Abstract
Background and aims: Genome wide scans in inflammatory bowel disease (IBD) have indicated various susceptibility regions with replication of 16cen (IBD1), 12q (IBD2), 6p (IBD3), 14q11 (IBD4), and 3p21. As no linkage was previously found on IBD regions 3, 7, 12, and 16 in Flemish IBD families, a genome wide scan was performed to detect other susceptibility regions in this population.
Methods: A cohort of 149 IBD affected relative pairs, all recruited from the Northern Flemish part of Belgium, were genotyped using microsatellite markers at 12 cM intervals, and analysed by Genehunter non-parametric linkage software. All families were further genotyped for the three main Crohn’s disease associated variants in the NOD2/CARD15 gene.
Results: Nominal evidence for linkage was observed on chromosomes 1 (D1S197: multipoint non-parametric linkage (NPL) score 2.57, p = 0.004; and at D1S305-D1S252: NPL 2.97, p = 0.001), 4q (D4S406: NPL 1.95, p = 0.03), 6q16 (D6S314: NPL 2.44, p = 0.007), 10p12 (D10S197: NPL 2.05, p = 0.02), 11q22 (D11S35-D11S927: NPL 1.95, p = 0.02) 14q11-12 (D14S80: NPL 2.41, p = 0.008), 20p12 (D20S192: NPL 2.7, p = 0.003), and Xq (DXS990: NPL 1.70, p = 0.04). A total of 51.4% of patients carried at least one NOD2/CARD15 variant. Furthermore, epistasis was observed between susceptibility regions 6q/10p and 20p/10p.
Conclusion: Genome scanning in a Flemish IBD population found nominal evidence for linkage on 1p, 4q, 10p12, and 14q11, overlapping with other genome scan results, with linkage on 14q11-12 supporting the IBD4 locus. The results further show that epistasis is contributing to the complex model of IBD and indicate that population heterogeneity is not to be underestimated. Finally, NOD2/CARD15 is clearly implicated in the Flemish IBD population.
Keywords: inflammatory bowel disease, genome scan, genetics
The inflammatory bowel diseases (IBD) Crohn’s disease (CD) and ulcerative colitis (UC) are chronic disorders of the gastrointestinal tract with a prevalence estimated at 2/1000.1,2 Although CD and UC are grouped under the unifying term IBD, clinical but also pathogenic differences exist. Both diseases are also characterised by different serological antibody expression: CD expresses ASCA (anti-Saccharomyces cerevisiae antibodies)3,4 whereas UC is associated with the occurrence of perinuclear antineutrophil cytoplasmic antibodies.5,6
Family studies,7–9 ethnic differences,10 and twin studies11–13 have underlined the contribution of an important genetic susceptibility in the pathogenesis of IBD. From these studies, a recurrence risk for siblings (lambda s (λs)) of 15 has been calculated for IBD overall (25 for CD and 10 for UC).7–9
The model of inheritance, which appears to best explain the data, is that CD and UC are a group of polygenic diseases sharing some, but not all, susceptibility genes: a model of genetic heterogeneity. This model can explain the clinical variability in disease presentation—a specific disease phenotype may result from the interaction between environmental factors with specific polymorphisms in a number of susceptibility loci. Apart from clinical differences, serological antibodies, genetic markers, and knockout mice models also support this heterogeneity concept.
Whereas the first two genome wide scans in IBD14,15 showed linkage on chromosome 16 and chromosomes 3–7 and 12, respectively, subsequent genome wide scans have shown at least nominal evidence for linkage on various chromosomes.16–20 A number of the implicated regions have been replicated: 16cen (the IBD1 locus),14,21–28 12q (the IBD2 locus),15,29–31 6p (the IBD3 locus),15,20,32 14q11–12 (the IBD4 locus),18,19 and recently 3p21.15,33
Successful identification of IBD1 as NOD2/CARD15,34,35 a gene involved in recognition of microbial lipopolysaccharides and nuclear factor κB activation, showed that this gene is only the first of many more genes underlying IBD susceptibility, and therefore greatly encouraged investigators in the field to continue the search for other genes.
Several explanations can account for the variation in results seen among the genome searches: genetic heterogeneity among populations, different phenotypes studied, insufficient power to detect linkage, or ethnic differences. Some genome wide searches have included a large group of Ashkenazi Jewish patients16,18 who are known to be genetically different from Caucasian patients and who are highly predisposed to the development of IBD. Others have restricted their genome wide analyses to only CD families.18,19
We, along with others, could not find evidence for linkage on IBD susceptibility regions located on chromosomes 3–7–12 and 16 in a smaller cohort of Belgian IBD affected sibling pairs.36,37 We were able to exclude regions on chromosomes 3, 7, and 12 in our Flemish sample with a LOD score of less than −2 for a locus with λs = 2. Additional IBD affected relative pairs have been collected for this report and a genome scan has been performed to identify IBD susceptibility regions in this Belgian population.
