutes.
PCR was used to amplify polymorphic portions of exon 8 of the ADH1C gene50 with specific primers.51 The reactions were assembled in a DNA free environment under a laminar flow hood by use of aerosol resistant barrier tips.
For determination of the ADH1C genotype, the method based on restriction fragment length polymorphism described by Groppi and colleagues51 was used. This is based on allele detection by Ssp I restriction enzyme digestion in combination with PCR directed mutagenesis, which creates a new Ssp I site. Primers 321 and 351 allow amplification of only exon 8 of the ADH1C gene and generate the Ssp I recognition sequence AATATT as an internal control outside of the tested region. Thus it is possible to distinguish between the ADH1C*1 allele with two fragments (67 and 63 bp), the ADH1C*2 allele with a 130 bp fragment, and their possible combinations.
The PCR product (7 μl) was mixed with 7.5 U of Ssp I (New England Biolabs, Beverly, Massachusetts, USA) in Ssp I NEB buffer (50 mM NaCl, 100 mM Tris HCl, 10 mM MgCl2, 0.025% Triton X-100, pH 7.5). Restriction digests were incubated at 37°C for 40 hours with addition of another 5 U of Ssp I restriction enzyme after an incubation period of 24 hours. Negative and positive controls were treated similarly. The whole volume of the digest mixed with 0.1 volume of 10× loading buffer was analysed on 4% Metaphor agarose gel (FMC Bioproducts, Maine, USA) by electrophoresis in 1×TBE buffer (7 V/cm) for 60 minutes. To determine the length of the restriction fragments, a molecular weight marker V (Roche, Mannheim, Germany) was used. After staining with ethidium bromide, photographs were taken under UV light in an Eagle Eye II device (Stratagene, Amsterdam, the Netherlands). Genotyping was performed by independent investigators, twice in both groups of samples, including positive (samples of known genotype) and negative controls (PCR reaction mixture without DNA, restriction digest without DNA).
Oral administration of ethanol to volunteers
Healthy volunteers were screened for the ADH1C genotype according to the method described above. Twenty one healthy volunteers (10 males, 11 females) with ADH1C*1,1 (n = 7; age 38 (8) years), ADH1C*1,2 (n = 7; age 36 (11) years), and ADH1C*2,2 (n = 7; age 34 (8) years) genotypes were examined. None had used any drugs or antiseptic mouthwashes for four weeks prior to the study. All participants were told to abstain from alcohol for at least 48 hours before the start of the study. One hour after a standard breakfast (consisting of one slice of bread with butter and marmalade and one cup of coffee with milk and sugar) ethanol (0.3 g/kg body weight) was administered orally to each volunteer.52 Ethanol diluted in orange juice was given as 5 g/100 ml concentration and administered orally within two minutes. Fifteen minutes prior to ethanol administration, volunteers received an oral antiseptic to reduce oral bacteria known to produce acetaldehyde from ethanol.53 Immediately following ethanol ingestion, volunteers rinsed their mouths carefully with water to remove local ethanol. Blood (3 ml) and saliva (3–5 ml) samples for determination of AA were collected at time 0 and after 10, 20, 40, 60, 100, 130, 160, and 240 minutes following ethanol ingestion. AA measurements were performed immediately at the end of the in vivo experiment.
In addition, 12 healthy Finnish men took part in the ad libitum study. Their age ranged from 18 to 35 years and their mean body weight was 78 (4) kg. This part of the study was approved by the ethics committee of the Department of Medicine, Helsinki University Central Hospital, and informed consent to participate in the study was obtained from all subjects. All studies started at 18:00 hours and volunteers were allowed to eat a light dinner two hours prior to the study. A commercially available paraffin wax chewing gum (Orion Diagnostics, Espoo, Finland) was used to stimulate production of saliva. After baseline saliva collection, each volunteer started ingesting ethanol in a standardised 10% v/v solution of absolute ethanol in orange juice ad libitum. To remove local ethanol, subjects rinsed their mouths three times with water. Subsequently, saliva samples were taken every 30 minutes for up to four hours. Breath ethanol levels were measured simultaneously with saliva collected using an alcometer (Lion Laboratories, Barry, UK) to monitor systemic ethanol concentrations.
Acetaldehyde determination
AA concentrations in blood and saliva were determined using high performance liquid chromatography (HPLC) with fluorescence detection after adduct formation with 2-diphenylacetyl-1,3-indandione-1-hydrazone (DHI), as reported earlier.53 DHI 30 mg were dissolved in 100 ml acetonitrile-methanol (80:20 v/v) by heating at 37°C for 10 minutes and storage at 4°C. Blood or saliva (1.0 ml) was added to 2.0 ml of DHI reagent in a glass tube, capped, and mixed, followed by addition of 30 μl of water. After cooling on ice for five minutes, the mixture was centrifuged at 2800 g for 10 minutes at room temperature. The supernatant was transferred and 30 μl 5 M HCl were added and vortexed. The vial was heated to 37°C for 10 minutes and then kept at room temperature for 20 minutes. After filtration of the prepared sample through a 0.45 μm filter, 200 μl were injected onto the HPLC column (Hypersil-Mos (c6) 250×5 mm; Medchrom, Germany). Calibration curves of AA in blood and saliva were also performed.
In the Finnish study, salivary AA concentrations were determined by gas chromatography, as described previously.54
Statistics
The allele frequencies of the various groups were analysed using Fisher’s exact test. Crude odds ratios were calculated by standard methods. Subsequently, unconditional logistic regression was undertaken to estimate the odds ratios after controlling for daily alcohol consumption (20–100 g, 101–150 g, 151–200, and >200 g), daily cigarette consumption (0, 1–10, 11–20, 21–30, 31–40, and >40), age, and sex.
ANOVA and the Student’s t test were used to detect significant differences in salivary AA concentrations between the ADH1C genotypes studied. Possible correlations were assessed using Pearson’s correlation coefficient. All statistical tests were two sided.
RESULTS
Patients with UADTC and excessive chronic alcohol consumption exhibited a significantly elevated ADH1C*1 allele frequency compared with age matched heavy drinkers without cancer (table 2 ▶). Increased ADH1C*1 allele frequency in cancer patients was seen in both males (60.7% v 52.9%; p = 0.20) and females (66.7% v 41.7%; p = 0.016). This increase in ADH1C*1 allele frequency in cancer patients was due to the increase in ADH 3*1 homozygotes. The highest ADH1C*1 allele frequency was found in patients with oral cancer and cancer of the larynx. Thirty eight per cent of subjects with oral cancer and 37% of those with laryngeal cancer were found to be homozygous for the ADH1C*1 allele whereas only 6% and 7%, respectively, were found to be homozygous for the ADH1C*2 allele. The associated crude odds ratios for all cancer cases versus alcoholic controls was 1.67 (95% confidence interval (CI) 1.13–2.47). A similar association (odds ratio 1.69 (95% CI 1.12–2.56)) appeared after controlling for other risk factors, including alcohol consumption, smoking, age, and sex. No difference in ADH1C genotype was observed between healthy controls and heavy alcohol consuming patients without cancer. ADH1C*1 allele frequency in healthy controls was 50%. Alcoholics without organ injury, patients with alcoholic cirrhosis of the liver, and alcoholic pancreatitis did not differ with respect to ADH1C genotype.
Table 2.
Genotype numbers and allele frequencies (%) of ADH1C in patients with upper aerodigestive tract cancer, healthy controls, and alcoholic control patients without cancer
| Group | n | Genotype | Allele (%) | ||||
| *1/*1 | *1/*2 | *2/*2 | *1 | *2 | |||
| 1 | Alcoholic cirrhosis | 39 | 8 | 23 | 8 | 50.0 | 50.0 |
| 2 | Alcoholic pancreatitis | 38 | 5 | 26 | 7 | 47.4 | 52.6 |
| 3 | Alcoholics without organ damage | 26 | 7 | 12 | 7 | 50.0 | 50.0 |
| 4 | All alcoholic controls without cancer | 103 | 20 | 61 | 22 | 49.0 | 51.0 |
| 5 | Laryngeal cancer | 41 | 15 | 23 | 3 | 64.6 | 35.4 |
| 6 | Oral cancer | 16 | 6 | 9 | 1 | 65.6 | 34.4 |
| 7 | Oropharyngeal cancer | 8 | 2 | 5 | 1 | 56.3 | 43.7 |
| 8 | Hypopharyngeal cancer | 22 | 7 | 11 | 4 | 56.9 | 43.1 |
| 9 | Oesophageal cancer | 20 | 7 | 10 | 3 | 60.0 | 40.0 |
| 10 | All cancers | 107 | 37 | 58 | 12 | 61.7 | 38.3 |
No significant difference in allele frequencies was detected between healthy controls and alcoholic controls without cancer (p = 0.90). However, allele frequency of the ADH1C*1/*1 allele was found to be significantly increased in all alcoholic cancer patients compared with all alcoholic non-cancer patients (p = 0.011).