SUBJECTS AND METHODS
Families
Eighty nine IBD affected families, living in the Flemish part of Belgium, were recruited. Place of birth was obtained from all patients, parents, and all four grandparents. In this context, it needs to be stressed that our study population has a very homogeneous ancestry, as 97% of all families were born and living in the northern Flemish part of Belgium for at least three generations, and their four grandparents had Flemish surnames. Only three families included members with a different origin. There were no Jewish patients in our dataset (table 1 ▶).
Table 1.
Characteristics of the study population
| Families | Affected | Unaffected | ASP (sibships) | Relative pairs | |
| CD | 76 | 186 | 110 | 106 (2 sibships: n = 61) | 129 |
| (3 sibships: n = 11) | |||||
| (4 sibships: n = 2) | |||||
| Mixed | 13 | 31 | 28 | 19 (2 sibships: n = 10) | 20 |
| (3 sibships: n = 3) | |||||
| Total | 89 | 217 | 138 | 125 | 149 |
Data show the number of families studied, number of affected and unaffected individuals, number of affected sibpairs (ASP) (sibships in parentheses), and total number of relative pairs.
CD, Crohn’s disease.
All families had two or more members affected. Seventy six families were pure CD affected. In 13 families, some relatives had CD (n = 18) while others had UC (n = 13) (mixed families). There were no pure UC families. All patients were seen at the gastroenterology unit of the University Hospital Gasthuisberg Leuven, Belgium. Confirmation of a diagnosis of CD or UC was obtained from review of the clinical, radiological, endoscopic, and histological data, according to well established criteria.38 In total, 217 patients were IBD affected: 204 (94%) were diagnosed with CD and 13 (6%) with UC. There was one affected mother of a CD affected sibling pair in whom differential diagnosis between CD and UC was impossible and who was diagnosed as indeterminate colitis. This family was analysed as mixed. In total, 149 relative pairs were genotyped. Among these, 125 were affected sibling pairs (106 CD only, 19 mixed CD-UC) and 24 were second degree relative pairs (four aunt/nephew; three aunt/niece; three uncle/nephew; five uncle/niece; nine second cousin pairs). Of the total study cohort, chromosomes 3, 7, 12, and 16 had previously been studied in 56 families.37 After obtaining informed consent, blood samples for DNA extraction were collected. DNA of both parents was available in 46% and of one parent in 20% of families. If one or both parents were absent, one or more unaffected siblings were genotyped to infer the missing parental genotype. Ethics agreement for the study was given by the Catholic University of Leuven.
Genotyping
DNA was extracted using a salting out procedure from whole venous blood and amplified by polymerase chain reaction (PCR) using 323 microsatellite markers spaced over the genome (Applied Biosystems ABI Prisms Linkage Mapping Set 1, PE Belgium SA/NV). Average spacing between the markers was 12.6 cM and was calculated from the Généthon human linkage map.39 Optimal conditions were obtained for each primer before amplification over ranging annealing temperatures (55–60°C) and MgCl2 concentrations (1.5–2.5 mmol). PCR reactions were done in 15 µl volumes containing 5 µl of DNA (10 ng/µl), 0.04 µl Taq polymerase (5 U/µl), 0.6 µl of each primer (66 ng/µl), 1.5 µl 10× buffer, 0.375 µl dNTPs (concentration 100 µMol), and MgCl2 in sterile H2O. Amplified fragments were detected using 6% acrylamide gels electrophoresed on 373A DNA sequencers (Applied Biosystems ABI). Semi-automated DNA fragment sizing was carried out using the Genescan software 3.1. Genotyping was done using Genotyper 2.1 (ABI).
In a second phase, all families were genotyped for the three identified single nucleotide polymorphisms (SNPs) in the IBD1 gene, NOD2/CARD15, and results obtained in this cohort of patients were compared with a control group consisting of 157 healthy hospital workers (94 females and 63 males).34,35 The missense mutation R702W (SNP8, GenBank accession No G67950) was genotyped by an allele specific PCR procedure with primers: 5′-ATC TGA GAA GGC CCT GCT CC-3′ (wild-type, forward), 5′-ATC TGA GAA GGC CCT GCT CT-3′ (mutated, forward), and 5′-CCC ACA CTT AGC CTT GAT G-3′ (reverse) (annealing temperature 58°C), followed by detection on a 2% ethidium bromide stained agarose gel. The missense mutation G908R (SNP12, GenBank accession No G67951) was genotyped by means of PCR-restriction fragment length polymorphism, using PCR-primers 5′-CCCAGCTCCTCCCTCTTC-3′ and 5′-AAGTCTGTAATGTAAAGCCAC-3′ (annealing temperature 55°C), followed by digestion by Hha1 (Gibco BRL, Merelbeke, Belgium) and agarose gel electrophoresis, resulting in a band of 380 bp (wild-type) or two bands of 138 and 242 bp (variant). For the frameshift mutation 1007fs (SNP13, GenBank accession No G67955), we refer to the method described by Ogura and colleagues.35