Ethanol intake and smoking habits were generally excessive in all alcoholic patients, regardless of whether or not they had cancer. No significant correlation was found between the amount of alcohol intake and ADH1C genotype (table 3 ▶) although cancer patients with more than 100 g of alcohol intake per day seemed to have a somewhat higher homozygosity for the ADH1C*1 allele (39.2%) compared with patients consuming 20–50 g of alcohol per day (28.5%).
Table 3.
Alcohol intake and ADH1C genotype in patients with upper aerodigestive tract cancer and controls
| No | Daily alcohol intake (g) | Genotype | Allele (%) | |||
| *1/*1 | *1/*2 | *2/*2 | *1 | *2 | ||
| 11 (8) | >200 | 2 (2) | 9 (2) | 0 (4) | 59.1 (37,5) | 40.9 (62.5) |
| 15 (31) | 150–200 | 5 (2) | 5 (24) | 5 (5) | 50.0 (45.2) | 50.0 (54.8) |
| 25 (64) | 100–150 | 12 (16) | 11 (35) | 2 (13) | 70.0 (52.4) | 30.0 (47.6) |
| 21 (0) | 50–100 | 7 | 11 | 3 | 59.5 | 40.5 |
| 35 (0) | 20–50 | 10 | 23 | 2 | 61.4 | 38.6 |
| 107 (103) | >20 | 37 (20) | 58 (61) | 12 (22) | 61.7 (49.0) | 38.3 (51.0) |
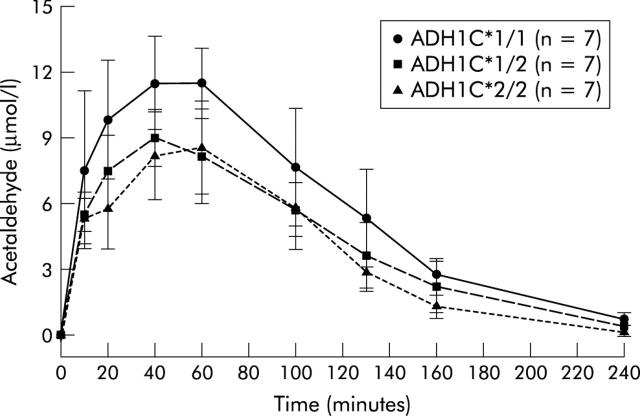
Salivary AA concentrations were significantly modulated by the ADH1C genotype in German as well as in Finnish volunteers. German volunteers homozygous for the ADH1C*1 allele had significantly higher salivary AA concentrations compared with those heterozygous for ADH1C or homozygous for the ADH1C*2 allele (fig 1 ▶). When salivary AA concentrations were compared in volunteers heterozygous for ADH1C and homozygous for ADH1C*2, no significant difference in salivary AA concentrations were detected.
Figure 1.
Effect of alcohol dehydrogenase 1C (ADH1C) polymorphisms on salivary acetaldehyde concentrations following oral ethanol intake (0.3 g/kg body weight). No significant difference in salivary acetaldehyde concentrations was found between subjects with ADH1C*1,2 and ADH1C*2,2. In contrast, subjects with ADH1C*1,1 had higher acetaldehyde concentrations than subjects with ADH1C*1,2 and ADH1C*2,2 when the areas under the acetaldehyde concentration-time curves were compared (p = 0.056 and p = 0.011, respectively). Peak acetaldehyde concentrations determined after 40 minutes were also significantly different (p = 0.033 and p = 0.017, respectively). Values are mean (SD).
Neither blood AA concentrations nor plasma ethanol concentrations were found to vary significantly between the different groups studied (data not shown). It should be noted that the ethanol dose of 0.3 g/body weight resulted in plasma ethanol concentrations not exceeding 30 mg/100 ml.
ADH1C genotyping revealed that four of the Finnish subjects were homozygous for the ADH1C*1 allele (ADH1C*1,1), six were heterozygous (ADH1C*1,2), and the remaining two were homozygous for the ADH1C*2 allele (ADH1C*2,2). No significant differences were observed between these three groups with respect to sex, age, mean weight, smoking habits, or alcohol consumption.