Statistical analysis
The Genehunter 1.2 program analysed the proportion of alleles shared identity-by-decent at a certain locus.40 The observed proportion of alleles shared was compared with the expected, and an excess of sharing of 2 versus 1 or 0 alleles was taken as evidence for linkage. Genehunter provides a non-parametric linkage (NPL) score from which the significance level (p value) is derived. Allele frequencies for each marker were calculated from the unrelated parents in the samples. The statistical probability of linkage (two point and multipoint) was calculated for IBD overall, for CD and mixed families individually, as well as for families expressing ASCA. With the current sample size, we could detect a locus with λs⩾2.5 at an NPL of 1.63 (p = 0.05) with a power of >80%.41 Epistatic interactions between susceptibility regions were tested in two ways. Firstly, we analysed each of the linkage regions, conditioned on the presence (positive NPL scores for that region) or absence (negative NPL scores) of linkage on each of the other loci with the Genehunter program.16 Therefore, in an analysis for chromosome A conditioned on the results for chromosome B, the criteria to stratify a family to the “linked to chromosome B subset” was a positive NPL score only at those markers within the observed chromosome B linkage peak. Similarly, epistasis between the NOD2/CARD15 gene and other susceptibility loci was tested by reanalysing each of the linkage regions only with those families showing at least one variant/no variant in the NOD2/CARD15 gene. Since each of the seven identified loci were stratified in these analyses based on the results of the other six loci, p values were corrected by multiplying them by 42. Both corrected and uncorrected p values are given. To further assess the significance of our results, we also reanalysed the data in a second step using the Genehunter-Twolocus software. This program performs non-parametric linkage analysis using two marker maps. Allele sharing is evaluated simultaneously at two disease loci. The position of the first disease locus on one chromosome is held fixed at one particular site on the first marker map (at the site of the observed linkage with the Genehunter program). The position of the second disease locus is then varied along a second marker map.42,43
RESULTS
The markers showing nominal evidence for linkage on two point analysis are listed in table 2 ▶. Strongest linkage on two point analysis was seen at D1S252 (156.7 cM; NPL 2.61; p = 0.004) and D20S192 (18.5 cM; NPL 2.60; p = 0.004). On chromosome 14, three consecutive markers on 14q11 showed nominally significant NPL scores and p values in the total cohort of IBD: D14S50 (12.4 cM; NPL 2.34; p = 0.009), D14S80 (26.5 cM; NPL 2.15; p = 0.016), and D14S49 (36.7 cM; NPL 2.44; p = 0.007) (table 2 ▶).
Table 2.
Markers showing non-parametric linkage (NPL) scores of >2 on two point analysis in the overall IBD group. NPL scores for CD only and mixed families are also given (Genehunter 1.2)
| Marker | Position (cM) | NPL | p Value | ||||
| IBD overall | CD only | Mixed | IBD overall | CD only | Mixed | ||
| D1S197 | 78.9 | 2.07 | 1.92 | 0.78 | 0.019 | 0.027 | 0.78 |
| D1S252 | 156.7 | 2.61 | 2.59 | 0.61 | 0.004 | 0.004 | 0.27 |
| D4S406 | 112 | 2.17 | 2.48 | 0.27 | 0.015 | 0.006 | 0.60 |
| D8S198 | 129.6 | 2.47 | 2.64 | 0.78 | 0.007 | 0.004 | 0.22 |
| D14S50 | 12.4 | 2.34 | 2.30 | 0.36 | 0.009 | 0.01 | 0.36 |
| D14S80 | 26.5 | 2.15 | 2.04 | 0.79 | 0.016 | 0.02 | 0.21 |
| D14S49 | 36.7 | 2.44 | 2.40 | 1.04 | 0.007 | 0.008 | 0.15 |
| D18S62 | 17.7 | 2.30 | 1.96 | 1.24 | 0.011 | 0.025 | 0.11 |
| D20S192 | 18.5 | 2.60 | 2.22 | 1.44 | 0.004 | 0.01 | 0.07 |
| D22S274 | 51.5 | 2.19 | 2.01 | 0.86 | 0.014 | 0.022 | 0.19 |
IBD, inflammatory bowel disease; CD, Crohn’s disease; NPL, non-parametric linkage.