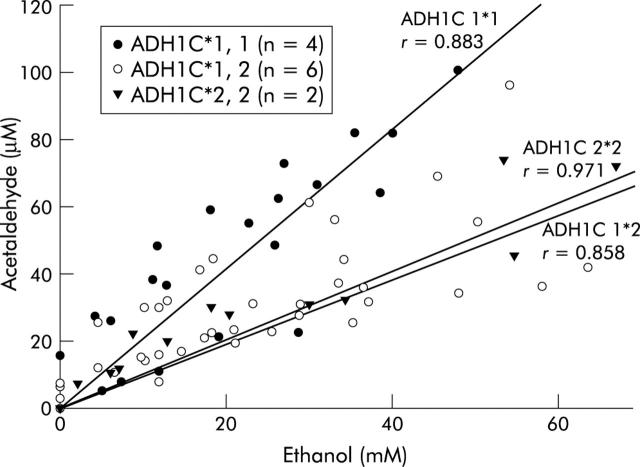
The mean amount of absolute alcohol consumed in all groups in four hours was 211 (29) g. A significant (p<0.001) correlation between salivary AA and ethanol levels was found in all subjects whereby subjects with the ADH1C*1,1 genotype produced significantly more salivary AA than the other genotypes (fig 2 ▶). Slopes in the curves in the different groups were: 2.06 (ADH1C*1,1), 0.95 (ADH1C*1,2), and 1.01 (ADH1C*2,2).
Figure 2.
Correlation between salivary acetaldehyde and ethanol in different alcohol dehydrogenase 1C (ADH1C) genotypes after alcohol intake ad libitum. (r = Pearson’s correlation coefficient). Subjects with ADH1C*1,1 had higher salivary acetaldehyde concentrations than subjects with ADH1C*1,2 and ADH1C*2,2.
DISCUSSION
The results of the present study suggest that heavy alcohol consumers homozygous for the ADH1C*1 allele have an increased risk of developing alcohol associated UADTC. In contrast with other studies investigating the effect of ADH1C genotype on the risk of UADTC, we compared for the first time well matched heavy drinkers with and without cancer. Our findings are in accordance with those reported by Harty and colleagues42 and Coutelle and colleagues.41 The epidemiological study by Harty et al was performed in Puerto Rico, a country with the highest rate of oral cancer in the world. Therefore, factors other than the ADH1C genotype may modulate ethanol associated oral and pharyngeal cancer, such as the type and contaminants of alcoholic beverages or additional dietary habits. It is interesting to note however that, comparable with our study, the risk associated with the ADH1C*1,1 genotype was greater for tumours in the oral cavity than for pharyngeal carcinoma. Data from France, although limited in number, were similar. In a case controlled study of 39 subjects, a 2.6-fold and 6.1-fold higher risk, respectively, of developing oropharyngeal and laryngeal cancer was found for individuals with the ADH1C*1,1 allele. Again, as in our study, this risk was found to be higher for the larynx than for the pharynx.
In contrast, other studies did not confirm an increased risk of UADTC in heavy drinkers with the ADH1C*1 allele.43–45 In the study of Bouchardy and colleagues,43 121 patients with oral cancer and 129 with laryngeal cancer were compared with 172 control subjects. However, the allele frequency for ADH1C*1 in their controls was much higher than in our study and was almost similar to the ADH1C*1 allele frequency observed in our cancer patients. Indeed, geographic variations in the ADH1C genotype of Europeans have been reported with surprisingly high ADH1C*1 allele frequencies in Southern France47 where the study of Bouchardy et al was performed. Thus the effect of the ADH1C*1 allele on alcohol associated carcinogenesis is particularly difficult to determine in such a population.
This may also be true for the study of Olshan and colleagues44 performed in North Carolina and for the study of Sturgis and colleagues45 carried out in Texas. The ethnicity of the patients in the North Carolina study is not specified but it seems that a mixed population was investigated as Spanish speaking patients had been integrated into the protocol. In both studies the ADH1C*1 allele frequency was relatively high in patients with UADTC (62% and 57.4%, respectively).
Subsequently, it should be mentioned in this context that a study by Freudenheim and colleagues55 investigated the effect of the ADH1C genotype on the risk of developing breast cancer, and their preliminary data support the hypothesis that the ADH1C*1 allele is a considerable risk factor for female breast cancer also, especially when ethanol consumption is high, as in our study.