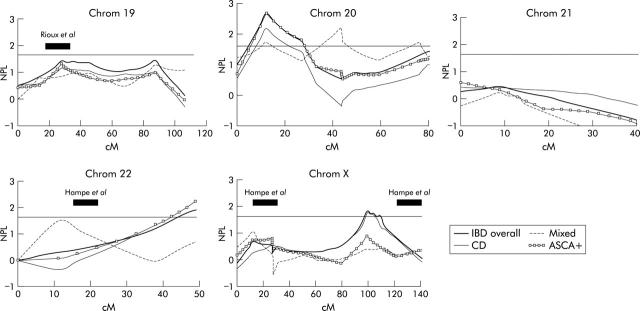
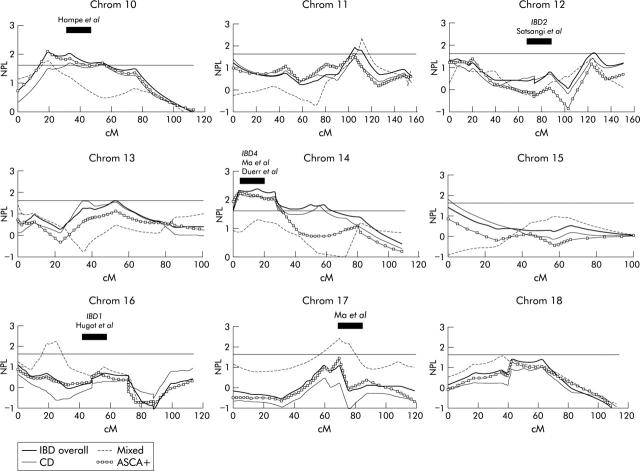
The results of multipoint non-parametric analysis for all chromosomes as well as for the CD and mixed subgroups are shown in figs 1 ▶–3 ▶. The multipoint curve for chromosome 1 showed two peaks: around D1S197 (78.9 cM; max NPL 2.57; p = 0.004), overlapping with the region found by Cho and colleagues16 and around D1S305 and D1S252 (156.7 cM; max NPL = 2.97; p = 0.001) (fig 1 ▶). On chromosome 4 at D4S406 (112 cM), a multipoint NPL in the CD subgroup of 2.21 (p = 0.013) was seen, overlapping partially with the region found by Hampe and colleagues17 (fig 1 ▶). On chromosome 6, maximum multipoint linkage was obtained around D6S314 (137 cM) on 6q16 (NPL = 2.44; p = 0.007) (fig 1 ▶). Chromosome 10 showed linkage around D10S197 (50.5 cM; NPL = 2.05; p = 0.02), again partially overlapping with the region found by Hampe and colleagues17 (fig 2 ▶). On 11q22, linkage was observed over 15 cM between D11S35 and D11S927 (104 cM; NPL 1.95, p = 0.02). Linkage was observed on 14q11–12 over 30 cM and was maximal around 14S80 (26.5 cM; NPL = 2.41; p = 0.008) (fig 2 ▶). This region coincides with the reported IBD-4 locus (Ma and colleagues18; Duerr and colleagues19). On chromosome 20, a peak of linkage was observed around D20S192 (18.5 cM; NPL = 2.71; p = 0.003) (fig 3 ▶). Earlier findings of linkage on Xq around DXS990 were confirmed (101 cM; NPL = 1.70; p = 0.04).42
Figure 1.
The results of multipoint non-parametric analysis (Genehunter 1.2) for chromosomes 1–9 are plotted as the non-parametric linkage (NPL) score against the genetic distance in centimorgan (cM). Results for inflammatory bowel disease (IBD) overall, Crohn’s disease (CD) only, mixed families, and anti-Saccharomyces cerevisiae antibody positive (ASCA+) families are shown. The reported regions of linkage from previous scans are indicated.
Figure 3.
The results of multipoint non-parametric analysis (Genehunter 1.2) for chromosomes 19–22 and chromosome X, and are plotted as the non-parametric linkage (NPL) score against the genetic distance in centimorgan (cM). Results for inflammatory bowel disease (IBD) overall, Crohn’s disease (CD) only, mixed families, and anti-Saccharomyces cerevisiae antibody positive (ASCA+) families are shown. The reported regions of linkage from previous scans are indicated.
Figure 2.
The results of multipoint non-parametric analysis (Genehunter 1.2) for chromosomes 10–18 are plotted as the non-parametric linkage (NPL) score against the genetic distance in centimorgan (cM). Results for inflammatory bowel disease (IBD) overall, Crohn’s disease (CD) only, mixed families, and anti-Saccharomyces cerevisiae antibody positive (ASCA+) families are shown. The reported regions of linkage from previous scans are indicated.
On chromosomes 8 (NPL = 2.12; p = 0.018), 9 (NPL = 2.77; p = 0.003), 16 (NPL = 2.25; p = 0.013), and 17 (NPL = 2.40; p = 0.008), significant NPL scores were seen only in mixed families (figs 1 ▶, 2 ▶). However, as this subgroup consisted of only 13 families and 20 affected relative pairs, no definitive conclusions should be drawn from these results. We noted that the region on chromosome 17 overlapped with the findings of the genome scan by Ma and colleagues18 although the latter genome scan only consisted of CD families. Our linkage curve on 17 for the CD only pairs, on the contrary, did not show significant linkage in this region.
Subsequently, all families were genotyped for the three main associated variants in the NOD2/CARD15 gene. Overall, 51.4% of CD patients carried at least one variant within NOD2/CARD15 compared with 31 cases (19.7%) in the control group (n = 157). Allele frequencies for Arg702Trp (16.7%), Gly908Arg (3.8%), and fs1007insC (12.7%) differed significantly from those observed in controls (6.4%, 1.9%, 2.3%, respectively; all p<0.01). There were 6.4% homozygotes and 7.3% compound heterozygotes observed in patients. Among the controls, two cases were found to be compound heterozygous.
ASCA was determined in all patients and relatives. For CD, 126/204 (62%) CD patients and 29/138 (21%) unaffected relatives were ASCA positive. Families expressing ASCA (71/89) were reanalysed for all chromosomes as a separate subgroup to see whether expression of this antibody is linked to certain chromosomal regions (figs 1 ▶–3 ▶). ASCA+ multipoint curves overall corresponded well with the curves of the CD subgroup and no single region associated with ASCA expression could be identified. This linkage curve correspondence reflects the close association between ASCA and the CD phenotype.