In vitro kinetic studies indicate that the enzyme encoded by the ADH1C*1 allele can metabolise ethanol 2.5 times faster than enzymes encoded by the ADH1C*2 allele.33 Therefore, it is believed that AA levels may be increased in individuals with the ADH1C*1 allele when they consume alcohol chronically. AA is a well known mutagen and carcinogen. Supporting evidence for AA as an important factor in alcohol associated carcinogenesis and its modulation by mutation of an important AA metabolising enzyme comes from Asia. Forty per cent of the Asian population have a mutation of ALDH2. These individuals cannot adequately metabolise AA to acetate and therefore AA accumulates and subsequently results in organ toxicity,56 including the development of UADTC.37,38 The recent observation of increased AA concentrations in the saliva of individuals with inactive ALDH2 following alcohol ingestion may contribute to elucidation of the increased risk for the development of these tumours at a site in direct contact with saliva.36
We report here similar observations for ADH1C. The increased AA concentrations in the saliva of individuals with the ADH1C*1,1 genotype may directly affect carcinogenesis in the upper aerodigestive tract and may be one explanation for their increased tumour risk. Contrary to σ-ADH, γ-ADH is not expressed in the oral mucosa39 and, therefore, changes in salivary AA levels cannot be explained by changes in the mucosal metabolism of ethanol due to ADH1C. As the ADH1C genotype did not affect blood AA concentrations, part of the AA found in saliva may be produced in the salivary glands, as in the case of ALDH2 deficient Asians.36,57 Indeed, there is some evidence that the salivary glands play a key role in UADTC. It has been shown that regenerative changes of the oral mucosa due to chronic ethanol consumption similar to those observed following chronic AA administration in rodents29 could be completely abolished following sialoadenectomy.58
However, AA can also be produced by oral bacteria or yeasts.53,59–62 It has been shown that the number and type of these bacteria are modulated by oral hygiene60 and smoking habits.61 Poor oral hygiene and heavy smoking are risk factors in the development of UADTC.
It should be emphasised that the ALDH2 mutation was not present in our Caucasian population and that the allele frequency of ADH1B*2, another allele which encodes for a rapid ethanol metabolising ADH enzyme, was found to be extremely low (1.2%) in our population.47 Therefore, it is unlikely that our data were affected by these two enzymes which by themselves may be responsible for modulation of AA levels. Our study was performed with age matched alcoholic controls and alcohol associated organ injury as no effect of the ADH1C polymorphism on the development of alcoholism or alcoholic cirrhosis of the liver was detected in Caucasians.47 These groups and the healthy controls had a comparable ADH1C genotype.47 It should be noted that the ADH1C*1,1 genotype is only one factor which increases the risk of developing UADTC in heavy drinkers. Excessive and long term alcohol consumption by itself, consumption of highly concentrated alcoholic beverages, smoking, certain dietary components, and type of food preparation, as well as poor oral hygiene are additional factors associated with a high risk of UADTC.
In conclusion, the ADH1C*1,1 genotype appears to increase considerably the risk of developing UADTC in heavy drinkers, particularly oral and laryngeal cancer. This risk seems to be due to elevated AA concentrations in the saliva of individuals with the ADH1C*1,1 genotype following alcohol consumption. These data provide further evidence for the carcinogenicity of AA, in humans also.
Acknowledgments
The authors thank Drs Vladimir Benes and Wilhelm Ansorge, Genomics Core Facility, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany, for making available their specific laboratory facilities. This study was supported by a grant from the German-Chinese Medical Association and the Volkswagen Foundation awarded to J Li.
Abbreviations
ADH, alcohol dehydrogenase
UADTC, upper aerodigestive tract cancer
ALDH, aldehyde dehydrogenase
AA, acetaldehyde
PCR, polymerase chain reaction
HPLC, high performance liquid chromatography
DHI, 2-diphenylacetyl-1,3-indandione-1-hydrazone
REFERENCES
- 1.Graham S, Dayal H, Rohrer T, et al. Dentition, diet, tobacco and alcohol in the epidemiology of oral cancer. J Natl Cancer Inst 1977;59:1611–16. [DOI] [PubMed] [Google Scholar]
- 2.Herety B, Moriaty M, Daly L. The role of tobacco and alcohol in the etiology of lung and larynx cancer. Br J Cancer 1982;46:961–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blot WJ, McLaughlin JK, Winn DM, et al. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res 1988;48:3282. [PubMed] [Google Scholar]
- 4.McLaughlin JK, Gridley G, Block G, et al. Dietary factors in oral and pharyngeal cancer. J Natl Cancer Inst 1988;80:1237–45. [DOI] [PubMed] [Google Scholar]
- 5.Merletti F, Bofetta P, Ciccione G, et al. Role of tobacco and alcoholic beverages in the etiology of cancer of the oral cavity/oropharynx in Torino, Italy. Cancer Res 1989;49:4919. [PubMed] [Google Scholar]
- 6.Bofetta P, Garfinkel L. Alcohol drinking and mortality among men enrolled in an American Cancer Society prospective study. Epidemiology 1990;1:342–8. [DOI] [PubMed] [Google Scholar]
- 7.Talamini R, Franceschi S, Barra S, et al. The role of alcohol in oral and pharyngeal cancer in nonsmokers, and of tobacco in nondrinkers. Int J Cancer 1990;46:391. [DOI] [PubMed] [Google Scholar]
- 8.Maier H, Dietz A, Zielinski D, et al. Risikofaktoren bei Patienten mit Plattenepithelkarzinomen der Mundhöhle, des Oropharynx, des Hypopharynx und der Larynx. Deutsch Med Wschr 1990;115:843–50. [DOI] [PubMed] [Google Scholar]
- 9.Thun MJ, Peto R, Lopez. et al. Alcohol consumption and mortality among middle aged and elderly U.S. adults. N Engl J Med 1997;337:1705–14. [DOI] [PubMed] [Google Scholar]
- 10.Wynder EL, Bross IJ. Etiological factors in mouth cancer: an approach to its prevention. BMJ 1957;1:389–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seitz HK, Matsuzaki S, Yokoyama A. Alcohol and cancer. Alcohol Clin Exp Res 2001;25:137–43S. [DOI] [PubMed] [Google Scholar]
- 12.Fang JL, Vaca CE. Development of a 32P-postlabelling method for the analysis of adducts arising through the reaction of acetaldehyde with 2′-deoxyguanosine-3′-monophosphate and DNA. Carcinogenesis 1995;16:2177–85. [DOI] [PubMed] [Google Scholar]
- 13.Fang JL, Vaca CE. Detection of DNA adducts of acetaldehyde in peripheral white blood cells of alcohol abusers. Carcinogenesis 1997;18:627–32. [DOI] [PubMed] [Google Scholar]
- 14.Matsuda T, Terashima I, Matsumoto Y, et al. Effective utilization of N2-ethyl-2′-deoxyguanosine triphosphate during DNA synthesis catalyzed by mammalian replicative DNA polymerases. Biochemistry 1999;38:929–35. [DOI] [PubMed] [Google Scholar]
- 15.Vaca CE, Fang JL, Schweda FK. Studies of the reaction of acetaldehyde with deoxynucleosides. Chem Biol Interact 1995;98:51–67. [DOI] [PubMed] [Google Scholar]
- 16.Espina N, Lima V, Lieber CS, et al. In vitro and in vivo inhibitory effect of ethanol and acetaldehyde on O6-methylguanine transferase. Carcinogenesis 1988;9:761–6. [DOI] [PubMed] [Google Scholar]
- 17.Wouterson RA, Appelmann LM, Van Garderen-Hoetmer A, et al. Inhalation toxicity of acetaldehyde in rats. III. Carcinogenicity study. Toxicology 1986;41:213–31. [DOI] [PubMed] [Google Scholar]
- 18.Feron VJ, Kuper CF, Spit BJ, et al. Glass fibers and vapor phase components of cigarette smoke as cofactors in experimental respiratory tract carcinogenesis. Carcinogen Com Surv 1985;8:93–118. [PubMed] [Google Scholar]
- 19.Helander A, Lindahl-Kiessling K. Increased frequency of acetaldehyde-induced sister chromatide exchanges in human lymphocytes treated with an aldehyde dehydrogenase inhibitor. Mutat Res 1991;264:103–7. [DOI] [PubMed] [Google Scholar]
- 20.Dellarco VL. A mutagenicity assessment of acetaldehyde. Mutat Res 1988;195:1–20. [DOI] [PubMed] [Google Scholar]
- 21.Obe G, Jonas R, Schmidt S. Metabolism of ethanol in vitro produces a compound which induces sister chromatid exchanges in human peripheral lymphocytes in vitro: acetaldehyde not ethanol is mutagenic. Mutat Res 1986;174:47–51. [DOI] [PubMed] [Google Scholar]
- 22.Ristow H, Seyfarth A, Lochmann ER. Chromosomal damages by ethanol and acetaldehyde in Saccharomyces cerevisiae as studied by pulsed field gel electrophoresis. Mutat Res 1995;326:165–70. [DOI] [PubMed] [Google Scholar]
- 23.Singh NP, Khan A. Acetaldehyde: genotoxicity and cytotoxicity in human lymphocytes. Mutat Res 1995;337:9–17. [DOI] [PubMed] [Google Scholar]
- 24.Grafstrom RC, Dypbukt JM, Sundquist K, et al. Pathobiological effects of acetaldehyde in cultured human epithelial cells and fibroblasts. Carcinogenesis 1994;15:985–90. [DOI] [PubMed] [Google Scholar]