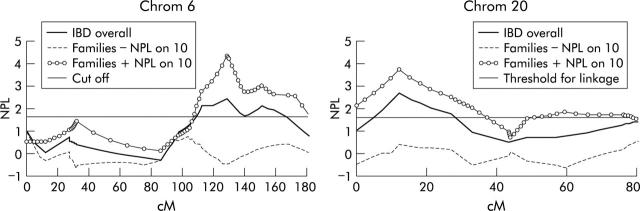
To test epistasis, all families were reanalysed for chromosomes 1, 4, 6, 10, 11, 20, and X, conditioned on the presence or absence of linkage on each of the other regions of linkage. Interaction between loci was observed at chromosomes 6 and 20, both when conditioned for linkage on chromosome 10 (fig 4 ▶): an NPL score of 4.30 was reached on 6q when analysing only those families with +NPL scores on chromosome 10 (uncorrected p<0.0001, corrected p = 0.0004). Also, an NPL score of 3.74 was observed on chromosome 20 when analysing only those families with +NPL scores on chromosome 10 (uncorrected p<0.0001, corrected p = 0.0004). The results obtained from the Genehunter-Twolocus output were very similar: significant NPL scores were obtained for 6q when conditioned on chromosome 10 (NPL 2.47; p = 0.006 uncorrected, p = NS corrected at 128 cM distance) and on chromosome 20 when conditioned on chromosome 10 (NPL 2.35; p = 0.009 uncorrected and NS corrected at 15 cM). Given the earlier reported epistasis between 1p and IBD1,16 the region harbouring the NOD2/CARD15 gene, those families carrying at least one NOD2/CARD15 variant, were reanalysed for regions on chromosomes 1, 4, 6, 10, 11, 20, and X. No arguments for epistatic interactions were seen.
Figure 4.
Epistasis between loci: shown are the non-parametric multipoint linkage (NPL) curves of chromosomes 6 and 20 for the overall inflammatory bowel disease (IBD) group, and conditioned on the presence (+) or absence (−) of linkage to the observed susceptibility region on chromosome 10. Results are plotted as NPL score against the genetic distance in centimorgan (cM).
DISCUSSION
Given the absence of linkage on chromosome 16, and even exclusion on chromosomes 3, 7, and 12 in a smaller Belgian dataset of IBD families,37 a genome wide search in a larger Belgian IBD population was performed to see if other linkages could be identified. Lander and Kruglyak have proposed a classification with thresholds of linkage for genome wide scans.44 Although none of the identified regions in our genome scan meet the Lander and Kruglyak criteria for significant (Lod >3.6, p = 2×10−5) or suggestive linkage (Lod>2.2, p = 7×10−4), several findings are noteworthy and deserve attention. Firstly, four of the susceptibility regions found in this genome scan coincided with regions found by other investigators. Intriguing is the fact that two of these regions—namely, on chromosomes 4 and 10—overlapped with findings from the European collaborative study.17 This study consisted of 353 affected sibling pairs originating from the UK, the Netherlands, and Germany mainly. The migration waves that took place in Europe between 8000 and 5000 BC consisted of a German-Austrian wave, of which the Flemish population (Northern Belgium) was the most Southern branch. The French and Walloons (Southern Belgium) descend from the Mediterranean wave. This reflects in the language differences between the (Northern) Germanic and (Southern) Romanic parts of Europe. Furthermore, within the Germanic populations, two subclusters can be distinguished based on phylogenetic tree analysis: a subcluster which is made up of Dutch, Danish, and English people and a subcluster of Austrian, Swiss, German, and Flemish people.45,46 The overlapping findings of our genome scan with Hampe et al’s fit into the model of common ancestry between these populations. However, the exact origin and distribution of the study population in the study of Hampe and colleagues17 are not entirely clear and it would be interesting to know whether the observed linkages were detected in all subpopulations. In this context, it needs to be stressed that our study population had a very homogeneous ancestry, as 97% of all families were born and living in the northern Flemish part of Belgium for at least three generations, and their four grandparents had Flemish surnames.
Again, no linkage was observed for IBD1 in our population after having almost doubled the number of affected pairs for the actual genome scan. However, the first underlying gene indicating susceptibility to CD was identified as NOD2/CARD15 and all families were genotyped for the three main associated variants within this gene. Overall, 51.4% of CD patients carried at least one variant within NOD2/CARD15; 6.4% were homozygotes and 7.3% compound heterozygotes. Although previously, and also in this genome scan, we did not observe linkage to the IBD1 region, the underlying NOD2/CARD15 gene is indeed implicated in the Flemish CD population. This finding emphasises once more the difficulty in replicating linkages in complex diseases. This is almost only possible with large collaborative approaches and hence more studies, such as the combined study on IBD1 by the International Genetics IBD Consortium,28 are warranted to analyse other underlying genes in IBD.
The positive results observed on 14q11–12 overlap with previously reported IBD linkages by two independent investigators18,19 and therefore further support the IBD4 locus on 14q11.
Earlier findings of linkage on Xq around DXS990 were also observed in this genome scan and further fine mapping within this region is ongoing.47 Finally, new linkages were observed in this genome scan—namely, on chromosomes 1q, 6q, and 20p12.
In this study, families expressing ASCA were analysed as a separate subgroup, independent of CD and mixed families. ASCA have been detected in a higher proportion of unaffected first degree relatives of CD patients, in contrast with healthy spouses,4,46 and intrafamilial aggregation of ASCA has been demonstrated,48,49 which indicates that ASCA expression is genetically modulated. However, in this genome scan, we could not identify a specific locus associated with ASCA expression. In contrast, the multipoint linkage curves of the ASCA+ subgroup coincided very well with those of the CD subgroup, further indicating the close relationship between this serological marker and the clinical CD phenotype. Similarly, mixed families were analysed as a separate subgroup as we demonstrated that serological antibody expression in mixed families differs from pure CD or UC families, being less stringently related to the phenotype.4 There was linkage observed on chromosomes 8, 9, 16, 17, and 20 with only these mixed families. However, given the small size of this subgroup, no definitive conclusions should be drawn from these results, and whether these mixed families represent a different entity needs to be established.
We further tested epistasis between the observed linkage regions and found interactions with chromosomes 6 and 20, both when conditioned for the identified susceptibility region on chromosome 10. It is believed that epistasis results from interactions among loci that contribute to the same biochemical or developmental pathways. All complex disorders probably include gene-gene interactions, at least to some extent.50,51 However, there is no agreement on the best way to look for locus-locus interactions and also how to correct for multiple comparisons. We therefore acknowledge that our findings remain exploratory until confirmed. Identification of epistasis however can help to lead us towards common gene pathways. Indeed, all observed regions of linkage harbour a number of interesting candidate genes (table 3 ▶).
Table 3.
Candidate genes in the regions of linkage observed in this genome scan
| Region | Marker | Candidate genes |
| 1p32 | D1S197 | VCAM-1, IL-12Rβ2 |
| 1p13-1q3 | D1S252-D1S305 | DAP-3, IL-6R, MUC-1 |
| 4q27-31 | D4S406 | IL-21, IL-15 |
| 6q16-27 | D6S314 | Schizophrenia susceptibility locus, IDDM-15 |
| 10p12-15 | D10S191-D10S197-D10S220 | MRC-1, IL-2R, IL-15Rα, transcription factors |
| 11q22 | D11S35-D11S927 | MMP-1,3,12,13,20, API1 and 2 |
| 14q11-12 | D14S80-D14S49-D14S50 | DAD-1, MMP-14, T-cell Ag receptor polypeptides |
| 20p12 | D20S192 | Signal regulatory proteins |
| Xq | DXS990 | IL-2Rγ; IL-13Rα2 |
VCAM-1, vascular cell adhesion molecule 1; IL, interleukin; DAP-3, death associated protein 3; MUC-1, mucin 1; MRC-1, mannose receptor C type 1; API1, apoptosis inhibitor 1; DAD-1, defender against cell death 1; MMP, matrix metalloproteinase.
In conclusion, this genome wide scan in a Belgian population of IBD affected families supports the existence of IBD4 on 14q11, has shown additional evidence for the existence of other susceptibility loci, and has revealed new regions, all in which lie a number of interesting candidates. The results further indicate that epistasis and gene-gene interactions are also present in IBD and that population heterogeneity is not to be underestimated.
Acknowledgments
This work was supported by a grant from the Funds for Scientific Research (FWO), Brussels, Belgium (to SV) and a grant from A Lazarri.
The authors would like to thank the IBD patients, their families, and the National Crohn’s and Colitis Foundation (CCV) for their collaboration. We would also like to thank the following physicians for their contribution of families and clinical data: Dr F Baert, Dr S Bourgeois, Dr L Van De Mierop, Dr F D’Heygere, Dr P Schurmans, Dr G D’Haens, Dr P Potvin, Dr G Deboever, Dr P Maes, Dr L Terriere, Dr Bertrand, Dr B De Schepper, Dr F Van De Mierop, Dr M Cabooter, Dr M Simoens, Dr Van Outryve, Dr L Vandeputte, Dr Mokabbem, Dr J Wyndaele, Dr R Milo, Dr N Büscher, Dr P Van Der Spek, Dr G Van Assche, Dr D Mendez de Leon, Dr R Kums, Dr D Walgraeve, Dr P Pelckmans, Dr M Ferrante, Dr J Callens, and Dr D Sprengers.
Abbreviations
IBD, inflammatory bowel disease
CD, Crohn’s disease
UC, ulcerative colitis
ASCA, anti-Saccharomyces cerevisiae antibodies
PCR, polymerase chain reaction
SNP, single nucleotide polymorphism
NPL, non-parametric linkage
REFERENCES
- 1.Calkins BM, Mendelhoff AI. The epidemiology of ideopathic inflammatory bowel disease. In: Kirsner JB, Shorter RG, eds. Inflammatory bowel disease. 4th ed. Baltimore: Williams & Wilkins, 1995:31–68.