- 25.Ile SM, Lambert B. Acetaldehyde induced mutation at the hprt locus in human lymphocytes in vitro. Environ Mol Mutagen 1990;16:57–63. [DOI] [PubMed] [Google Scholar]
- 26.Kruysse A, Feron VJ, Til HP. Repeated exposure to acetaldehyde vapor. Studies in Syrian golden hamsters. Arch Environm Health 1975;30:449–52. [DOI] [PubMed] [Google Scholar]
- 27.Appelman LM, Woutersen RA, Feron VJ. Toxicity of acetaldehyde in rats. Acute and subacute studies. Toxicology 1982;23:293–307. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman BT, Crawford GD, Dahl R, et al. Mechanism of acetaldehyde mediated growth inhibition: delayed cell cycle progression and induction of apoptosis. Alcohol Clin Exp Res 1995;19:434–40. [DOI] [PubMed] [Google Scholar]
- 29.Homann N, Kärkkäinen P, Koivisto T, et al. Effect of acetaldehyde on cell regeneration and differentiation of the upper gastrointestinal mucosa. J Natl Cancer Inst 1997;89:1692–7. [DOI] [PubMed] [Google Scholar]
- 30.Seitz HK, Simanowski UA, Garzon FT, et al. Possible role of acetaldehyde in ethanol-related rectal cocarcinogenicity in the rat. Gastroenterology 1990;98:406–13. [DOI] [PubMed] [Google Scholar]
- 31.Simanowski UA, Suter PM, Russell RM, et al. Enhancement of ethanol induced rectal mucosal hyperregeneration with age in F344 rats. Gut 1994;35:1102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.International Agency for Research on Cancer. Working group on the evaluation of the carcinogenic risk of chemicals in humans. Acetaldehyde. IARC Monograph 1985;36:101–32. [PubMed] [Google Scholar]
- 33.Bosron WF, Li TK. Genetic polymorphism of human liver alcohol and aldehyde dehydrogenases and their relationship to alcohol metabolism and alcoholism. Hepatology 1986;6:502–10. [DOI] [PubMed] [Google Scholar]
- 34.Iwahashi K, Suwaki H. Ethanol metabolism, toxicity and genetic polymorphism. Addict Biol 1998;3:249–59. [DOI] [PubMed] [Google Scholar]
- 35.Mizoi Y, Yamamoto K, Ueno Y, et al. Involvement of genetic polymorphism of alcohol and aldehyde dehydrogenases in individual variation of alcohol metabolism. Alcohol Alcohol 1994;29:707–10. [PubMed] [Google Scholar]
- 36.Väkeväinen S, Tillonen J, Agarwal D, et al. High salivary acetaldehyde after a moderate dose of alcohol in ALDH2-deficient subjects: strong evidence for the local carcinogenic action of acetaldehyde. Alcohol Clin Exp Res 2000;24:873–7. [PubMed] [Google Scholar]
- 37.Yokoyama A, Muramatsu T, Ohmori T, et al. Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis 1998;19:1383–87. [DOI] [PubMed] [Google Scholar]
- 38.Yokoyama A, Muramatsu T, Ohmori T, et al. Multiple primary esophageal and concurrent upper aerodigestive tract cancer and the aldehyde dehydrogenase-2 genotype of Japanese alcoholics. Cancer 1996;77:1986–90. [DOI] [PubMed] [Google Scholar]
- 39.Yin SJ, Liao CS, Wu CW, et al. Human stomach alcohol and aldehyde dehydrogenases: comparison of expression pattern and activities in alimentary tract. Gastroenterology 1997;112:766–75. [DOI] [PubMed] [Google Scholar]
- 40.Seitz HK, Egerer G, Simanowski UA, et al. Human gastric alcohol dehydrogenase activity: effect of age, sex and alcoholism. Gut 1993;34:1433–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coutelle C, Ward PJ, Fleury B, et al. Laryngeal and oropharyngeal cancer, and alcohol dehydrogenase 3 and glutathion S-transferase M1 polymorphism. Hum Genet 1997;99:319–25. [DOI] [PubMed] [Google Scholar]
- 42.Harty LC, Caporaso NE, Hayes RB, et al. Alcohol dehydrogenase 3: genotype and risk of oral cavity and pharyngeal cancers. J Natl Cancer Inst 1997;89:1698–705. [DOI] [PubMed] [Google Scholar]
- 43.Bouchardy C, Hirvonen A, Coutelle C, et al. Role of alcohol dehydrogenase 3 and cytochrome P-4502E1 genotypes in susceptibility to cancer of the upper aerodigestive tract. Int J Cancer 2000;87:734–40. [PubMed] [Google Scholar]
- 44.Olshan AF, Weissler MC, Watson MA, et al. Risk of head and neck cancer and the alcohol dehydrogenase 3 genotype. Carcinogenesis 2001;22:57–61. [DOI] [PubMed] [Google Scholar]
- 45.Sturgis EM, Dahlstrom KR, Guan Y, et al. Alcohol dehydrogenase 3 genotype is not associated with risk of squamous cell carcinoma of the oral cavity and pharynx. Cancer Epidemiol Biomarkers Prev 2001;10:273–5. [PubMed] [Google Scholar]
- 46.American Psychiatric Association. Diagnostic and statistical manual of mental disorders, 3rd edn., Washington, DC: American Psychiatric Association 1987:165–75.