- 2.Binder V . Genetic epidemiology in inflammatory bowel disease. Dig Dis 1998;16:351–5. [DOI] [PubMed] [Google Scholar]
- 3.Sendid B , Colombel JF, Jacquinot PM, et al. Specific antibody response to oligomannosidic epitopes in Crohn’s disease. Clin Diag Lab Immunol 1996;3:219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vermeire S , Peeters M, Vlietinck R, et al. Anti-Saccharomyces cerevisiae antibodies (ASCA), phenotypes of IBD and intestinal permeability: a study in familial IBD. Inflamm Bowel Dis 2001;7:8–15. [DOI] [PubMed] [Google Scholar]
- 5.Duerr RH, Targan SR, Landers CJ, et al. Anti-neutrophil cytoplasmic antibodies in ulcerative colitis: comparison with other colitides/diarrhoeal illnesses. Gastroenterology 1992;100:1590–6. [DOI] [PubMed] [Google Scholar]
- 6.Satsangi J , Landers CJ, Welsh KI, et al. The presence of anti-neutrophil antibodies reflects clinical and genetic heterogeneity within inflammatory bowel disease. Inflamm Bowel Dis 1998;4:18–26. [DOI] [PubMed] [Google Scholar]
- 7.Orholm M , Munkholm P, Langholz E, et al. Familial occurrence of inflammatory bowel disease. New Engl J Med 1991;324:84–8. [DOI] [PubMed] [Google Scholar]
- 8.Satsangi J , Rosenberg W, Jewell DP. The prevalence of inflammatory bowel disease in relatives of patients with Crohn’s disease. Eur J Gastroenterol Hepatol 1994;6:413–16. [Google Scholar]
- 9.Peeters M , Nevens H, Baert F, et al. Familial aggregation in Crohn’s disease: increased age adjusted risk and concordance in clinical characteristics. Gastroenterology 1996;111:597–603. [DOI] [PubMed] [Google Scholar]
- 10.Yang H , McElree C, Shanahan F, et al. Familial empirical risks for inflammatory bowel disease: differences between Jews and non-Jews. Gut 1993;34:517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tysk C , Lindberg E, Jarnerot G, et al. Ulcerative colitis and Crohn’s disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut 1988;29:990–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orholm M , Binder V, Sorensen TI, et al. Concordance of inflammatory bowel disease among Danish twins. Results of a nationwide study. Scand J Gastroenterol 2000;35:1075–81. [DOI] [PubMed] [Google Scholar]
- 13.Thompson NP, Driscoll R, Pounder RE, et al. Genetic versus environment in inflammatory bowel disease: results of a British twin study. BMJ 1996;312:95–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugot JP, Laurent-Puig P, Gower-Rousseau C, et al. Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature 1996;379:821–3. [DOI] [PubMed] [Google Scholar]
- 15.Satsangi J , Parkes M, Louis E, et al. Two-stage genome wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet 1996;14:199–202. [DOI] [PubMed] [Google Scholar]
- 16.Cho JH, Nicolae DL, Gold LH, et al. Identification of novel susceptibility loci for inflammatory bowel disease on chromosomes 1p, 3q, and 4q: evidence for epistasis between 1p and IBD 1. Proc Natl Acad Sci USA 1998;95:7502–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hampe J , Schreiber S, Shaw SH, et al. A genomewide analysis provides evidence for novel linkages in inflammatory bowel disease in a large European cohort. Am J Hum Genet 1999;64:808–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Y , Ohmen JD, Li Z, et al. A genome wide search identifies potential new susceptibility loci for Crohn’s disease. Inflamm Bowel Dis 1999;5:271–8. [DOI] [PubMed] [Google Scholar]
- 19.Duerr RH, Barmada MM, Zhang L, et al. High-density genome scan in Crohn disease shows confirmed linkage to chromosome 14q11–12. Am J Hum Genet 2000;66:1857–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rioux JD, Silverberg MS, Daly MS, et al. Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet 2000;66:1863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohmen JD, Yang HY, Yamamoto KK, et al. Susceptibility locus for inflammatory bowel disease on chromosome 16 has a role in Crohn’s disease, but not in ulcerative colitis. Hum Mol Genet 1996;5:1679–83. [DOI] [PubMed] [Google Scholar]
- 22.Parkes M , Satsangi J, Lathrop GM, et al. Susceptibility loci in inflammatory bowel disease. Lancet 1996;348:1588. [DOI] [PubMed] [Google Scholar]
- 23.Mirza MM, Lee J, Teare D, et al. Evidence of linkage of the inflammatory bowel disease susceptibility locus on chromosome 16 (IBD1) to ulcerative colitis. J Med Genet 1998;35:218–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brant SR, Fu Y, Fields CT, et al. American families with Crohn’s disease have strong evidence for linkage to chromosome 16 but not chromosome 12. Gastroenterology 1998;115:1056–61. [DOI] [PubMed] [Google Scholar]
- 25.Cavanaugh JA, Callen DF, Wilson SR, et al. Analysis of Australian Crohn’s disease pedigrees refines the localization for susceptibility to inflammatory bowel disease on chromosome 16. Ann Hum Genet 1998;62:291–8. [DOI] [PubMed] [Google Scholar]