- 47.Borras E, Coutelle C, Rosell A, et al. Genetic polymorphism of alcohol dehydrogenase in Europeans: the ADH2*2 allele decreases the risk for alcoholism and is associated with ADH3*1. Hepatology 2000;31:984–9. [DOI] [PubMed] [Google Scholar]
- 48.Couzigou B, Fleury B, Groppi A, et al. Genotyping study of alcohol dehydrogenase class I polymorphism in French patients with alcoholic cirrhosis. Alcohol Alcohol 1990;25:623–6. [DOI] [PubMed] [Google Scholar]
- 49.Poupon RE, Nalpas B, Coutelle C, et al. The French Group for Research on Alcohol and Liver. Polymorphism of alcohol dehydrogenase, alcohol and aldehyde dehydrogenase activities: implication in alcoholic cirrhosis in white patients, Hepatology 1992;15:1017–22. [DOI] [PubMed] [Google Scholar]
- 50.Xu Y, Carr LG, Bosron WF, et al. Genotyping of human alcohol dehydrogenases at the ADH2 and ADH3 loci following DNA sequence amplification. Genomics 1988;2:209–14. [DOI] [PubMed] [Google Scholar]
- 51.Groppi A, Begueret J, Iron A. Improved methods for genotype determination of human alcohol dehydrogenase (ADH) at ADH2 and ADH3 loci using polymerase chain reaction-directed mutagenesis. Clin Chem 1990;36:1765–8. [PubMed] [Google Scholar]
- 52.Oneta CM, Simanowski UA, Martinez M, et al. First pass metabolism of ethanol is strikingly influenced by the speed of gastric emptying. Gut 1998;43:612–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rideout JM, Lim CK, Peters TJ. Assay of blood acetaldehyde by HPLC with fluorescence detection of its 2-diphenylacetyl-1,3-indandione-1-azine derivatives. Clin Chim Acta 1986;161:29–35. [DOI] [PubMed] [Google Scholar]
- 54.Homann N, Jousimies-Somer H, Jokelainen K, et al. High acetaldehyde levels in saliva after ethanol consumption: methodolical aspects and pathogenetic implications. Carcinogenesis 1997;18:1739–43. [DOI] [PubMed] [Google Scholar]
- 55.Freudenheim JL, Ambrosone CB, Moysich KB, et al. Alcohol dehydrogenase 3 genotype modification of the association of alcohol consumption with breast cancer risk. Cancer Causes Control 1999;10:369–77. [DOI] [PubMed] [Google Scholar]
- 56.Enomoto N, Takase S, Takada N, et al. Alcoholic liver disease in heterozygotes of mutant and normal aldehyde dehydrogenase-2 genes. Hepatology 1991;13:1071–5. [PubMed] [Google Scholar]
- 57.Väkeväinen S, Tillonen J, Salaspuro M. 4-Methylpyrazole decreases salivary acetaldehade levels in ALDH2 deficient subjects but not in subjects with normal ALDH2. Alcohol Clin Exp Res 2001;25:829–34. [PubMed] [Google Scholar]
- 58.Simanowski UA, Suter PM, Stickel F, et al. Esophageal epithelial hyperregeneration following chronic ethanol ingestion: effect of age and salivary gland function. J Natl Cancer Inst 1993;85:2030–3. [DOI] [PubMed] [Google Scholar]
- 59.Jokelainen K, Heikkonen E, Roine R, et al. Increased acetaldehyde production by mouthwashings from patients with oral cavity, laryngeal, or pharyngeal cancer. Alcohol Clin Exp Res 1996;20:1206–10. [DOI] [PubMed] [Google Scholar]
- 60.Homann N, Tillonen J, Rintamäki H, et al. Poor dental status increases the acetaldehyde production from ethanol in saliva. A possible link to the higher risk of oral cancer among alcohol consumers. Oral Oncol 2001;37:153–8. [DOI] [PubMed] [Google Scholar]
- 61.Homann N, Tillonen J, Meurman JH, et al. Increased salivary acetaldehyde levels in heavy drinkers and smokers: a microbiological approach to oral cavity cancer. Carcinogenesis 2000;21:663–8. [DOI] [PubMed] [Google Scholar]
- 62.Tillonen J, Homann N, Rautio M, et al. Role of yeasts in the salivary acetaldehyde production from ethanol among risk groups for ethanol associated oral cavity cancer. Alcohol Clin Exp Res 1999;23:1409–15. [PubMed] [Google Scholar]