- 26.Curran ME, Lau KF, Hampe J, et al. Genetic analysis of inflammatory bowel disease in a large European cohort supports linkage to chromosomes 12 and 16. Gastroenterology 1998;115:1–7. [DOI] [PubMed] [Google Scholar]
- 27.Annese V , Latiano A, Bovio P, et al. Genetic analysis in Italian families with inflammatory bowel disease supports linkage to the IBD1 locus—a GISC study. Eur J Hum Genet 1999;7:567–73. [DOI] [PubMed] [Google Scholar]
- 28. IBD Genetics Consortium. International collaboration provides convincing linkage replication in complex disease through analysis of a large pooled dataset: Crohn’s disease and chromosome 16. Am J Hum Genet 2001;68:1165–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duerr RH, Barmada MM, Zhang L, et al. Linkage and association between inflammatory bowel disease and a locus on chromosome 12. Am J Hum Genet 1998;63:95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkes M , Barmada MM, Satsangi J, et al. The IBD2 locus shows linkage heterogeneity between ulcerative colitis and Crohn disease. Am J Hum Genet 2000;67:1605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H , Ohmen JD, Ma Y, et al. Additional evidence of linkage between Crohns disease and a putative locus on chromosome 12. Genet Med 1999;1:194–8. [DOI] [PubMed] [Google Scholar]
- 32.Hampe J , Shaw SH, Saiz R, et al. Linkage of inflammatory bowel disease to human chromosome 6p. Am J Hum Genet 1999;65:1647–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hampe J , Lynch NJ, Daniels S, et al. Fine mapping of the chromosome 3p susceptibility locus in inflammatory bowel disease. Gut 2001;48:191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucin-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001;411:599–603. [DOI] [PubMed] [Google Scholar]
- 35.Ogura Y , Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001;411:603–6. [DOI] [PubMed] [Google Scholar]
- 36.Rioux JD, Daly MJ, Green T, et al. Absence of linkage between inflammatory bowel disease and selected loci on chromosomes 3,7,12 and 16. Gastroenterology 1998;115:1062–5. [DOI] [PubMed] [Google Scholar]
- 37.Vermeire S , Peeters P, Vlietinck R, et al. Exclusion of linkage of Crohn’s disease to previously reported regions on chromosomes 12, 7 and 3 in the Belgian population indicates genetic heterogeneity. Inflamm Bowel Dis 2000;6:165–70. [DOI] [PubMed] [Google Scholar]
- 38.Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol 1989; (suppl 24) :2–6. [DOI] [PubMed]
- 39.Dib C , Faure S, Fizames C, et al. A comprehensive genetic map of the human genome based on 5.264 microsatellites. Nature 1996;380:152–4. [DOI] [PubMed] [Google Scholar]
- 40.Kruglyak L , Daly MJ, Reeve Daly MP, et al. Parametric and non-parametric linkage analysis: a unified multipoint approach. Am J Hum Genet 1996;58:1347–63. [PMC free article] [PubMed] [Google Scholar]
- 41.Risch N . Linkage strategies for genetically complec traits. II. The power of affected relative pairs. Am J Hum Genet 1990;46:229–41. [PMC free article] [PubMed] [Google Scholar]
- 42.Strauch K , Fimmers R, Kurz T, et al. Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: application to mite sensitization. Am J Hum Genet 2000;66:1945–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cordell HJ. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum Mol Genet 2002;11:2463–8. [DOI] [PubMed] [Google Scholar]
- 44.Lander E , Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 1995;11:241–7. [DOI] [PubMed] [Google Scholar]
- 45.Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton, New Jersey: Princeton University Press, 1994.
- 46.Cann RL. Genetic clues to dispersal in human populations: retracing the past from the present. Science 2001;291:1742–8. [DOI] [PubMed] [Google Scholar]
- 47.Vermeire S , Satsangi J, Parkers M, et al. Evidence for a susceptibility locus on the X chromosome in Crohn’s disease. Gastroenterology 2001;120:834–40. [DOI] [PubMed] [Google Scholar]
- 48.Sendid B , Quinton JF, Charrier G, et al. Anti Saccharomyces cerevisiae mannan antibodies in familial Crohn’s disease. Am J Gastroenterol 1998;93:1306–10. [DOI] [PubMed] [Google Scholar]
- 49.Sutton CL, Yang H, Rotter JI, et al. Familial expression of anti Saccharomyces cerevisiae mannan antibodies in affected and unaffected relatives of patients with Crohn’s disease. Gut 2000;46:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoltenberg SF, Burmeister M. Recent progress in psychiatric genetics—some hope but no hype. Hum Mol Genet 2000;9:927–35. [DOI] [PubMed] [Google Scholar]
- 51.Gray-McGuire C , Moser KL, Gaffney PM, et al. Genome scan of human systemic lupus erythematous by regression modeling: evidence of linkage an epistasis at 4p16–15.2. Am J Hum Genet 2000;67:1460–9. [DOI] [PMC free article] [PubMed] [Google Scholar]